Abstract
With a new serotype (H17) of hemagglutinin (HA) recently being discovered, there are now 17 serotypes (H1 to H17) of influenza A viruses in total. It is believed that HA is initially expressed as a precursor of HA0 and then cleaved into HA1 and HA2, forming a disulfide bond-linked complex, for its full function. Structural data show that a loop structure exists in the cleavage site between HA1 and HA2, and this flexible loop is crucial for the efficient cleavage of HA0. Here, the crystal structures of H16 (a low-pathogenicity avian influenza virus) in their HA0 form (H16HA0) have been solved at 1.7-Å and 2.0-Å resolutions. To our surprise, an α-helix element in the cleavage site which inserts into the negatively charged cavity with the key residue R329 hidden behind the helix was observed. In vitro trypsin cleavage experiments demonstrated inefficient cleavage of H16HA0 under both neutral and low-pH conditions. The results provide new insights into influenza A virus pathogenicity; both the relatively stable α-helix structure in the flexible cleavage loop and inaccessibility of the cleavage site likely contribute to the low pathogenicity of avian influenza A virus. Furthermore, compared to all of the HAs whose structures have been solved, H16 is a good reference for assigning the HA subtypes into two groups on the basis of the three-dimensional structure, which is consistent with the phylogenetic grouping. We conclude that in light of the current H16HA0 structure, the natural α-helix element might provide a new opportunity for influenza virus inhibitor design.
INTRODUCTION
There are three types of influenza viruses, i.e., influenza A, B, and C viruses. Among them, influenza A viruses are the major pathogens responsible for the seasonal flu and occasional pandemics (16, 18, 30). Influenza A viruses can be subtyped according to the antigenic properties of their two membrane-embedded envelope glycoproteins, hemagglutinin (HA) and neuraminidase (NA). After the recent discovery of a new virus genome subtype identified from bat, H17N10 (43), there are currently 17 HA subtypes (H1 to H17) and 10 NA subtypes (N1 to N10) known. HA plays a pivotal role in virus entry and fusion, and NA contributes to virus release (24, 31, 35, 36, 42). Under the current model, HA is initially synthesized as a precursor, HA0, and subsequently cleaved into HA1 and HA2 by host enzymes for its full functions (36). Initiation of virus infection involves binding of multiple HAs to host cell receptors that contain either terminal α-2,6-linked or α-2,3-linked sialic acid moieties for virus entry (36). After internalization of the virus by endocytosis, subsequent membrane-fusion events within the endosomal pathway in the infected cell require the cleavage of the HA0 precursor into the mature HA1/HA2 form, a complex of the two disulfide-linked subunits. Cleavage is essential for infectivity because it activates the potential of HA to undergo a low-pH-induced, irreversible conformational change in endosomes (21, 23, 48).
Structural biology contributes a great deal to the elucidation of the molecular mechanism of influenza virus entry and fusion (36, 41), which follows the general mechanism of enveloped virus entry and fusion as a whole (4, 15, 36, 46). HAs can be classified into two groups, group 1 (H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, and H17) and group 2 (H3, H4, H7, H10, H14, and H15), on the basis of their primary sequences (14). Since the first HA structure was solved in 1981 (49), 7 out of 17 HA crystal structures have been reported thus far, including those of H1, H2, H3, H5, H7, H9, and H14, and their common structural characteristics have been delineated (19, 25, 33, 34, 38).
It is believed that the amino acid sequence of the cleavage site modulates tissue tropism and systemic spread of influenza viruses, affecting their pathogenicity (27). For HAs of most subtypes, the cleavage sites contain a single arginine (R329), and cleavage occurs extracellularly via specific trypsin-like proteases that are present only in the intestinal and respiratory mucosal surfaces of the host (2). However, for some highly pathogenic avian strains of the H5 and H7 subtypes, the cleavage sites are rich in basic amino acids, which can be cleaved by a family of more widespread intracellular proteases that are found in multiple organs (39), resulting in systemic infections and increased virulence, especially in birds and small mammals (20). This difference in pathogenicity correlates with structural differences at the HA0 cleavage site (4).
Both the HA0 and HA1/HA2 complex structures for H1 and H3 have been solved, and the structural basis for cleavage, in which a labile loop from the cleavage site is released after cleavage, has been established (4, 13, 38, 49). The first uncleaved HA0 structure (H3HA0) was determined using a mutant HA (R329Q) from the A/Hong Kong/68 H3 virus to prevent its cleavage into HA1 and HA2 (4). This structure reveals that the cleavage site forms a loop and projects out from the surface of the molecule. A negatively charged cavity is found adjacent to the loop, and the fusion peptide inserts into the cavity after HA0 cleavage and is possibly guided by an electrostatic force generated when the positively charged amino terminus of HA2 is formed (4). The second uncleaved HA0 structure (18HA0) was determined by using the baculovirus expression system to produce the recombinant HA0 precursor protein of the 1918 H1N1 pandemic virus (38). The 18HA0 structure displays a different conformation of the cleavage loop, which abuts the glycoprotein surface and is less exposed than in the H3HA0 structure. This conformational difference is believed to provide an explanation for the different tryptase susceptibilities of wild-type H3HA0 and 18HA0 (38). However, as only two HA0 structures with different loop conformations are known so far, the true scenario for the transition between HA0 and the HA1/HA2 complex is still debatable.
In this study, we report the HA0 structures of the H16 subtype virus. The H16 subtype virus was first reported in 2005 from black-headed gulls (11) in Sweden and has since primarily been detected in gulls and shorebirds with low pathogenicity (22). We also showed that the HA gene from the H16 serotype (H16HA0) displayed resistance to trypsin in neutral and low-pH environments, which has never been observed before. A unique α-helix conformation in the cleavage site was observed, with the key cleavage residue R329 being hidden behind the helix. These structural features could prevent access of the protease to the enzymatic site, revealing that not only the flexibility but also the accessibility of the loop contribute to the influenza virus pathogenicity. Moreover, using H16 as a reference structure, HAs were clustered into the same two groups on the basis of three-dimensional (3D) structures and primary sequences (14).
MATERIALS AND METHODS
Cloning.
On the basis of H3 numbering (47), cDNA corresponding to residues 11 to 329 (HA1) and 1 to 176 (HA2) of the ectodomain of HA from A/Black-Headed Gull/Sweden/2/99 (H16N3) was cloned into the baculovirus transfer vector pFastBac1. In a fashion similar to that for the constructs used for expression of the 1918 and 2009 influenza virus HAs for structural studies (51), the construct also incorporated a C-terminal thrombin cleavage site, a foldon sequence (12), and a His6 tag at the extreme C terminus to enable convenient protein purification. The HA16-V327G mutation was introduced by overlapping PCR technology.
Transfection and virus amplification were performed according to the Bac-to-Bac baculovirus expression system manual (Invitrogen). Recombinant HA protein secreted into the cell culture medium contains additional plasmid-encoded residues at both the N terminus (ADGIQ) and C terminus (RLVPRGSPGSGYIPEAPRDGQAYVRKDGEWVLLSTFLGHHHHHH, where the sequence in italics is the thrombin site, the foldon sequence is underlined, and the His6 tag is in bold) (37).
Expression and purification.
Suspension cultures of Trichoplusia ni (Hi5) cells (Invitrogen) were cultured in Sf-900 II SFM serum-free medium (GIBCO). The chambers were incubated at 28°C, and after 2 days, cells were removed by centrifugation (6,000 rpm, 60 min). Soluble HA proteins were recovered from the cell supernatant by metal affinity chromatography using Ni-nitrilotriacetic acid (NTA) resin (GE Healthcare). Fractions containing HA proteins were pooled and dialyzed against 10 mM Tris-HCl (pH 8.0) and 40 mM NaCl and then subjected to ion-exchange chromatography (IEX) using a Mono Q 4.6/100 PE column (GE Healthcare).
For mammalian cell expression, pcDNA4.0 vectors containing the HA ectodomain-encoding sequences were transfected into HEK293T cells using polyethyleneimine I (PEI) in a 1:2 ratio (μg DNA to μl PEI). At 5 h posttransfection, the transfection mixture was replaced by CD293 expression medium (Invitrogen) supplemented with l-glutamine. Tissue culture supernatants were harvested at 3 to 6 days posttransfection. HA proteins were purified by metal affinity chromatography using Ni-NTA resin (GE Healthcare). HA protein expression and secretion were confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE).
For crystallization and structural studies, IEX-purified H16HA and HA16-V327G proteins were subjected to thrombin digest (3 U/mg protein overnight at 4°C) and purified by gel filtration chromatography using a Superdex 200 100/300 GL column (GE Healthcare), and 20 mM Tris-HCl (pH 8.0)–50 mM NaCl was used as the running buffer. Protein eluting as a trimer was concentrated to 10 mg/ml. A typical yield of pure protein was approximately 10 mg per preparation.
Crystallization.
H16HA and HA16-V327G crystals were grown by the hanging-drop vapor diffusion method with a reservoir solution (0.2 ml) of 0.1 M imidazole (pH 7.0) and 20% (vol/vol) Jeffamine ED-2001 polyether diamine (pH 7.0). Crystallization drops were set up at 20°C, with 1 μl of protein mixed with an equal volume of reservoir solution.
Data collection, processing, and structure determination.
Crystals were flash-cooled at 100 K under a stream of cold nitrogen. X-ray diffraction data for H16HA were collected using National Synchrotron Radiation Research Center (NSRRC) beamline 13C1 in Taiwan, and diffraction data for HA16-V327G were collected at SSRF beamline BL17U. The data were processed and scaled using the HKL2000 program package (45). Data collection and processing statistics are summarized in Table 1. The structures were solved by the molecular replacement (MR) method using the Phaser program (32) from the CCP4 program suite (1) and the H2 structure (Protein Data Bank [PDB] accession number 2WR0) as a search model. Model building and refinement were performed using the COOT (10) and REFMAC5 (29) programs, respectively. The stereochemical quality of the final model was assessed with the program PROCHECK (28).
Table 1.
Data collection and refinement statistics
| Parametera | Value(s) forb: |
|
|---|---|---|
| H16HA | HA16-V327G | |
| Data collection statistics | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 69.33, 240.94, 69.67 | 69.07, 240.78, 69.48 |
| α, β, γ (°) | 90.00, 119.79, 90.00 | 90.00, 119.68, 90.00 |
| Resolution (Å) | 50–1.70 (1.76–1.70) | 50–2.00 (2.07–2.00) |
| Rmerge (%) | 0.079 (0.343) | 0.093 (0.211) |
| I/σ〈I〉 | 27.4 (4.5) | 9.2 (4.3) |
| Completeness (%) | 98.5 (91.6) | 99.9 (100.0) |
| Redundancy | 3.8 (3.6) | 3.8 (3.9) |
| Refinement statistics | ||
| Resolution (Å) | 50–1.70 | 50–2.00 |
| No. of reflections | 200,026 | 132,070 |
| Rwork/Rfree | 0.1597/0.1922 | 0.1932/0.2282 |
| No. of atoms | ||
| Protein | 12,210 | 12,174 |
| Ligand/ion | ||
| Water | 1,980 | 1,568 |
| B factors | ||
| Protein | 19.4 | 19.5 |
| Ligand/ion | ||
| Water | 31.7 | 27.9 |
| RMSD | ||
| Bond length (Å) | 0.027 | 0.008 |
| Bond angle (°) | 1.904 | 1.182 |
Rmerge = Σ I – 〈I〉 /ΣI × 100, where I is the intensity of a reflection, σ represents background intensity, and 〈I〉 is the average intensity; Rwork was calculated from 95% of reflection data for structure validation; Rfree was calculated from 5% of randomly selected data for cross-validation; B factor is called the temperature factor.
Values in parentheses are for the highest-resolution shell.
Trypsin susceptibility assays.
Tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin was added to each sample at a final ratio of 40:1 (wt/wt, HA/trypsin), and digestion was performed at 37°C for 0, 10, 20, 40, 60, and 120 min. In the low-pH assay, sodium acetate was used for pHs over the range from 5.0 to 5.5 (8). The reaction mixtures were thoroughly mixed, centrifuged at 12,000 rpm for 30 s, and allowed to incubate at 37°C for 30 min. After incubation, the reaction mixtures were equilibrated to room temperature. Trypsin was added to all samples except the controls, and samples were digested for 1 h at 37°C. At each time point, the digestion was stopped by adding a 5× SDS loading buffer containing dithiothreitol and boiling at 100°C for 5 min. Samples were then loaded onto 12% SDS-PAGE gels for analysis (5).
Protein structure accession numbers.
The crystal structures were deposited in PDB under accession numbers 4F23 and 4FIU.
RESULTS
Overall structures of H16HA0 and H16HA grouping based on 3D structures.
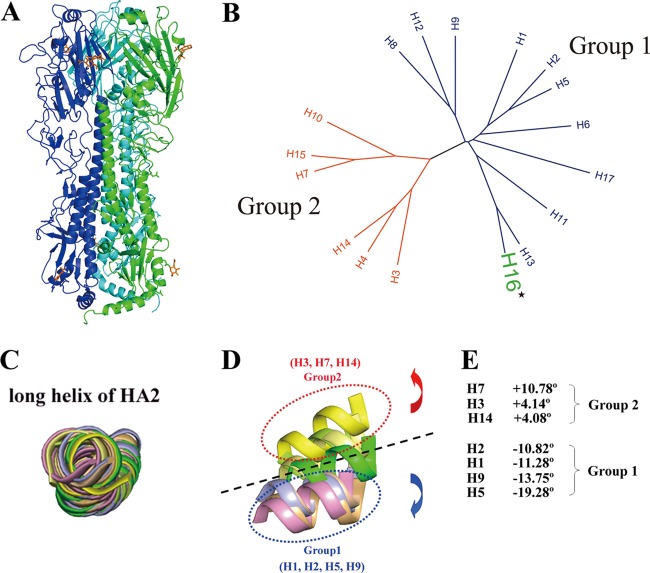
The ectodomain of the HA gene from the H16 serotype (H16HA) influenza virus A/Black-Headed Gull/Sweden/2/99 (H16N3) was cloned and expressed in a baculovirus expression system, as described previously (51). H16HA was crystallized at pH 7.0, and its structure was solved by MR to a high resolution of 1.7 Å using the H2 structure (PDB accession number 2WR0) as a search model (Table 1). A mutant with V327G of H16HA (HA16-V327G), representing some natural isolates of H16 subtype, has also been solved at a resolution of 2.0 Å (Table 1). Each of the two structures has only one molecule in the asymmetric unit. The crystal structure shows that H16HA exists as a classical homotrimer with two distinct domains: a stem domain and a globular head domain (Fig. 1A). The membrane-proximal stem domain is composed of HA2 and two segments of HA1, residues 1 to 55 and 275 to 329. The C-terminal half of HA2 from three HA molecules forms a triple-stranded α-helical coiled coil as the homotrimer core surrounded by the N-terminal half of the HA2 and HA1 segments. This region also contains the cleavage site where host enzymes normally cut HA0 into a disulfide bond-linked HA1 and HA2 complex. On top of the stem domain, the membrane-distal globular domain contains the receptor binding subdomain and the vestigial esterase subdomain. There are eight potential N-linked glycosylation sites, i.e., N20, N21, N46, N169, N170, N291, N474, and N483, but only two of them (N169 and N483) are visible in our structure. For glycosylation site N169, two N-acetylglucosamine (NAG) sugar monomers are observed, and only one NAG sugar monomer is observed in the N483 glycosylation site. Further analysis revealed that the H16HA structure solved here is a noncleaved HA0 structure (H16HA0).
Fig 1.
Overall structure of H16H0 and HA grouping based on 3D structures. (A) Overview of the H16HA0 trimer, represented as a ribbon diagram. For clarity, each monomer has been colored differently (A, green; B, blue; C, cyan). Carbohydrates observed in the electron density maps are colored orange. (B) Phylogenetic tree of the 17 HA subtypes. The 17 HA subtypes can be divided phylogenetically into two groups on the basis of their full-length sequences: group 1 (blue) and group 2 (red). H16, highlighted in green, belongs to group 1. (C to E) Superimpositions of the long α helix (residues 76 to 126) of HA2 reveal the displacements of the HA1 globular subdomain in seven HA subtype structures. The 190 helix (residues 188 to 195) in the receptor binding domain is used as a representative of the globular subdomain of each HA subtype. H1 is colored light blue, H2 is wheat, H3 is lemon, H5 is light orange, H7 is yellow, H9 is pink, H14 is pale yellow, and H16 is green. When we use H16 as a reference, the group 1 members rotate downward with negative-angle degrees (from −10.82° to −19.28°), while the group 2 members rotate upward with positive-angle degrees (from 4.08° to 10.78°).
The superimposition of other HA structures onto the H16HA monomer by means of their HA2 domains (root mean square deviations [RMSDs] in Table 2) indicated that H16HA is most closely related to the human 18H1 subtype (RMSD = 1.225), whereas the human H3 subtype and the avian H14 subtype are the most divergent (RMSDs = 2.004 and 2.005, respectively). On the basis of their HA1 domains, H16HA is the most closely related to the swine H9 subtype (RMSD = 1.680), whereas the human H3 subtype is the most divergent (RMSD = 2.594).
Table 2.
Comparison of RMSDs for individual domainsa
| Group | Subtype | PDB accession no. | RMSD (Å) |
Rotation (°) | |||
|---|---|---|---|---|---|---|---|
| HA2 domain | HA1 domain | HA1 R region | HA1 E region | ||||
| 1 | Hu-H1 | 1RD8 | 1.225 | 2.248 | 0.973 | 1.324 | −11.28 |
| 1 | Hu-H1 | 3AL4 | 1.578 | 2.034 | 0.939 | 0.940 | −8.73 |
| 1 | Hu-H2 | 2WRC | 1.511 | 2.004 | 0.817 | 0.966 | −10.82 |
| 1 | Hu-H5 | 2FK0 | 1.298 | 2.441 | 1.279 | 0.933 | −19.28 |
| 1 | Av-H5 | 1JSM | 1.501 | 2.045 | 1.013 | 1.133 | −20.29 |
| 1 | Sw-H9 | 1JSD | 1.673 | 1.680 | 0.955 | 1.867 | −13.75 |
| 2 | Hu-H3 | 2HMG | 2.004 | 2.594 | 1.212 | 5.577 | +4.14 |
| 2 | Tu-H7 | 1TI8 | 1.862 | 2.549 | 1.087 | 3.894 | +10.78 |
| 2 | Av-H14 | 3EYJ | 2.005 | 2.226 | 1.215 | 4.934 | +4.08 |
Individual domains were superimposed separately with the H16HA counterparts. Hu, human; Av, avian; Sw, swine; Tu, turkey. Accession or identification numbers for viruses whose genes/proteins were used in this work are as follows: A/South Carolina/1/18 (H1N1), PDB accession no. 1RD8; A/California/04/2009(H1N1), PDB accession no. 3AL4; A/Singapore/1/1957 (H2N2), PDB accession no. 2WRC; A/Viet Nam/1203/04 (H5N1), PDB accession no. 2FK0; A/Duck/Singapore/3/97 (H5N3), PDB accession no. 1JSM; A/Swine/Hong Kong/9/98 (H9N2), PDB accession no. 1JSD; A/Hong Kong/1/68 (H3N2), PDB accession no. 2HMG; A/Turkey/Italy/8000/02 (H7N3), PDB accession no. 1TI8; and A/mallard/Astrakhan/263/1982 (H14N5), PDB accession no. 3EYJ.
The phylogenetic tree in Fig. 1B shows that H16 belongs to group 1. Previously solved HA structures demonstrate that there are group-specific features at sites where extensive conformational changes occur for HA activation. In addition to local variations in structure, there are significant differences in the rigid body orientation of head subdomains between HAs in the groups. The superimposition of H16 with solved structures by means of the long central α helices of HA2 revealed that the head subdomain of H16 may represent a third group when it is compared with the head subdomains of the already defined group 1 and group 2 (Fig. 1C and D), which is consistent with the phylogenetic groups. If H16 is placed in ground zero, members of group 2 are above H16 and members of group 1 are below it. Using H16 as the origin of the coordinates, group 2 members rotate by positive-angle degrees from approximately 4° to 11° (Fig. 1E) and group 1 members rotate by negative-angle degrees from approximately −11° to −19° (Fig. 1E). Thus, as one of the most recently identified HA subtypes, H16 is an excellent reference from which to group the HA subtypes on the basis of their 3D structures.
Inefficient cleavage of H16HA0.
For most of the HA subtypes, the cleavage site is a single arginine residue, and cleavage occurs by some trypsin-like enzymes, such as TMPRSS2 and HAT (2). Here, we used trypsin to measure the susceptibility of the cleavage site in H16HA0 compared to other baculovirus-expressed HA0 proteins, including 18HA0 (38) and QH05HA0 (H5HA from the highly pathogenic H5N1 influenza A virus isolated from Qinghaihu Lake in China) (26). To our surprise, H16HA0 displayed inefficient cleavage at neutral pH (pH 8.0), and with the incubation time extended to 2 h, more than half of the H16HA0 remained uncleaved (Fig. 2C). In contrast, the cleavage of 18HA0 and QH05HA0 was nearly complete after 10 min (Fig. 2A and B).
Fig 2.
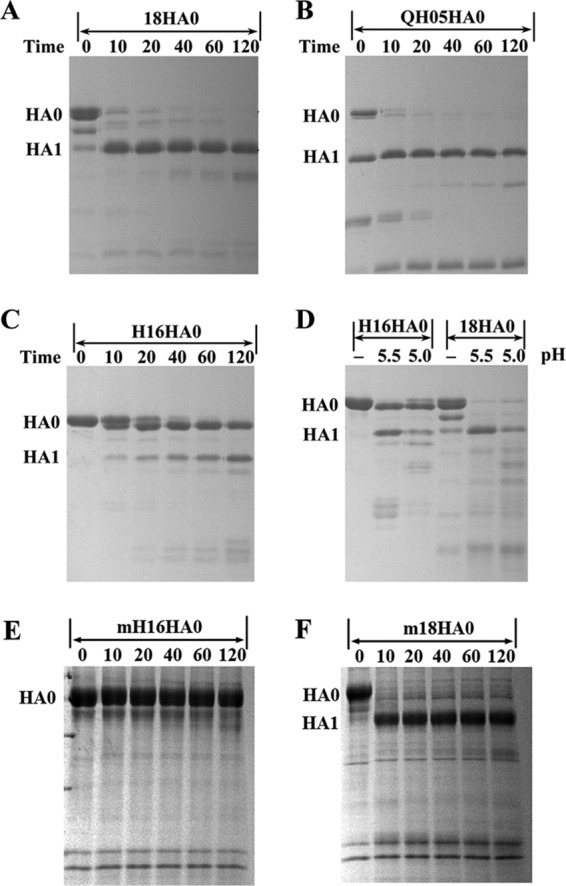
Enzymatic susceptibilities of different HA0s to trypsin. Enzymatic assay with time course of from 0 to 120 min at neutral pH (8.0) for 18HA0 (A), QH05HA0 (B), and H16HA0 (C), which were produced in a baculovirus expression system. The temperature of trypsin incubation is 37°C. It is clear that QH05HA0 is a mixture of HA0 and HA1/HA2 forms when purified from the baculovirus expression system. (D) Enzymatic assay of H16HA0 and 18HA0 at low pH (5.5 and 5.0). H16HA0 is clearly resistant to the trypsin treatment. (E and F) To put this enzymatic resistance in a biological perspective, enzymatic assays of mammalian cell-expressed H16HA0 (E) and 18HA0 (F) (mH16HA0 and m18HA0, respectively) were performed. H16HA0 exhibits similar resistance to trypsin digestion, whereas 18HA0 displays similar trypsin susceptibility.
Newly synthesized viral proteins are exported to the cell surface by way of the Golgi complex, where the pH becomes more acidic during progression through the secretory pathway (7, 17). Taking this into account, we also measured the trypsin susceptibility of H16HA0 under low-pH conditions. After incubation at low fusion pHs (pH 5.5 or 5.0) for 1 h, H16HA0 still behaved as a trypsin-resistant trimer, almost half of which remained in precursor form. In contrast, the cleavage of 18HA0 was nearly complete under the same reaction conditions (Fig. 2D). These results imply that this protease-resistant feature could potentially preclude H16HA0 from undergoing an irreversible conformational change at the low pH of endosomes that is required for membrane fusion.
To put this inefficient cleavage in a biological perspective, we produced the H16HA0 and 18HA0 proteins in mammalian 293T cells, which can be infected by influenza virus. Trypsin susceptibility assays were applied to these mammalian cell-derived proteins. The H16HA0 protein exhibited a similar resistance to trypsin digestion as the protein derived from a baculovirus expression system (produced in insect cells). The control mammalian cell-derived 18HA0 protein also exhibited trypsin susceptibility similar to that derived from a baculovirus expression system. This confirms that both the mammalian cell-expressed and insect cell-expressed HAs show similar trypsin resistance characters.
Special features of the H16HA0 cleavage site: an α helix.
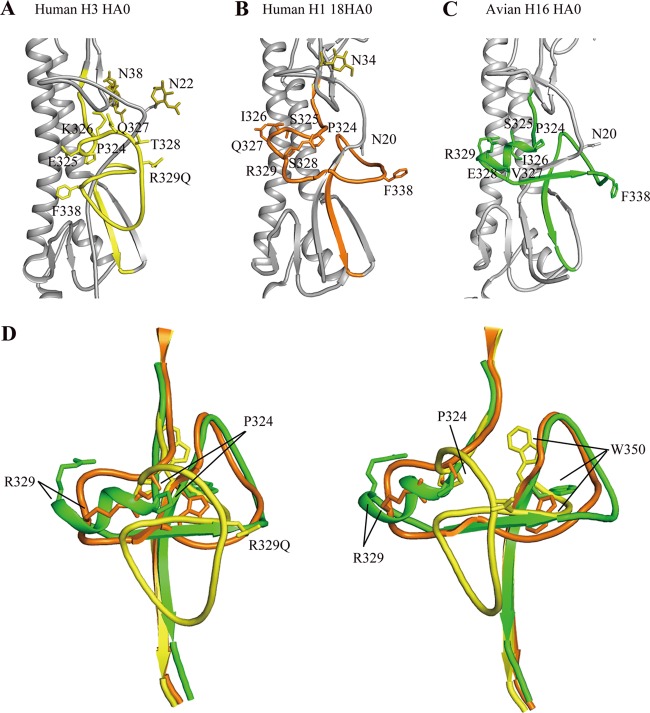
The HA precursor HA0 is cleaved by host enzymes into HA1 and HA2 between R329 and G330 (R/G site) in the cleavage site (36). Thus far, only two HA0 structures have been reported, the R329Q mutant HA0 of the H3 subtype and 18HA0 of the H1 subtype (4, 38). The R329Q mutation makes the loop uncleavable, and 18HA0 was produced in the baculovirus expression system as a noncleaved form (38). Most in vitro-prepared HAs, including those made using the baculovirus expression system, are purified in the cleaved H1/H2 complex form (19, 33, 49, 51). In comparison to the uncleaved H3 and H1 structures, the cleavage site of the H16HA0 structure was more similar to that in H1 than that in H3 (Fig. 3). The H3 cleavage loop projects out from the glycoprotein surface, exposing it to potential proteases, whereas the H16HA0 cleavage site abuts the HA surface like it does in the 18HA0 structure. As discussed by Wilson and colleagues (38), the different cleavage loop conformations in the H3HA0 and 18HA0 structures may be influenced, in part, by nearby glycosylation sites. In both 18HA0 and H16HA0, the similar N20 glycosylation sites may preclude the cleavage loop from protruding. In contrast, the equivalent glycosylation site (N22) in H3HA0 is farther from the cleavage site loop and, thus, may exert less influence on its conformation.
Fig 3.
Structural comparison of the H16HA0 cleavage site with other HA0s. HA2 domains for human H3HA0 and human H1 18HA0 were aligned with H16HA0. The cleavage sites are colored yellow for human H3HA0 (A), orange for human 18HA0 (B), and green for avian H16HA0 (C). (D) Overlay of the cleavage sites of H3HA0, 18HA0, and H16HA0. The two views differ by a rotation of 90° about the 3-fold vertical axis. For H3HA0, the cleavage site forms a loop structure that projects from the glycoprotein surface, while for 18HA0, the cleavage site forms a loop structure that abuts the glycoprotein surface. Extraordinarily, in H16HA0, the cleavage site forms an α-helix structure that has not been observed before.
Both the H3HA0 and 18HA0 cleavage sites form loop-like structures. However, a unique structural feature of the H16HA0 cleavage site was observed; it forms a short α-helix structure. This solid α-helix element starts at P324 and consists of five residues: S325, I326, V327, E328, and R329 (Fig. 3). The equivalent residues in H3HA0 are E325, K326, Q327, T328, and R329Q, while the equivalent residues in 18HA0 are S325, I326, Q327, S328, and R329 (Fig. 3). Compared to the cleavage site sequences of H3HA0 and 18HA0, H16HA0 displays the most significant difference in the P-3 residue at position 327 (valine). The hydrophobic residue V327 in H16HA0 is distinct from the hydrophilic Q327 residues in both H3HA0 and 18HA0. To date, 17 different H16 virus strains have been reported, and the most polymorphic amino acid in the cleavage site occurs at position 327, such as G, V, S, N, and D, except in one special strain, which has an insertion mutation (Fig. 4). Seven out of 17 virus strains have residue G at the position 327. Thus, we replaced the residue V with G and solved the crystal structure of the mutated HA (HA16-V327G) to test the special feature in the cleavage site. Similarly, in the HA16-V327G structure, the cleavage site still forms an α-helix structure and the loop has inserted into the negatively charged cavity (Fig. 4). Therefore, this α-helix structure in the cleavage site with an inefficient cleavage property is the common property among different H16 virus strains.
Fig 4.
Sequence alignment of the cleavage sites from different H16 virus strains and a similar α-helix conformation in the cleavage site of the HA16-V327G structure. (A) Sequence alignment of the cleavage sites from 17 previously reported H16 virus strains. The most polymorphic amino acids occur at position 327, such as G, V, N, S, and D. (B) Conformation of the cleavage site in the HA16-V327G structure. Although residue V327 is replaced by glycine (G), the cleavage site still forms an α-helix structure element.
Hidden key residue R329.
For HAs of most subtypes, the HA1 and HA2 polypeptide chains are cleaved at a single arginine residue (R329), which is eliminated from the C terminus of HA1 by virion-associated carboxypeptidase after cleavage (48). For some members of the H5 and H7 subtypes, the HA1 and HA2 polypeptide chains are separated by polybasic residues that are inserted at the cleavage site (36).
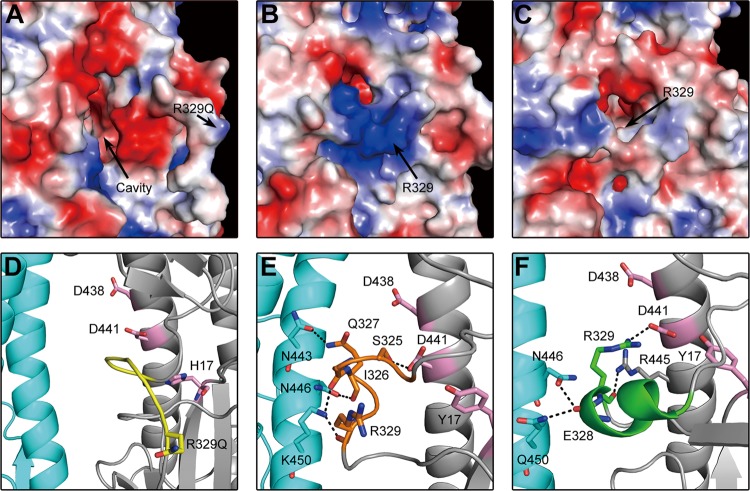
In the R329Q H3HA0 mutant, there is a deep cavity adjacent to the cleavage site loop, and the R329Q residue is far from the cavity, which contains ionizable residues, including residues D109 and D112 on the long helix of HA2 and H17 of HA1 (Fig. 5A and D). However, in the 18HA0 structure, the different conformation of the cleavage loop shows R329 covering the electronegative cavity (Fig. 5B and E). This cavity is filled in the cleaved HA1/HA2 structure by HA2 residues 1 to 10, a nonpolar segment of the fusion peptide (38). In the H16HA0 structure, the unique α helix inserts into the electronegative cavity (Fig. 5C and F), and extraordinarily, the key R329 residue points toward the electronegative cavity, hiding behind the helix. Thus, the R329 residue in the H16HA0 structure is less exposed than the equivalent counterpart in the 18HA0 structure, which points toward the solvent. This special structural feature blocks the access of potential proteases to the R329 residue, resulting in inefficient cleavage.
Fig 5.
Different positions of key R329 residues in the cleavage site. (A to C) Electrostatic diagrams of cleavage sites show that the key R329 residue is far from the negatively charged cavity in H3HA0 (A), R329 covers the negatively charged cavity in 18HA0 (B), and R329 is buried in the negatively charged cavity in H16HA0 (C). (D to F) Detailed interaction between the cleavage site and the negatively charged cavity and adjacent subunit in different HA0s. The cleavage site is colored yellow in H3HA0 (D), orange in 18HA0 (E), and green in H16HA0 (F). The main residues in the negatively charged cavity are colored pink. The hydrogen bonds are shown as black dashed lines.
In the cleaved HA structures at neutral pH, the N-terminal HA2 fusion peptide inserts into the electronegative cavity and makes up to five hydrogen bonds from its backbone amide groups to conserved HA2 ionizable residues (D109 and D112) (4). In the H16HA0 structure, the unique α helix also inserts into the same electronegative cavity (including residues Y17, D438, and D441). The R329 residue forms a strong salt bridge with D441 (equivalent to D109 in the HA2 chain) and a hydrogen bond with R445 (Fig. 5F). It also forms two hydrogen bonds between the backbone carbonyl group of E328 and residues N446 and Q450 from the adjacent subunit. In contrast, the 18HA0 cleavage loop does not penetrate as far into this cavity and makes only one hydrogen bond, between the loop and the conserved acidic residues (S325 to D441) (Fig. 5E). In addition, the 18HA0 cleavage loop forms four hydrogen bonds with adjacent subunit residues (Q327 bonds to N443 and N446, and I326 and R329 bond to K450). As in H3HA0, the cleavage loop is far from the cavity and forms no contacts (Fig. 5D).
Putative receptor binding site.
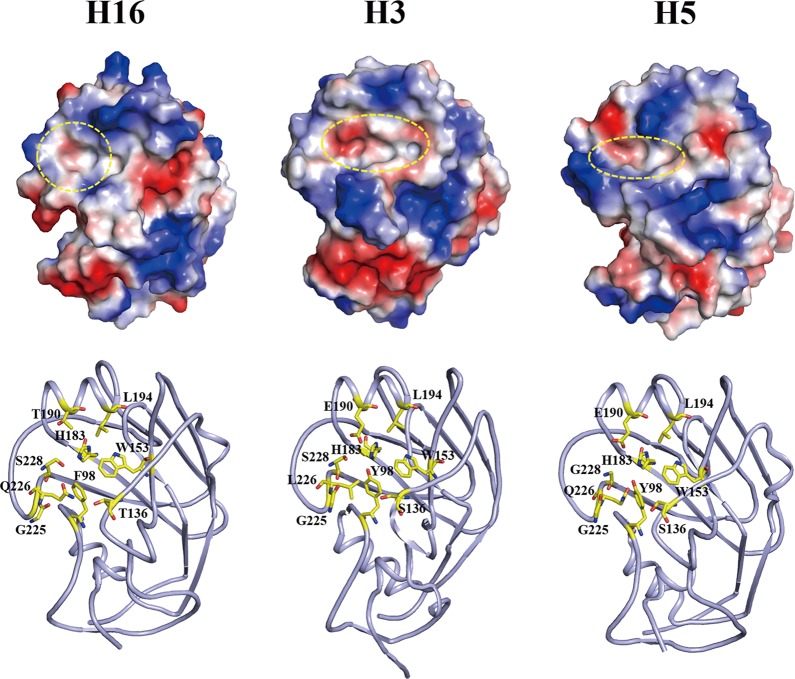
Influenza virus infection is initiated by the binding of the virus HA to the host receptor that contains either terminal α-2,6-linked or α-2,3-linked sialic acid moieties for virus entry (14). Avian viruses preferentially bind to α-2,3-linked sialic acid receptors, whereas the human-adapted viruses are specific for the α-2,6-linked sialic acid receptors. The HA receptor binding site is a shallow pocket in the head domain, formed by two parts, an edge part and a base part. The edge part is formed by three secondary elements, the 130 loop, the 190 helix, and the 220 loop, while the base part is formed by four conserved residues, Y98, W153, H183, and Y195 (14). In H2, H3, and H5 HAs, residues 226 and 228 have been linked to receptor specificity (14). Viruses with Q226 and G228 are specific for the α-2,3-linked sialic acid receptors, whereas viruses with L226 and S228 preferentially bind to receptors with α-2,6 linkage. In H16HA, the receptor binding site is a round cavity and less negatively charged, due to residue T190 (Fig. 6). In contrast, in the representative H5 and H3 HAs, the receptor binding sites are oval-shaped cavities with different sizes and partially negatively charged, due to residue E190 (Fig. 6). In particular, the cavity is narrower in avian H5 than in human-adapted H3. H16HA possesses a broader cavity than avian H5, and the avian-specific residue Q226 is maintained. On the basis of these findings, H16 likely binds to both the α-2,6- and α-2,3-linked receptors, and evaluation of this likelihood should be pursued in the near future.
Fig 6.
Structural comparison of the HA receptor binding site. (Top) Electrostatic-potential maps of the head domains of the H16 structure, human H3 structure (PDB accession number 1HA0), and avian H5 structure (PDB accession number 2FK0) were generated by the PyMOL program. The shape of the receptor binding site is marked by a yellow circle. (Bottom) Cartoon diagrams of the receptor binding site are shown with key residues that determine the specificity. Clearly, the H16HA0 receptor binding site possesses a round cavity, whereas the H3 and H5 HAs have an oval-shaped cavity. The H5 HA has a narrower cavity than the H3 HA, which is unfavorable for the human receptor binding. A broader cavity and avian-specific residue Q226 indicate that H16HA0 likely binds to both the human and avian receptors.
Conserved hydrophobic groove for cross-reactive anti-HA2 antibodies in H16.
The hemagglutinin of influenza virus is the major target for vaccine design (50). Recently, some cross-reactive anti-HA2 neutralizing antibodies have been identified to neutralize a wide spectrum of influenza A viruses by binding to highly conserved epitopes in the stem region of HA (5, 8, 9, 40). Among these antibodies, FI6 has been reported to bind to H16HA (5). The crystal structure of the FI6-09H1 complex revealed that the antibody targets a shallow hydrophobic groove on the F subdomain of the HA, where the sides of the groove are formed by the residues from the A helix of HA2 (including L38, T41, I45, and I48) and parts of two strands of HA1 (including V40 and T318) and the HA2 turn (including W21), encompassing residues 18 to 21 (Fig. 7A). In H16HA, due to the different conformation of the HA2 turn, the corresponding region of W21 is replaced by several hydrophobic residues, P15, G16, L17, and I18 (Fig. 7B). Then, W21 locates below those hydrophobic residues, close to residue K38. Thus, a similar hydrophobic core is formed with different residue components, a finding which explains why the FI6 antibody could bind to H16HA.
Fig 7.
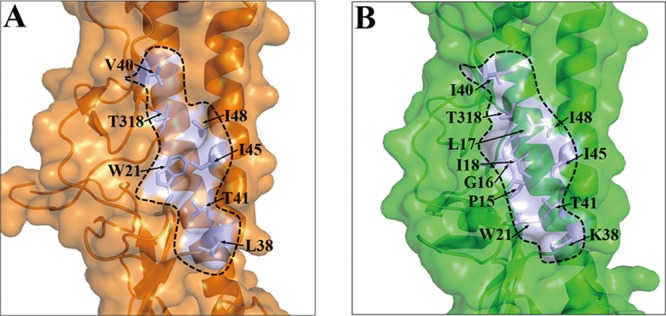
The conserved hydrophobic groove in H16HA0 reveals the structural basis of binding with the broadly neutralizing antibody FI6. Surface representations of the F subdomains of 09H1 HA (A) and H16HA0 (B) with selected side chains that contribute to the conserved hydrophobic groove are shown. The approximate boundaries of the hydrophobic grooves are indicated by the black lines. Although the residues contributing to the hydrophobic groove are moderately different between 09H1 and H16, similar hydrophobic grooves guarantee the binding potential by the FI6 antibody.
DISCUSSION
In this study, the crystal structure of the H16 subtype HA of influenza A virus was solved in its HA0 form. To our surprise, the cleavage site that would create the HA1/HA2 complex displays an α-helical structure and hides in the negatively charged cavity, in contrast to the labile loop conformations found in other known structures, and more importantly, this unusual structure is correlated with its inefficient cleavage by trypsin. The inefficient cleavage is implicated in low pathogenicity, and early experimental infections with H16N3 influenza virus in chickens resulted in a limited infection and mild pathology (44), supporting this notion. Influenza virus pathogenesis is correlated with several factors, but the sequence of the HA cleavage site is a major determinant of disease severity (27). For most of the HA subtypes, the cleavage sites contain a single arginine residue (R239) that can be recognized by specific extracellular trypsin-like proteases. For the highly pathogenic H5N1 or H7N7 viruses, the cleavage sites usually contain polybasic amino acids that can be more generally recognized by enzymes in multiple tissues, therefore causing damage to multiple tissues (6). Here, we observed a relatively stable helix structure and inaccessibility by the enzyme in the H16HA0 loop, correlating with its low pathogenicity.
Phylogenetically, there are two groups of HAs that are grouped on the basis of their primary sequences: group 1 contains H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, and H17; and group 2 contains H3, H4, H7, H10, H14, and H15 (14). The members of the two phylogenetic groups of HA are characterized by group-specific structural features at sites where their sequences differ, such as interhelix loop conformations and globular subdomain orientations, as well as by differences in their response to compounds that inhibit conformational changes and membrane fusion activity (19, 33, 34). When we compared the H16HA0 structure with the previously determined HA structures of different subtypes, we found that the H16HA0 globular subdomain orientation does not belong to either group 1 or group 2 but can separate group 1 from group 2 as a reference protein. Previous studies report that the H16 subtype viruses are gull specific and rarely exist in other species. Thus, to some extent, the H16 subtype viruses may be more closely related to ancient influenza viruses. It is possible that the HA protein of H16 subtype viruses may retain some structural features derived from ancient influenza viruses, while in other subtypes, the HA molecules might have evolved as a result of adaptation to different hosts.
The first HA0 structure was determined using a mutant H3 subtype HA protein containing an R329Q substitution, derived from the A/Hong Kong/68 virus (4). The structure of HA0 provides explanations for the correlation of precursor cleavage efficiency with pathogenesis. The P-4 residue is packed against the HA surface in the first HA0 structure and would not be accessible for binding to a recognition site, as furin enzyme activity appears to require. However, in some H5 and H7 subtype HAs (HA1/HA2 complex structures), four polybasic residues are inserted in the cleavage sites, and these insertions project the site into the solution, exposing the inserted P-4 residues as a recognition site for furin protease. The second HA0 structure was determined using the baculovirus expression system to generate uncleaved 1918 pandemic virus HA0 protein. This 18HA0 structure displays a different conformation of the cleavage loop, which abuts the glycoprotein surface instead of projecting from the surface like the first H3 subtype HA0 structure. Due to this structural feature, the 18HA0 protein is tryptase resistant but susceptible to trypsin protease (38). Interestingly, our insect cell-derived and mammalian cell-derived H16HA0 protein is resistant to trypsin protease under both neutral and low-pH conditions. The key residue R329 hides behind the helix and forms a strong salt bridge with the D441 residue in the cavity. This structural feature precludes protease accessibility and explains why H16HA0 is resistant to trypsin protease. Our results provide new insight into the correlation of cleavage site conformation and enzyme accessibility with virus pathogenicity.
Virus fusion and, hence, entry require cleavage of HA0 into the HA1/HA2 form. One question was raised: how are H16 viruses are activated and spread in gulls? We believe that two reasonable possibilities might answer the question. First, the cleavage of H16HA0 might happen efficiently, but only in highly restricted specialized cells in the gull. Second, the cleavage of H16HA0 might happen more broadly, but just at a very low level in all tissues. This question remains to be answered in the future.
Three structural conformations of HA have now been defined: the HA0 precursor, cleaved HA, and the low-pH-induced fusion-state HA (3, 4, 49). Potential targets for antiviral compounds exist in these conformations and their interconversion events. The transition of HA0 to the cleaved HA1/HA2 conformation may be blocked by inhibition of the cleavage enzymes or by the binding of an inhibitor into the cavity revealed in the first H3 subtype HA0 structure to prevent the insertion of the fusion peptide. Wiley and colleagues proposed that a charged peptide or mimetic might bind in the pocket on HA0, block the formation of the cleaved HA conformation, and, hence, block infectivity (4). According to this strategy, our H16HA0 structure with insertion of an α-helix element into the cavity provides a new concept for drug design; i.e., this α-helix element could be developed as a peptide drug active against influenza virus infection.
ACKNOWLEDGMENTS
We thank Ruiting Guo, Chunchi Chen, and the staff at National Synchrotron Radiation Research Center (NSRRC) beamline 13C1 in Taiwan for their help with data collection.
This work was supported by grants from the Ministry of Science and Technology of China (MOST; project 973; grant no. 2011CB504703; http://www.most.gov.cn/eng/). G.F.G. is a leading principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (NSFC; grant no. 81021003; http://www.nsfc.gov.cn/Portal0/default106.htm).
The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
G.F.G., X.L., and Y.S. conceived the research; X.L., F.G., and J.Q. performed the protein expression, purification, crystallization, structure determination, and trypsin susceptibility assays; G.F.G., Y.S., H.X., F.G., M.W., J.Q., and X.L. performed the data analysis; and G.F.G. and Y.S. wrote the manuscript.
We declare that we have no conflict of interest.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Anonymous 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 2. Bottcher E, et al. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43 [DOI] [PubMed] [Google Scholar]
- 4. Chen J, et al. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417 [DOI] [PubMed] [Google Scholar]
- 5. Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 6. de Jong MD, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demaurex N, Furuya W, D'Souza S, Bonifacino JS, Grinstein S. 1998. Mechanism of acidification of the trans-Golgi network (TGN). In situ measurements of pH using retrieval of TGN38 and furin from the cell surface. J. Biol. Chem. 273:2044–2051 [DOI] [PubMed] [Google Scholar]
- 8. Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ekiert DC, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 11. Fouchier RA, et al. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frank S, et al. 2001. Stabilization of short collagen-like triple helices by protein engineering. J. Mol. Biol. 308:1081–1089 [DOI] [PubMed] [Google Scholar]
- 13. Gamblin SJ, et al. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842 [DOI] [PubMed] [Google Scholar]
- 14. Gamblin SJ, Skehel JJ. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 285:28403–28409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao GF. 2007. Peptide inhibitors targeting virus-cell fusion in class I enveloped viruses, p 226–246 In Torrence PF. (ed), Combating the threat of pandemic influenza: drug discovery approaches. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 16. Gao GF, Sun Y. 2010. It is not just AIV: from avian to swine-origin influenza virus. Sci. China Life Sci. 53:151–153 [DOI] [PubMed] [Google Scholar]
- 17. Grabe M, Oster G. 2001. Regulation of organelle acidity. J. Gen. Physiol. 117:329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan Y, et al. 2010. The emergence of pandemic influenza viruses. Protein Cell 1:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2002. H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. EMBO J. 21:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jang H, et al. 2009. Highly pathogenic H5N1 influenza virus can enter the central nervous system and induce neuroinflammation and neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 106:14063–14068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klenk HD, Rott R, Orlich M, Blodorn J. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426–439 [DOI] [PubMed] [Google Scholar]
- 22. Krauss S, et al. 2007. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3:e167 doi:10.1371/journal.ppat.0030167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lazarowitz SG, Choppin PW. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68:440–454 [DOI] [PubMed] [Google Scholar]
- 24. Liu C, Eichelberger MC, Compans RW, Air GM. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69:1099–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J, et al. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc. Natl. Acad. Sci. U. S. A. 106:17175–17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J, et al. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309:1206. [DOI] [PubMed] [Google Scholar]
- 27. Medina RA, Garcia-Sastre A. 2011. Influenza A viruses: new research developments. Nat. Rev. Microbiol. 9:590–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morris AL, MacArthur MW, Hutchinson EG, Thornton JM. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345–364 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 30. Neumann G, Noda T, Kawaoka Y. 2009. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Palese P, Tobita K, Ueda M, Compans RW. 1974. Characterization of temperature sensitive influenza virus mutants defective in neuraminidase. Virology 61:397–410 [DOI] [PubMed] [Google Scholar]
- 32. Read RJ. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57:1373–1382 [DOI] [PubMed] [Google Scholar]
- 33. Russell RJ, et al. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287–296 [DOI] [PubMed] [Google Scholar]
- 34. Russell RJ, et al. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 105:17736–17741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sauter NK, et al. 1989. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry 28:8388–8396 [DOI] [PubMed] [Google Scholar]
- 36. Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 37. Stevens J, et al. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410 [DOI] [PubMed] [Google Scholar]
- 38. Stevens J, et al. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870 [DOI] [PubMed] [Google Scholar]
- 39. Stieneke-Grober A, et al. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sui J, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y, et al. 2010. In silico characterization of the functional and structural modules of the hemagglutinin protein from the swine-origin influenza virus A (H1N1)-2009. Sci. China Life Sci. 53:633–642 [DOI] [PubMed] [Google Scholar]
- 42. Takemoto DK, Skehel JJ, Wiley DC. 1996. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217:452–458 [DOI] [PubMed] [Google Scholar]
- 43. Tong S, et al. 2012. A distinct lineage of influenza A virus from bats. Proc. Natl. Acad. Sci. U. S. A. 109:4269–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tonnessen R, Valheim M, Rimstad E, Jonassen CM, Germundssond A. 2011. Experimental inoculation of chickens with gull-derived low pathogenic avian influenza virus subtype H16N3 causes limited infection. Avian Dis. 55:680–685 [DOI] [PubMed] [Google Scholar]
- 45. Vaguine AA, Richelle J, Wodak SJ. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191–205 [DOI] [PubMed] [Google Scholar]
- 46. Wang X, Zhang GWH, Wang M, Gao GF. 2004. The molecular mechanism of enveloped virus-cell membrane fusion. Prog. Biochem. Biophys. 31:482–491 [Google Scholar]
- 47. Weis WI, Brunger AT, Skehel JJ, Wiley DC. 1990. Refinement of the influenza virus hemagglutinin by simulated annealing. J. Mol. Biol. 212:737–761 [DOI] [PubMed] [Google Scholar]
- 48. Wiley DC, Skehel JJ. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev. Biochem. 56:365–394 [DOI] [PubMed] [Google Scholar]
- 49. Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366–373 [DOI] [PubMed] [Google Scholar]
- 50. Xuan C, et al. 2011. Structural vaccinology: structure-based design of influenza A virus hemagglutinin subtype-specific subunit vaccines. Protein Cell 2:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang W, et al. 2010. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell 1:459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]