Abstract
We constructed a herpes simplex virus 2 (HSV-2) bacterial artificial chromosome (BAC) clone, bHSV2-BAC38, which contains full-length HSV-2 inserted into a BAC vector. Unlike previously reported HSV-2 BAC clones, the virus genome inserted into this BAC clone has no known gene disruptions. Virus derived from the BAC clone had a wild-type phenotype for growth in vitro and for acute infection, latency, and reactivation in mice. HVEM, expressed on epithelial cells and lymphocytes, and nectin-1, expressed on neurons and epithelial cells, are the two principal receptors used by HSV to enter cells. We used the HSV-2 BAC clone to construct an HSV-2 glycoprotein D mutant (HSV2-gD27) with point mutations in amino acids 215, 222, and 223, which are critical for the interaction of gD with nectin-1. HSV2-gD27 infected cells expressing HVEM, including a human epithelial cell line. However, the virus lost the ability to infect cells expressing only nectin-1, including neuronal cell lines, and did not infect ganglia in mice. Surprisingly, we found that HSV2-gD27 could not infect Vero cells unless we transduced the cells with a retrovirus expressing HVEM. High-level expression of HVEM in Vero cells also resulted in increased syncytia and enhanced cell-to-cell spread in cells infected with wild-type HSV-2. The inability of the HSV2-gD27 mutant to infect neuronal cells in vitro or sensory ganglia in mice after intramuscular inoculation suggests that this HSV-2 mutant might be an attractive candidate for a live attenuated HSV-2 vaccine.
INTRODUCTION
Herpes simplex virus 2 (HSV-2) infects persons throughout the world (28, 29, 61) and is the primary agent of recurrent genital herpes (4, 9, 18). During primary infection, HSV-2 penetrates nerve endings, and viral nucleocapsids travel centripetally to neuronal cell bodies and establish latent infection in sensory neurons. Certain conditions such as UV light, fever, and trauma trigger virus reactivation. Immunosuppressive medication impairs the immune response to the virus and increases the likelihood of symptomatic disease and virus shedding. While antiviral therapy can block virus replication, reduce symptoms associated with HSV-2 infection, and reduce recurrences (26, 62), these drugs cannot clear latent virus. In addition to genital herpes and disease involving various organs, HSV-2 genital infection is associated with a 3-fold increased risk of HIV acquisition among both men and women in the general population (14). At present no vaccine is licensed to prevent HSV-2 infection or disease.
Genetic engineering of viruses to enable site-directed mutagenesis has become a powerful tool for studying viral pathogenesis and for creating candidate vaccines. HSV-2 is a large, complex virus with a DNA genome 154.7 kbp in length, encoding more than 80 proteins, a family of latency-associated transcripts (LAT), and 18 microRNAs (20, 50–52, 56). However, due to the large size of the HSV-2 genome, its high-GC content, and only a single complete genome sequence (12) from a virus strain that is not widely used in laboratories, HSV-2 mutagenesis has been laborious. Traditionally, HSV-2 mutants are constructed by cotransfecting Vero cells with full-length HSV-2 virion DNA and a DNA fragment harboring the desired mutation, screening for low-efficiency homologous recombination of the two molecules, and performing multiple cycles of PCR screening and plaque purification (23, 59). The procedure is time-consuming and not always successful.
The use of bacterial artificial chromosomes (BACs) to clone large fragments of DNA was developed during the sequencing of the human genome (43). BAC vectors contain four genes from the Escherichia coli factor F for self-replication and maintain only one or two copies of the BAC in each E. coli cell. BAC vectors can stably maintain a foreign DNA insert up to 300 kbp in length, which is sufficient to accommodate each of the known herpesvirus genomes. Once cloned into a BAC vector, the herpesvirus genome can be mutated in E. coli, and the desired recombinant clone can be selected using prokaryotic genetic screening methods. This approach has been used in several human herpesviruses, including HSV-1 (17, 27, 48), HSV-2 (34, 41), varicella-zoster virus (VZV) (39, 53, 63, 64), Epstein-Barr virus (EBV) (10, 42), cytomegalovirus (CMV) (3, 7), human herpesvirus 6 (HHV-6) (2, 49), and Kaposi's sarcoma-associated herpesvirus (KSHV) (65), and greatly simplifies herpesvirus mutagenesis. The HSV-2 BAC clones that have been reported are not optimal for pathogenesis studies in animals because the virus backbones in these BAC clones have one or more genes deleted (34, 41).
Both HSV-1 and HSV-2 infect a variety of cells including epithelial cells, fibroblasts, lymphocytes, and neurons. Cell surface receptor expression is one of the important factors that determine the susceptibility of the cell to HSV infection. HVEM (also known as HveA, TNFRSF14, LIGHTR, and CD270) and nectin-1 (also known as HveC, PVRL1, and CD111) are the two major cellular receptors for both HSV-1 and HSV-2. When individually expressed in cell lines resistant to HSV entry, such as CHO or B78H1 cells, HVEM or nectin-1 rendered the cells susceptible to HSV entry (15, 38). HVEM is abundantly expressed in lymphocytes, epithelial cells, and fibroblasts but is not thought to be expressed in neurons (44). In contrast, nectin-1 is highly expressed in neurons, epithelial cells, and fibroblasts.
HSV glycoprotein D (gD) binds to cellular receptors HVEM and nectin-1. Based on the structure of gD bound to its receptors, the amino terminus of gD is critical for interacting with HVEM (6), while a sequence in the carboxyl half of gD interacts with nectin-1 (11). By pseudotyping gD null viruses with gD proteins harboring deletion or point mutations, genetic studies showed that mutations in the amino terminus of gD impaired virus entry into cells expressing only HVEM, while mutations of amino acids 38, 215, 222, and 223 of gD impaired entry into cells expressing only nectin-1 (8, 30).
We report that we have constructed an HSV-2 BAC clone (which contains the full HSV-2 genome in a BAC vector) and generated a virus from the DNA that results in acute and latent infection and reactivation in animals in a manner similar to that of wild-type (WT) virus. A gD mutant was constructed from this HSV-2 BAC clone, which maintains the ability to infect an epithelial cell line but is severely impaired for infecting human neuronal cell lines and Vero cells and cannot infect sensory ganglia when the virus is inoculated intramuscularly in mice. This mutant virus might serve as a candidate vaccine with impaired neurovirulence.
MATERIALS AND METHODS
Viruses and cell lines.
HSV-2 strain R519, a plaque-purified version of strain 333 which does not form syncytia (unpublished data), was propagated in Vero cells. The mouse melanoma cell line B78H1 is resistant to HSV infection; B78H1-A10 and B78H1-C10 cell lines stably express human HVEM and nectin-1, respectively (35), and were provided by Gary Cohen (University of Pennsylvania). Vero cells, human retinal pigment epithelial cells (ARPE-19), and human neuronal cell lines SK-N-SH and SH-SY5Y were purchased from ATCC.
Plasmids.
pWC132 (which expresses enhanced green fluorescent protein [eGFP]), pWC155 (a BAC vector expressing GFP), and pWC205 (which expresses Cre recombinase) were provided by Peter Barry (University of California at Davis) (7). pWC155 and pWC205 are referred to as pBAC and pCre, respectively, elsewhere in this paper. To create plasmid p37LGL38, a portion of UL37 from HSV-2 strain R519 (corresponding to nucleotides 83956 to 84776 of HSV-2 strain HG52) with a PstI site and a loxP site was obtained by PCR and inserted into pWC132 upstream of the eGFP expression cassette, while a portion of UL38 from HSV-2 strain R519 (corresponding to nucleotides 84777 to 85541 of HSV-2 strain HG52) with BamHI and EcoRI sites and a loxP site was obtained by PCR and inserted downstream of the eGFP expression cassette (Fig. 1). pgalK contains a bacterial galK expression cassette (60) and was provided by Neal Copland (National Cancer Institute). pBEC10 contains the human HVEM gene (38) and was provided by Patricia Spear (Northwestern University). pCMV-gD2 contains the HSV-2 gD2 open reading frame from strain 333 inserted into pcDNA3 (Invitrogen). pCMV/HveAs containing HVEM cDNA from Vero cells (13) was provided by Konstantin G. Kousoulas (Louisiana State University).
Fig 1.
Schematic diagram of the steps for constructing the HSV2-BAC38 system. Vero cells were cotransfected with virion DNA from wild-type HSV-2 strain R519 and plasmid p37LGL38 resulting in insertion of the loxP-eGFP-loxP cassette into the region between UL37 and UL38 yielding HSV2-eGFP38. The eGFP expression cassette and one of the loxP sites were removed from the genome of HSV2-eGFP38 by cotransfecting Vero cells with virion DNA of HSV2-eGFP38 and plasmid pCre (expressing Cre recombinase) resulting in HSV2-loxP38. To generate HSV-2 with a BAC vector inserted between UL37 and UL38, Vero cells were cotransfected with BAC vector pBAC and pCre and then infected with HSV2-loxP38 resulting in HSV2-BAC38. Circular virion DNA from HSV2-BAC38-infected Vero cells was obtained by Hirt extraction and used to transform E. coli strain DH10B to produce BAC clone bHSV2-BAC38, which contains the HSV-2 genome in the BAC vector. To reconstitute virus from bHSV2-BAC38, BAC clone DNA was cotransfected with pCre into Vero cells, and GFP-negative plaques were selected as HSV2-ΔBAC38. Recombinant viruses were plaque purified three times before proceeding to the next step, and their clone numbers are shown in parentheses following their names. Figures are not drawn to scale. LTR, long terminal repeat; STR, short terminal repeat; UL and US, unique long and short regions, respectively.
Construction of HSV2-eGFP38, HSV2-loxP38, and HSV2-BAC38.
To construct HSV-2 strain R519 with an loxP-eGFP-loxP cassette inserted between UL37 and UL38 (nucleotides 84776 and 84777 corresponding to HSV-2 strain HG52), Vero cells were cotransfected with 1 μg of HSV-2 strain R519 virion DNA and 5 μg of plasmid p37LGL38 by calcium phosphate precipitation as described previously (59). Cells were harvested 3 days later, and cell-free virus was plaque purified three times by selecting GFP-positive plaques. The resulting virus, termed HSV2-eGFP38, contains eGFP bounded by loxP sites (Fig. 1).
HSV2-loxP38, which has a single loxP site flanked by HindIII and EcoRI sites inserted in between UL37 and UL38, was generated by transfecting Vero cells with 5 μg of pCre (which expresses Cre recombinase) using Lipofectamine 2000 (Invitrogen) and infecting the cells with 6 × 104 PFU of HSV2-eGFP38 1 day after the transfection. Two days later the cells were harvested, and cell-free virus was plaque purified by selecting GFP-negative plaques to obtain HSV2-loxP38 (Fig. 1).
HSV2-BAC38 was obtained by cotransfecting Vero cells with 3 μg of the BAC vector, pBAC, and 2 μg of pCre. One day later the transfected cells were infected with 106 PFU of HSV2-loxP38. Virus was plaque purified by selecting GFP-positive plaques. The resulting virus, HSV2-BAC38, contains a DNA cassette, loxP-eGFP-BAC vector-loxP, between HSV-2 UL37 and UL38 (Fig. 1).
Generation of BAC clone bHSV2-BAC38.
Circular viral genomic DNA of HSV2-BAC38 was isolated from Vero cells very early during infection when the viral genome was replicating as a circular episome. Vero cells in 100-mm dishes were infected with 108 PFU of HSV2-BAC38 in 2.5 ml of medium for 1 h; the medium was removed and replaced with fresh medium for 2 h, and circular viral genomes were isolated from infected cells by Hirt DNA extraction (19). Briefly, culture medium was removed from the dishes, the cells were washed with phosphate-buffered saline (PBS), 0.6 ml of Hirt extraction buffer (0.6% sodium dodecyl sulfate [SDS], 10 mM EDTA, pH 7.5) was added, and the cells were incubated at room temperature for 20 min. Lysates were prepared by scraping the cells, transferring the cell lysate to a 15-ml tube, and adding 150 μl of 5 M NaCl. The tube was inverted 20 times, incubated at 4°C overnight, and centrifuged at 17,000 × g for 30 min at 4°C. The supernatant containing circular viral genomes was treated with 10 μg of RNase A at 37°C for 15 min, extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 10 μl of H2O. The resulting DNA was used to transform E. coli strain DH10B by electroporation, and BAC-containing bacterial clones were selected on a Luria-Bertani (LB) medium plate with 12.5 μg/ml chloramphenicol. The resulting BAC clone, termed bHSV2-BAC38, is a circular form of HSV2-BAC38 (Fig. 1). The galK-negative E. coli strain SW102 (obtained from Neil Copland, National Cancer Institute) was transformed with bHSV2-BAC38, and the resulting bacterial clone was termed bHSV2-BAC38-S1.
Generation of HSV2-ΔBAC38 from bHSV2-BAC38.
Vero cells were cotransfected with 4 μg of BAC clone DNA, isolated from bHSV2-BAC38-S1, and 2 μg of pCre using calcium phosphate precipitation (59). Three days later the cells were harvested, and cell-free virus was prepared. In order to avoid selecting a single clone which might have acquired a cryptic mutation(s), multiple GFP-negative plaques were plaque purified three times in Vero cells, and the resulting clones were pooled to generate the virus stock HSV2-ΔBAC38.
Generation of mutations in the gD gene using bHSV2-BAC38 and the galK selection system.
A DNA fragment consisting of a galK expression cassette (60) flanked by HSV-2 DNA gD sequences (corresponding to nucleotides 141684 to 143733 and 141758 to 141807 of HSV-2 HG52) was generated by PCR. A mixture of 10 ng of pgalK, 0.1 μM each of primers gDgalkF and gDgalKR (with sequences corresponding to galK and HSV-2 gD) (Table 1), and 1× PCR buffer (Expanded High Fidelity PCR; Roche) was denatured at 96°C for 5 min, held at 60°C for 30 s, during which time 5 μl of deoxynucleoside triphosphate (dNTP)/Expanded High Fidelity polymerase mixture was added to each reaction mixture, and then heated to 72°C for 1.5 min. This hot-start cycle was followed by 30 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, followed by one cycle of 72°C for 7 min. The PCR product was digested with 20 U of DpnI at 37°C for 1 h to remove plasmid template, and the 1.3-kb gD-galK DNA fragment was isolated by agarose gel purification and resuspended in H2O to 5 ng/μl.
Table 1.
BAC recombineering, sequencing, and PCR oligonucleotides
| Oligonucleotide name | Sequence (5′ to 3′) | Length (no. of bases) |
|---|---|---|
| gDgalKf | CCCGGCAGCGTGCCTCACCTCGAAGGCCTACCAACAGGGCGTGACGGTCGCCTGTTGACAATTAATCATCGGCA | 74 |
| gDgalKr | GGCGATTTTTAAGCTGTATAGGGCGACGGTGCGCTGGTTTTCGGGGATAATCAGCACTGTCCTGCTCCTT | 70 |
| D215G/R222N/F223If | CCCGGCAGCGTGCCTCACCTCGAAGGCCTACCAACAGGGCGTGACGGTCGGCAGCATCGGGATGCTCCCCAACATTATCCCCGAAAACCAGCGCACCGTCGCCCTATACAGCTTAAAAATCGCC | 124 |
| D215G/R222N/F223Ir | GGCGATTTTTAAGCTGTATAGGGCGACGGTGCGCTGGTTTTCGGGGATAATGTTGGGGAGCATCCCGATGCTGCCGACCGTCACGCCCTGTTGGTAGGCCTTCGAGGTGAGGCACGCTGCCGGG | 124 |
| gDF101 | GCCCCGCTGGAGCTACTATGACA | 23 |
| gDF103 | CATAAGCTGCATTGCGAACGACTA | 24 |
| gDF106 | GGACGAGGCCCGAAAGCACAC | 21 |
| gDR101 | TCGACGGGATGTGCCAGTTTG | 21 |
| gDR103 | CCACACGGGCCCAGAGGTA | 19 |
| gDR106 | TTCGGGGATAAAGCGGGGTAG | 21 |
| NotIF1 | GTCGCGGCCGCCACCATGGAGCCTCCTGGAGACTGG | 34 |
| XhoIR2 | AGTCTCGAGGTCAGTGGTTTGGGCTCCTCCCCGTG | 33 |
| sHveAF1 | GTCGCGGCCGCCACCATGGAGCCTCCTGGAGGTTGG | 36 |
| sHveAR2 | AGTCTCGAGGTCAGCTCCTCCCAGTGAATGCGGG | 34 |
The underlined nucleotides indicate the sites of mutations.
To generate a bacterial clone containing galK inserted in gD, bacterial clone bHSV2-BAC38-S1 was cultured at 30°C overnight with shaking at 250 rpm in 5 ml of LB medium with 12.5 μg/ml of chloramphenicol. The next day, 0.5 ml of the overnight culture was inoculated into 25 ml of LB medium with 12.5 μg/ml chloramphenicol and incubated at 30°C for 2.5 to 3 h until the optical density at 600 nm (OD600) was 0.6. Ten milliliters of the culture was transferred to a 50-ml tube, and expression of λ recombination proteins was induced by heat shocking the cells in a 42°C water bath for 15 min and then chilling them on ice. The bacteria were then washed three times with cold (1°C) H2O, and the pellet was resuspended in 50 to 75 μl of cold H2O and kept on ice. Twenty-five microliters of these cells was mixed with 2.5 μl of the gD-galK DNA fragment and subjected to electroporation. Cells were then washed twice in M9 medium and plated onto M63 medium plates with chloramphenicol and galactose as the sole carbon sources. The plates were incubated at 30°C for 3 days, and bacterial colonies were picked and streaked two successive times on M63 plates with galactose and chloramphenicol. The resulting bacterial clones containing bHSV2-BAC38 with galK inserted in gD (bHSV2-BAC38-gD/galK+) (Fig. 2) were confirmed by PCR with primers gDF101 and gDR101 (Table 1).
Fig 2.
Generation of an HSV-2 mutant with point mutations in gD using the bHSV2-BAC38 clone and galK selection system. The galK expression cassette flanked by a portion of HSV-2 gD was inserted into the gD gene of bHSV2-BAC38 via homologous recombination in E. coli SW102, and transformants were plated on minimal medium with galactose as the sole carbon source to support the growth of galK+ clones. The resulting bacterial clone, containing bHSV2-BAC-gD/galK+, was transformed with a DNA fragment of HSV-2 gD containing mutations D215G/R222N/F223I, and transformants were plated on minimal medium with glycerol and 2-deoxy-d-galactose (DOG) which supports the growth of galK-deficient clones. The resulting galK mutant clone, bHSV2-BAC38-gD27, was cotransfected with plasmid pCre into B78H1-A10 cells resulting in HSV2-gD27 which has the D215G/R222N/F223I mutations in gD and no BAC vector sequences.
To generate bHSV2-BAC38 with point mutations in codons 215, 222, and 223 of gD, 10 μg each of oligonucleotides D215G/R222N/F223If and D215G/R222N/F223Ir (which contain point mutations in the gD codons) (Table 1) was annealed by heating to 98°C for 3 min and cooling slowly to room temperature. SW102 containing bHSV2-BAC38-gD/galK+ was heat shocked and washed with cold H2O as described in the previous paragraph, and the cells were electroporated with 1 μl of the annealed oligonucleotides containing mutations in gD. After electroporation the cells were incubated in 10 ml of LB medium for 4.5 h, washed twice with M9 medium, and plated on M63 minimal medium plates with chloramphenicol, 2-deoxy-galactose, and glycerol as the carbon source. After incubation at 30°C for 3 days, colonies were picked and streaked two successive times on plates with M63 minimal medium, chloramphenicol, 2-deoxy-galactose, and glycerol. The resulting BAC clone was termed bHSV2-BAC38-gD27 (Fig. 2).
Generation of HSV2-gD27 mutant virus from bHSV2-BAC38-gD27.
B78H1-A10 cells in six-well plates were cotransfected with 3 μg of bHSV2-BAC38-gD27 DNA and 1 μg of plasmid pCre using calcium phosphate precipitation; 3 days later the cells were harvested. The resulting virus, HSV2-gD27, was plaque purified three times, and sequencing confirmed that the gD gene contained the desired mutations. Restriction endonuclease digestions showed that the mutant virus had no genome rearrangements. HSV2-gD27 was propagated in B78H1-A10 cells and titrated in B78H1-A10 and ARPE-19 cells.
Generation of rescued virus from HSV2-gD27.
Vero cells were cotransfected with HSV2-gD27 virion DNA (obtained from B78H1-A10-infected cells) and pCMV-gD2 (which contains the wild-type HSV-2 gD open reading frame from HSV-2 strain 333), and three cycles of plaque purification were performed in B78H1-C10 cells. Rescued virus HSV2-gD27R (where R indicates rescue) was selected, and sequencing confirmed that that the gD gene had a wild-type sequence; restriction endonuclease digestions showed no genome rearrangements. HSV2-gD27R was propagated in B78H1-A10 cells and titrated in B78H1-A10 and ARPE-19 cells.
Restriction endonuclease mapping of virion and BAC clone DNA.
Virion DNA was isolated from HSV-infected Vero or ARPE-19 cells as described previously (59). bHSV2-BAC38 DNA was purified from overnight bacterial cultures (incubated at 37°C for DH10B cells or 30°C for SW102 cells) using a BAC-MAX DNA Purification Kit (Epicentre Biotechnologies). A total of 1.6 μg of DNA was digested with 5 U of restriction enzyme AgeI or BamHI at 37°C overnight, and the digestion products and a 1-kb DNA ladder (Ferma Scientific) were subjected to electrophoresis in a 0.5% agarose–Tris-borate-EDTA (TBE) gel containing 0.2 μg/ml ethidium bromide.
One-step growth curve and virus infectivity assays.
To compare the replication kinetics of HSV2-eGFP38, HSV2-lox38, HSV2-BAC38, and HSV2-ΔBAC38 clones, a one-step growth curve assay was performed. Briefly, six-well plates were seeded with 5 × 105 Vero cells/well 1 day before infection. The next day each well was infected with 3 × 103 PFU of virus and incubated at 37°C. Cells were harvested at 0, 3, 7, 20, and 24 h after infection; cell-free virus was prepared by subjecting the samples to three cycles of freeze-thawing, and the supernatant was recovered. Ten-fold serial dilutions were prepared from each sample, and 500 μl of the diluted virus was added to one well of a six-well plate with Vero cells. After incubation for 1 h at 37°C, the inoculum was removed, and Vero cell growth medium with 0.03% human immunoglobulin was added. After incubation for 2 days at 37°C, the plates were fixed and stained with crystal violet, and plaques were counted.
To compare the infectivity of wild-type HSV-2, HSV2-gD27, and HSV2-gD27R, viruses were propagated and titrated in B78H1-A10 cells. Infectivity was determined by plaque assays in relevant cell lines. The titers for each virus in each cell line were then normalized to their titers in B78H1-A10 cells.
Generation and characterization of Vero-A1 and SKNSH-A cell lines expressing human HVEM and of Vero-sA1 and B78H1-sA1 cells expressing simian HVEM.
Retrovirus expressing human or simian HVEM were generated by inserting the HVEM open reading frame (obtained by PCR from pBEC10 using primers NotIF1 and XhoIR2 for human HVEM and from pCMV/HveAs using primers sHveAF1 and aHveAR2 for simian HVEM encoded by Vero cells) (Table 1) into the NotI and XhoI restriction sites of retrovirus vector pCCMP-IRES-GFP (where IRES is internal ribosome entry site) (21). The resulting plasmids, pCCMP-HVEM and pCCMP-HVEMs, express human and simian HVEM, respectively, and GFP. Retrovirus particles were produced by cotransfecting 293T cells with pCCMP-HVEM or pCCMP-HVEMs, pMD-gagpol, and pHDM-G plasmids. Vero-A1, Vero-A4, Vero-A16, Vero-A64, Vero-A 256, and Vero-A1024 cells were generated by transducing Vero cells in six-well plates with 1 ml of undiluted and 4-, 16-, 256-, and 1,024-fold diluted human HVEM-expressing retrovirus, respectively, in the presence of Polybrene (6 μg/ml). SKNSH-A cells were generated by transducing SK-N-SH cells with 1 ml of the human HVEM-expressing retrovirus in the presence of Polybrene. Vero-sA1 and B78H1-sA1 cells were generated by transducing Vero and B78H1 cells with simian HVEM-expressing retrovirus in the presence of Polybrene. Expression of HVEM in the cells was confirmed by immunoblotting with goat anti-human HVEM antibody (directed against a peptide near the C terminus of HVEM) (sc-7765; Santa Cruz) or rabbit anti-human HVEM R140 (a gift from Claude Krummenacher at the University of Pennsylvania) and by flow cytometry for GFP (which was a surrogate marker for HVEM). Plaque morphology in cells infected with HSV was observed at 2 and 3 days postinfection using an Olympus IX51 microscope.
Analysis of HSV2-mutants in animal models.
All animal studies were conducted under a protocol approved by the Animal Care and Use Committee of the National Institute of Allergy and Infectious Diseases. For virulence testing of HSV-2 mutants, 6-week-old female BALB/c mice were subcutaneously injected with 0.1 ml of 20 mg/ml medroxyprogesterone acetate (Depo-Provera; Pfizer) to increase their susceptibility to HSV-2 intravaginal infection. Five days later, 10 mice per group were intravaginally infected with 104 PFU of HSV per mouse and monitored daily for signs of infection and mortality. Survival curves were analyzed using a permutation log rank test.
For testing latent infection of HSV-2 mutants, 6-week-old C57BL/6 mice were anesthetized and inoculated with virus by corneal scarification. Each cornea was scratched 16 times with a 25-gauge needle, followed by infection with 1,500 PFU of HSV-2 in 5 μl to each eye bilaterally. Mice received 0.5 ml of 1% human IgG (Gamunex-C; Talecris Biotherapeutics) intraperitoneally on the day of infection to reduce mortality associated with HSV-2 infection. Mice were euthanized 50 days later, both trigeminal ganglia were collected, and DNA was isolated with an ArchivePure DNA Cell/Tissue Kit (5 Prime). The HSV-2 DNA copy number in ganglion DNA was determined by TaqMan PCR as described previously (58), using primers and probes specific for the HSV-2 gG and mouse β-actin genes. The quantity of gG DNA was normalized to that of β-actin in each sample, and the results were analyzed by t test.
To determine the ability of viruses to reactivate, C57BL/6 mice were sacrificed 40 days after corneal infection, and trigeminal ganglia were cultured individually on Vero cell monolayers in the presence of 0.08% N,N′-hexamethylene-bis-acetamide. Vero cell monolayers were monitored daily for the formation of cytopathic effect (CPE) for up to 8 days, and the results were analyzed using Fisher's exact test.
To analyze neuroinvasiveness of viruses in mice, 6-week-old female BALB/c mice were injected intramuscularly with 16,000 PFU of HSV2-gD27 or 1,600 PFU of HSV2-ΔBAC38. Both viruses were titrated in B78H1-A10 cells; 1,600 PFU of HSV2-ΔBAC38 in B78H1-A10 cells is equivalent to 24,000 PFU in Vero cells. At various times after infection, lumbosacral dorsal root ganglia (DRG) from 5 mice were collected and homogenized in 500 μl of ARPE-19 cell culture medium with a Minilys homogenizer (Cayman Chemical Company) at 5,000 rpm for 20 s twice, and titers of infectious virus in homogenates were determined in ARPE-19 cells by plaque assay.
RESULTS
Construction and molecular characterization of an HSV-2 BAC clone, bHSV2-BAC38, for HSV-2 mutagenesis in bacteria.
We constructed a BAC clone, bHSV2-BAC38, for mutagenesis of HSV-2 in bacteria and characterized the HSV intermediates and viruses derived from the BAC clone bHSV2-BAC38. Three HSV-2 clones were precursors used to facilitate the cloning of HSV2-BAC38. First, eGFP and two loxP sequences were inserted into HSV-2 R519 (a plaque-purified virus obtained from HSV-2 strain 333) (Fig. 1, WT HSV-2) between the putative TATA boxes of HSV-2 UL37 and UL38 to generate HSV2-eGFP38 (Fig. 1). Second, eGFP and one of the loxP sequences was excised from the viral genome to obtain HSV2-loxP38 (Fig. 1). Third, a BAC vector sequence with eGFP and an loxP sequence was inserted into HSV2-loxP38 by contransfecting pBAC and pCre to yield HSV2-BAC38 (Fig. 1). A BAC clone, bHSV2-BAC38 (Fig. 1) that is an episomal form of the HSV2-BAC38 genome was obtained by transforming E. coli with circular viral DNA isolated from Vero cells infected with HSV2-BAC38. This BAC clone was subsequently used to generate HSV-2 mutants in bacteria.
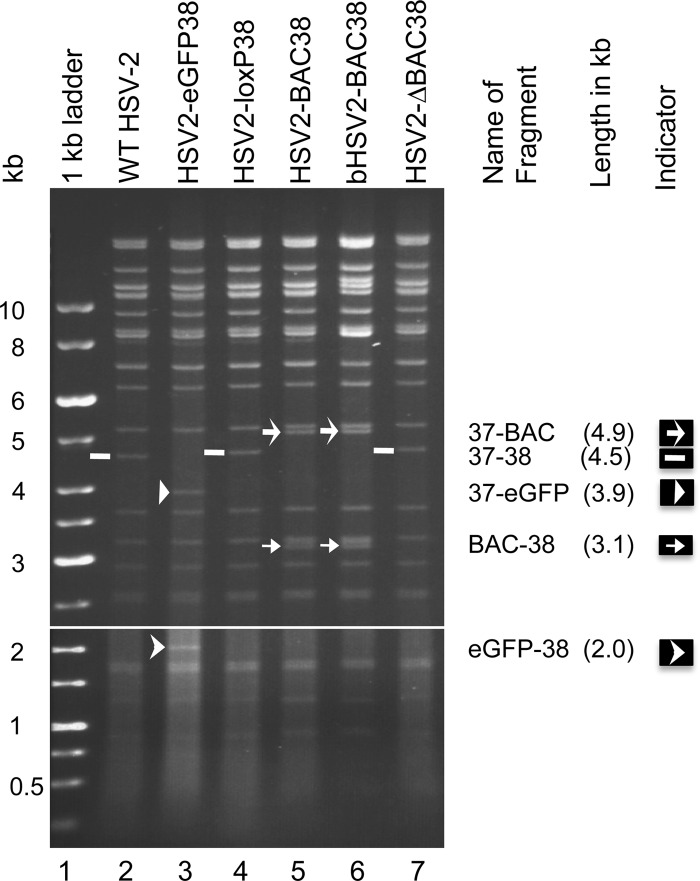
From beginning with the parental virus, HSV-2 R519, to ending with the BAC clone, bHSV2-BAC38, the multiple steps of molecular cloning might have introduced mutations in the HSV-2 backbone of bHSV2-BAC38. To ensure that the virus backbone in bHSV2-BAC38 is wild type and that the phenotypes of mutant viruses generated from the BAC clone are due to the specific mutations inserted in targeted gene(s), we reconstituted HSV-2 virus from bHSV2-BAC38 by cotransfecting Vero cells with bHSV2-BAC38 DNA and pCre plasmid to produce virus without the BAC vector. The reconstituted virus was designated HSV2-ΔBAC38. The genome of HSV2-ΔBAC38 should be identical to that of HSV2-loxP38 if no mutation and genome rearrangement occurred during the cloning process; both viruses should have a 34-bp loxP site insertion in the intergenic region between UL37 and UL38 compared to the wild-type virus. Restriction endonuclease digestion of virion DNA from HSV2-loxP38, HSV2-ΔBAC38, and wild-type R519 with AgeI showed identical bands (Fig. 3, lanes 2, 4, and 7) indicating that no large genome rearrangements occurred. Restriction endonuclease digestion of HSV2-eGFP38, HSV2-BAC38, and BAC plasmid DNA from bHSV-BAC38 showed that each clone had the expected inserted DNA (Fig. 3, lanes 3, 5, and 6). The new bands in bHSV-BAC38 were due to the joined termini of the viral genome in the circular BAC plasmid DNA; the other bands were identical to the wild-type virus R519. The virus-BAC vector junctions were confirmed by sequencing.
Fig 3.
Restriction endonuclease mapping of recombinant HSV-2 virion and BAC plasmid DNA. Virion DNA was isolated from infected Vero cells, and bHSV2-BAC38 DNA was purified from bacteria. DNA (1.6 μg) was digested with restriction enzyme AgeI overnight, and the digestion products and 1-kb DNA ladder were separated by electrophoresis in a 0.5% agarose gel at 1.7V/cm. The DNA bands were visualized at 8 h (bottom panel) and 22 h (top panel).
HSV2-ΔBAC38 replicates to similar titers as wild-type virus in Vero cells.
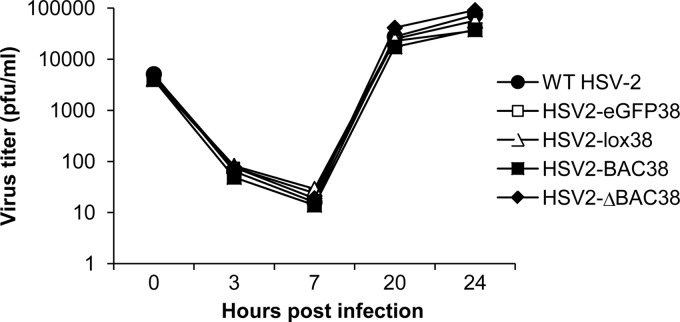
Virus derived from the BAC clone with a single loxP sequence (HSV2-ΔBAC38) was compared with wild-type HSV-2 R519 and to virus having an insertion of loxP (HSV2-loxP38), loxP-eGFP-loxP (HSV2-eGFP38), or loxP-eGFP-BAC vector-loxP (HSV2-BAC38) in a one-step growth curve in Vero cells. All of the viruses produced plaques of similar sizes, and the replication kinetics of all of the viruses were nearly identical (Fig. 4). Therefore, insertion of DNA sequences between UL37 and UL38 did not alter replication of HSV-2 in Vero cells in vitro.
Fig 4.
Replication of wild-type and recombinant HSV-2 with various inserts in Vero cells. Vero cell monolayers in 12-well culture plates were infected with 104 PFU of virus/well (multiplicity of infection, 0.05) and incubated at 37°C with 5% CO2. Cells were harvested at times as indicated on the horizontal axis, and viruses were titrated on Vero cells.
Virulence of HSV2-ΔBAC38 is comparable to that of wild-type virus in mice, while HSV2-BAC38 is attenuated.
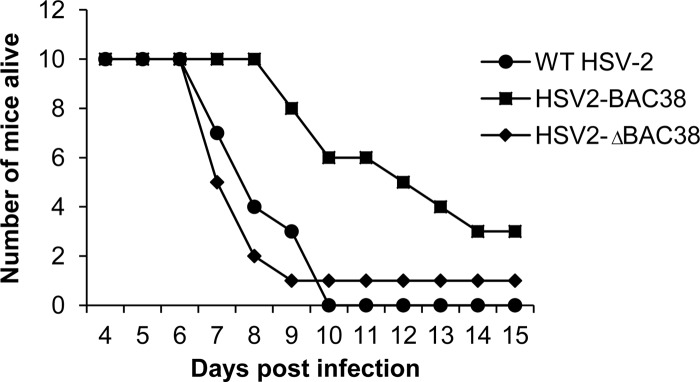
To determine if virus derived from the BAC clone is not impaired for virulence in mice, we infected animals intravaginally with 104 PFU of HSV2-ΔBAC38, wild-type HSV-2 R519, or HSV2-BAC38 (which contains the BAC vector). Animals infected with HSV2-ΔBAC38 or wild-type virus R519 began to die at day 7 after infection, and ≥90% of mice were dead by day 10 (Fig. 5). In contrast, animals infected with HSV2-BAC38 did not begin to die until day 9 after infection, and maximum mortality (70%) was not reached until day 14. The difference in survival between animals infected with HSV2-ΔBAC38, reconstituted from bHSV2-BAC38 and with the BAC vector sequence removed, and those infected with wild-type HSV-2 was not significant (P = 0.5). In contrast, the difference in survival between animals infected with HSV2-BAC38, which contains the BAC vector, and either wild-type HSV-2 or HSV2-ΔBAC38 was significant (P = 0.002). These data indicate that insertion of the BAC vector into the HSV-2 genome partially attenuated the virus in the mouse vaginal model of infection.
Fig 5.
Virulence of wild-type and recombinant HSV-2 in the mouse genital herpes model. Six-week-old female BALB/c mice were treated with 2 mg/mouse of medroxyprogesterone acetate subcutaneously. Five days later mice were infected intravaginally with 104 PFU of virus and monitored daily for signs of infection and mortality. Log rank test statistics was performed, and the P value was 0.002 for WT HSV-2 versus HSV2-BAC38, 0.508 for WT HSV-2 versus HSV2-ΔBAC38, and 0.015 for HSV2-BAC38 versus HSV2-ΔBAC38.
Latent infection and reactivation of HSV2-ΔBAC38 in mice are similar to those of wild-type virus.
HSV-2 latency and reactivation have been studied in mice in both the eye (31, 37) and genital models. To determine if HSV2-ΔBAC38 establishes latency similar to wild-type virus, C57BL/6 mice were infected with 1,500 PFU of virus in each eye after corneal scarification, and trigeminal ganglia were harvested 50 days after infection. Real-time PCR showed that the latent viral DNA load in ganglia in mice infected with HSV2-ΔBAC38 was comparable to that in mice infected with wild-type virus (P = 0.38) (Fig. 6A).
Fig 6.
Latent infection and reactivation of wild-type HSV-2 and HSV2-ΔBAC38 in mouse trigeminal ganglia. Six-week-old C57BL/6 mice were infected with 3,000 PFU of the indicated virus following corneal scarification and received 0.5 ml of 1% human IgG intraperitoneally. (A) Mice were euthanized 50 days later, DNA was isolated from both trigeminal ganglia, and the latent viral DNA load was determined by real-time quantitative PCR using primers and a probe specific for the HSV-2 gG gene. The amount of amplifiable HSV-2 DNA was normalized to the amount of amplifiable β-actin DNA in each sample. The ratios of the group medians of the gG/β-actin value from each group to the group medians from wild-type virus-infected animals (P = 0.38, t test) are shown. (B) The ability of HSV2-ΔBAC38 virus to reactivate from latently infected ganglia was assessed by explant cocultivation. C57BL/6 mice were infected by corneal scarification as described for panel A. Seven mice from each group were sacrificed 40 days postinfection, and trigeminal ganglia (TG) were removed and cultured individually with Vero cell monolayers in the presence of 0.08% N,N′-hexamethylene-bis-acetamide. The cultures were monitored daily for the formation of CPE for up to 8 days, and the percentages of trigeminal ganglia that yielded cultures with CPE were compared and analyzed with Fisher's exact test (P = 1.00).
To compare the ability of HSV2-ΔBAC38 to reactivate from latency in mice with that of wild-type virus, latently infected mouse trigeminal ganglia were explant cocultivated with Vero cells. HSV-2 reactivated from 85% of trigeminal ganglia of mice infected with each virus (P = 1) (Fig. 6B). Thus, both establishment of and reactivation from latency were similar in mice infected with HSV2-ΔBAC38 and wild-type virus. Therefore, virus obtained from bHSV-2-BAC has a wild-type phenotype in vitro and in vivo, and virus mutants generated from the BAC clone should have phenotypes related to the mutations and not due to the intermediate BAC cloning steps.
Construction and molecular characterization of HSV-2 with mutations D215G, R222N, and F223I in gD.
We used the HSV-2 BAC clone bHSV2-BAC38 and the bacterial galK selection system to generate an HSV-2 mutant. We inserted point mutations in the HSV-2 gD gene to construct an HSV-2 gD mutant that could use HVEM, but not nectin-1, as an entry receptor. First, we inserted a galK expression cassette into the gD gene of bHSV2-BAC38 (Fig. 2, top) to generate BAC clone bHSV2-BAC38-gD/galK+ with a galK expression cassette inserted between amino acids 214 and 224 of gD (Fig. 2, second panel). Next, we replaced the galK gene in bHSV2-BAC38-gD/galK+ with a portion of the gD gene with point mutations in amino acids 215, 222, and 223 to yield bHSV2-BAC38-gD27 (Fig. 2, third panel). To obtain virus without BAC vector sequence from bHSV2-BAC38-gD27, B78H1-A10 cells (mouse melanoma cells expressing human HVEM) were cotransfected with bHSV2-BAC38-gD27 and plasmid pCre, and the virus obtained was plaque purified and termed HSV2-gD27 (Fig. 2, bottom). A rescued virus, with the mutations in gD repaired, was constructed by cotransfecting Vero cells with HSV2-gD27 virion DNA and plasmid pCMV-gD2 (which contains the gD open reading frame of HSV-2 strain 333). Virus progeny was plaque purified in B78H1-C10 cells (mouse melanoma cells expressing human nectin-1) and designated HSV2-gD27R.
To confirm that the genomes of the HSV-2 gD mutant and the rescued virus did not have major genome rearrangements, virion DNA was prepared from HSV2-ΔBAC38, HSV2-gD27, and HSV2-gD27R and digested with BamHI. As expected, identical patterns of bands were present since the mutations in gD do not change BamHI sites in the virus (Fig. 7). PCR products encompassing the entire gD gene were amplified from virion DNAs of HSV2-gD27 and HSV2-gD27R and sequenced. The former virus had the expected gD mutations, the latter virus did not have these mutations, and the rest of the gD sequence in both viruses was wild type.
Fig 7.
Restriction endonuclease mapping of the HSV2-ΔBAC38 virus (derived from bHSV2-BAC38), HSV2-gD27, and its rescued virus HSV2-gD27R. Virion DNA was digested with BamHI and processed as described in the legend to Fig. 3.
HSV2-gD27 infects cells expressing HVEM but cannot infect cells expressing only nectin-1, including neuronal cell lines.
To determine if the mutations in gD altered the ability of HSV-2 to infect cells expressing nectin-1, HSV2-ΔBAC38, HSV2-gD27, and HSV2-gD27R were propagated and titrated in B78H1-A10 cells and used to infect mouse melanoma cell lines B78H1 (resistant to HSV infection), B78H1-A10 (expressing human HVEM), and B78H1-C10 (expressing human nectin-1). HSV2-ΔBAC38 and HSV2-gD27R produced few plaques in B78H1 cells but grew well in both B78H1-A10 and B78H1-C10 cells (Fig. 8A). In contrast, HSV2-gD27 lost the ability to infect B78H1-C10 cells but still infected B78H1-A10 cells. Infection with HSV2-gD27 propagated in B78H1-A10 cells yielded 1.7 × 105 plaques in B78H1-A10 cells but no plaques in B78H1-C10 cells. Thus, the mutations in HSV2-gD27 (D215G, R222N, and F223I) impaired the ability of HSV-2 to infect cells that express only nectin-1 by ≥10,000-fold.
Fig 8.
Infectivity of HSV2-gD27 in cell lines engineered to express human HSV receptors in epithelial and neuronal cell lines and in ganglia of mice. HSV2-ΔBAC38 (which served as the wild-type HSV-2 control), HSV2-gD27, and the rescued virus HSV2-gD27R were propagated in B78H1-A10 cells. The virus titers in all cell lines were determined by plaque assay and compared with their own titers in B78H1-A10 cells. (A) Viral titers of HSV-2 gD and control viruses in mouse melanoma cell line B78H1 (resistant to HSV infection), B78H1-A10 (expressing human HVEM), and B78H1-C10 (expressing human nectin-1). (B) Viral titers of HSV2-gD27 mutant and control viruses in the human epithelial cell line ARPE-19 and neuronal cell line SK-N-SH. (C) Mice were injected intramuscularly with 16,000 PFU of HSV2-gD27 or 1,600 PFU of HSV2-ΔBAC38 in the thigh. On days 2, 3, 4, and 5 postinfection, lumbosacral dorsal root ganglia were harvested and homogenized. The titer of HSV-2 in the homogenates was determined in ARPE-19 cells by plaque assay.
Since epithelial cells express HVEM and nectin-1 while neurons express abundant nectin-1 but undetectable HVEM, we tested the infectivity of HSV2-gD27 in a human epithelial cell line (ARPE-19) and in human neuronal cell lines SK-N-SH and SH-SY5Y. The infectivities of HSV2-ΔBAC38, HSV2-gD27, and HSV2-gD27R were similar in B78H1-A10 and ARPE-19 (a human epithelial cell line) cells (Fig. 8B). However, while HSV2-ΔBAC38 and HSV2-gD27R infected SK-N-SH neuronal cells to similar levels, no plaques were observed with HSV2-gD27. The mutations in HSV2-gD27 impaired the ability of HSV-2 to infect the neuronal cells by at least 10,000-fold. Similar results were seen in SH-SY5Y neuronal cells (data not shown).
HSV2-gD27 cannot infect Vero cells unless the cells are transduced with a vector expressing HVEM.
To our surprise, HSV2-gD27 failed to infect Vero cells (Fig. 9A), an African green monkey kidney cell line widely used in laboratories for growing HSV. To investigate the mechanism of resistance of human neuronal cells and simian Vero cells to the infection with HSV2-gD27, cell lysates of SK-N-SH and Vero cells were immunoblotted with goat anti-human HVEM antibody. HVEM was not detected in either SK-N-SH or Vero cells (Fig. 9C). Therefore, we transduced SK-N-SH and Vero cells with a retrovirus expressing human HVEM to produce SKNSH-A and Vero-A1 cells, respectively. Immunoblotting detected HVEM in both SKNSH-A and Vero-A1 cells (Fig. 9C).
Fig 9.
Infection of Vero and SK-N-SH cell lines expressing exogenous human or simian HVEM by HSV2-gD27 and control viruses. (A) Vero and SK-N-SH cells were transduced with retrovirus expressing human HVEM and designated Vero-A1 and SKNSH-A cells, respectively. The ratios of titers of HSV2-gD27 and control viruses (determined by plaque assay) in various cell lines to titers in B78H1-A10 cells are shown. (B) Vero and B78H1 cells were transduced with simian HVEM and designated Vero-sA1 and B78H1-sA1, respectively. The ratios of titers of HSV2-gD27 and control viruses in different cell lines to titers in B78H1-A10 cell are shown. (C) Expression of human HVEM was determined by immunoblotting. Cell lysates were subjected to PAGE and transferred to membranes, and expression of human HVEM in Vero-A1 and SKNSH-A cells was detected with goat anti-human HVEM. (D) Expression levels of human and simian HVEM in various cells were detected with rabbit anti-human HVEM. Membranes in panels C and D were incubated with antibody to β-actin as a cellular protein control.
Expression of human HVEM in SKNSH-A and Vero-A1 cells did not significantly increase the titers of HSV2-ΔBAC38 and HSV2-gD27R compared with their titers in parental SK-N-SH and Vero cells (Fig. 9A). In contrast, expression of human HVEM in SKNSH-A and Vero-A1 cells rendered the cells susceptible to HSV2-gD27 infection, resulting in numbers of plaques comparable to those for infection with HSV2-ΔBAC38 and HSV2-gD27R (Fig. 9A). Thus, the resistance of Vero and human neuronal cells to infection of HSV2-gD27 is due to insufficient expression of functional HVEM in these cells.
To further study the dependency of HSV2-gD27 infection on HVEM in Vero cells, we generated a series of Vero cell lines expressing different levels of HVEM. Vero-A1, Vero-A4, Vero-A16, Vero-A256, and Vero-A1024 were generated by infecting Vero cells with increasing dilutions of retrovirus expressing HVEM. The level of GFP in the various cell lines, a surrogate marker for expression of HVEM which was inserted in the retrovirus vector upstream of the GFP gene, was inversely proportional to the dilution of retrovirus used to generate the cell lines (Fig. 10A). We confirmed this finding by observing that the level of HVEM was negatively correlated with the dilution of retrovirus used to generate the Vero-A1 and -A16 cell lines (Fig. 10B). The level of HVEM expression was positively correlated with the number of plaques observed in Vero-A cells infected with HSV2-gD27 (Fig. 10C).
Fig 10.
HVEM expression in Vero-A cell lines and complementation of the HSV2-gD27 mutant. Vero cells expressing different amounts of human HVEM were generated by transducing Vero cells with serial 4-fold dilutions of retrovirus expressing human HVEM and GFP and designated Vero-A1, Vero-A4, etc. (A) GFP expression in retrovirus-transduced cell lines, which served as a surrogate marker for HVEM expression from the retrovirus vector. (B) The level of HVEM expression in cell lines was determined by Western blotting with goat anti-human HVEM. β-Actin served as a cellular protein control. (C) Virus titers of HSV2-ΔBAC38 (wild-type control) and HSV2-gD27 in HVEM-transduced Vero-A cell lines. (D) Plaque morphologies of HSV2-ΔBAC38 and HSV2-gD27 in Vero and Vero-A1 cells expressing human HVEM.
Since HSV2-gD27 did not infect Vero cells, it is possible that these cells express relatively low levels of HVEM or that HSV2-gD27 cannot use Vero cell HVEM effectively. Therefore, we determined whether overexpression of Vero cell HVEM in Vero cells could increase the susceptibility of the cells to HSV2-gD27 infection. Vero cells were transduced with retrovirus containing Vero cell (simian) HVEM cDNA, and the resulting cells were designated Vero-sA1. Expression of simian HVEM in Vero-sA1 cells was detected by immunoblotting with a rabbit polyclonal anti-human HVEM antibody (R140) (Fig. 9D). Even though Vero-sA1 overexpressed simian HVEM, the level of HSV2-gD27 infection in these cells was more than 10,000-fold less than infection of Vero-A1 cells which express human HVEM (Fig. 9B). To study this effect further, we then transduced B78H1 mouse cells, which are resistant to HSV infection, with retrovirus expressing simian HVEM to produce B78H1-sA1 cells. While B78H1-sA1 cells expressed more HVEM than B78H1-A10 cells (which express human HVEM) based on immunoblotting (Fig. 9D), wild-type HSV-2 and HSV2-gD27 infected B78H1-sA1 at levels of only 4.5% and 0%, respectively, compared with the levels observed with infection of B78H1-A10 cells (Fig. 9B). Therefore, we conclude that simian HVEM is much less effective in mediating HSV-2 infection than human HVEM.
Overexpression of HVEM in Vero cells increases syncytia and cell-to-cell spread in cells infected with wild-type HSV-2.
Examination of plaques in cells infected with HSV2-ΔBAC38 showed that the virus resulted in larger, more syncytial plaques in Vero-A1 cells than in Vero cells (Fig. 10D). Similar results were seen after infection with HSV-2 strain R519 (data not shown). Large plaques with syncytia were not observed with HSV infection of other cell lines expressing human or simian HVEM. These results indicate that overexpression of HVEM increases fusion between infected cells and increases cell-to-cell spread of wild-type HSV-2. In contrast, infection of Vero-A1 cells with HSV2-gD27 did not result in large, syncytial plaques. HSV2-gD27 produced smaller plaques than wild-type virus in all other cell lines in which the gD mutant was able to produce plaques. These data indicate that amino acids 215, 222, and 223 of gD are important not only for binding to nectin-1 but also are involved in fusion and cell-to-cell spread of virus.
HSV2-gD27 is attenuated for neuroinvasiveness in mice.
Since HSV2-gD27 is severely impaired for infection of neuronal cell lines in vitro, we determined whether the virus had reduced neurovirulence in a mouse model. Mice were infected intramuscularly in the thigh with 16,000 PFU of HSV2-gD27 or 1,600 PFU of wild-type HSV-2 (HSV2-ΔBAC38). Mice infected with HSV2-gD27 showed no signs of disease, while animals infected with wild-type virus lost hair at the injection site, and 20% had hind limb paralysis in the leg that was inoculated with virus. On days 2, 3, 4, and 5 postinfection, lumbosacral dorsal root ganglia were collected and homogenized, and HSV-2 titers were determined in ARPE-19 cells. No virus was recovered from any of the mice infected with HSV2-gD27, while virus was detected in DRG homogenates in 60% of mice infected with wild-type virus (Fig. 8C). These data indicate that the HSV2-gD27 has reduced neuroinvasiveness in mice.
DISCUSSION
While HSV-2 is associated with morbidity at a level similar to if not greater than that of HSV-1, less information is known about HSV-2. At present there is only a single published complete genome sequence available (strain HG52) (12) for HSV-2 compared to four for HSV-1 (strains 17 [32, 33, 40], F, H129 [47], and KOS [28a; K. M. Payne, D. A. Russell, and P. R. Kinchington, GenBank accession no. JQ780693]). While three HSV-1 BAC clones (which have no viral genes deleted or mutated in the viral backbone) are available for HSV-1 mutagenesis studies (17, 27, 48), the HSV-2 BAC clones that have been reported are not optimal for pathogenesis studies in animals because the virus backbones in these BAC clones have one or more genes deleted (34, 41). Therefore, we developed a BAC clone that could be used to generate virus with minimal extraneous sequences and that would be inserted into a site in the genome that should not affect virus replication or pathogenesis. Since insertion of BAC vector sequences into other herpesviruses can reduce the virulence of the virus (1, 57), we designed the HSV2-BAC so that infectious HSV-2 can be easily reconstituted from our BAC clone, bHSV2-BAC38, with only a 34-bp loxP sequence remaining between UL37 and UL38 but no BAC vector sequence. Virus derived from the BAC clone replicated identically to wild-type virus in vitro, and acute and latent infection in mice as well as reactivation from latency was virtually indistinguishable from that of wild-type virus. This validation was essential to ensure that BAC clone bHSV2-BAC38 is appropriate for generating mutations in HSV-2 genes for elucidating their functions in cell culture, in acute and latent infection, and in reactivation.
HSV gD is essential for virus entry, binds to several cellular receptors including HVEM and nectin-1, and is involved in virus-cell membrane fusion (8, 16, 25, 45, 54). To construct a mutant HSV-2 that is unable to use nectin-1 as a receptor but is still able to use HVEM, we mutated three amino acids in gD (D215, R222, and F223) that were reported to be critical for its interaction with nectin-1 but not with HVEM (5, 11, 30). The cell tropism of our HSV2-gD27 is very similar to that of two previously reported constructs: a gD null virus transiently complemented with recombinant gD bearing the same three amino acid mutations (30) as our HSV2-gD27 and an HSV-1 mutant harboring mutations R222N/F223I or A3C/Y38C in gD (8, 55). All of these mutants can infect cell lines engineered to express HVEM but are impaired for infecting cells that express nectin-1 in the absence of HVEM. However, when these mutants are tested in cell lines that naturally express HVEM, the infectivity of the mutants is not always predictable. The gD null virus transiently complemented with recombinant D with three mutations is impaired for infecting IMR-5 and A431 cells that express HVEM (24, 30), while the HSV-1 gD mutant with A3C/Y38C mutations is not able to infect a human tumor cell line, HT29, that also expresses HVEM (55).
To our surprise, we found that our HSV2-gD27 mutant was not able to infect Vero cells. Vero cells, an African green monkey kidney cell line, encode an ortholog of HVEM that differs from the human protein by 41 amino acids, of which 17 amino acids are nonconservative changes compared with the human sequence (13). However, CHO cells expressing Vero cell HVEM are susceptible to HSV-1 infection, while CHO cells not expressing the protein are not infected (13), indicating that the simian protein is able to mediate entry of wild-type HSV. Krummenacher et al. detected low levels of HVEM on the surface of Vero cells by flow cytometry (24). We could not reliably detect HVEM in Vero cells with two different polyclonal antibodies to HVEM using immunoblotting, but HVEM was detected in Vero cells transduced with retrovirus vectors expressing simian or human HVEM (Fig. 9). Nectin-1 has been detected on the surface of Vero cells (24), but unlike with other cell types, nectin-1 is not downregulated after incubation of Vero cells with cells expressing gD (46). Nectin-1 from Vero cells can function as a receptor for HSV in cells that cannot otherwise support HSV infection, and anti-nectin-1 antibodies can block HSV entry into Vero cells (36).
We found that over expression of human HVEM in Vero cells rendered the cells susceptible to infection with the HSV2-gD27 mutant. We postulated several reasons why Vero cells are resistant to infection by HSV2-gD27. First, the level of simian HVEM in Vero cells may be insufficient to support the entry of HSV2-gD27. Second, the sequence of simian HVEM differs sufficiently from human HVEM such that simian HVEM might not support entry of the HSV gD mutant. Third, Vero cells may lack a functional coreceptor required for HVEM-mediated entry of HSV2-gD27 which can be compensated by overexpression of human HVEM. The observation that overexpression of simian HVEM in Vero-sA1 and B78H1-sA1 cells resulted in a very modest increase in wild-type HSV-2 and HSV2-gD27 infection supports the hypothesis that simian HVEM is less effective than human HVEM in mediating HSV-2 infection.
We found that overexpression of HVEM in Vero cells resulted in increased syncytia and enhanced cell-to-cell spread in cells infected with wild-type HSV but not with the HSV2-gD27 mutant. In addition, HSV2-gD27 exhibited a small-plaque phenotype in all the cell types that were susceptible to infection, including B78H1-A10 cells that express only human HVEM (data not shown). These data imply that amino acids 215, 222, and 223 are involved in cell-to-cell spread. These findings are supported by reversal of the small-plaque phenotype in Vero-A1 cells infected with gD27R, the rescued virus.
Manoj and colleagues postulated that HSV gD mutants that do not interact with nectin-1 might be nonneurotropic in animals (30). Studies of nectin-1 knockout mice indicate an important role for nectin-1 in neuronal infection by HSV (22). Since the HSV2-gD27 mutant did not cause signs of infection in mice after intramuscular injection and since no infectious virus could be recovered from dorsal root ganglia after infection, we conclude that the nectin-1 interaction domain of gD is critical for neurotropism of HSV-2 in vivo and that HSV2-gD27 is a promising vaccine candidate.
ACKNOWLEDGMENTS
This research was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases.
We thank Peter Barry, Anthony V. Nicola, Patricia G. Spear, Neal Copland, Gary H. Cohen, Roselyn J. Eisenberg, Claude Krummenacher, and Konstantin G. Kousoulas for sharing reagents and Jing Qin for help with statistical analyses.
Footnotes
Published ahead of print 19 September 2012
REFERENCES
- 1. Adler H, Messerle M, Koszinowski UH. 2001. Virus reconstituted from infectious bacterial artificial chromosome (BAC)-cloned murine gammaherpesvirus 68 acquires wild-type properties in vivo only after excision of BAC vector sequences. J. Virol. 75:5692–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borenstein R, Frenkel N. 2009. Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Proc. Natl. Acad. Sci. U. S. A. 106:19138–19143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borst EM, Hahn G, Koszinowski UH, Messerle M. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brugha R, Keersmaekers K, Renton A, Meheus A. 1997. Genital herpes infection: a review. Int. J. Epidemiol. 26:698–709 [DOI] [PubMed] [Google Scholar]
- 5. Carfi A, et al. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. D Biol. Crystallogr. 58:836–838 [DOI] [PubMed] [Google Scholar]
- 6. Carfi A, et al. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169–179 [DOI] [PubMed] [Google Scholar]
- 7. Chang WL, Barry PA. 2003. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J. Virol. 77:5073–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Connolly SA, et al. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 79:1282–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corey L, Adams HG, Brown ZA, Holmes KK. 1983. Genital herpes simplex virus infections: clinical manifestations, course, and complications. Ann. Intern. Med. 98:958–972 [DOI] [PubMed] [Google Scholar]
- 10. Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Di Giovine P, et al. 2011. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 7:e1002277 doi:10.1371/journal.ppat.1002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dolan A, Jamieson FE, Cunningham C, Barnett BC, McGeoch DJ. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foster TP, Chouljenko VN, Kousoulas KG. 1999. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology 258:365–374 [DOI] [PubMed] [Google Scholar]
- 14. Freeman EE, et al. 2006. Herpes simplex virus 2 infection increases HIV acquisition in men and women: systematic review and meta-analysis of longitudinal studies. AIDS 20:73–83 [DOI] [PubMed] [Google Scholar]
- 15. Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 16. Gianni T, Amasio M, Campadelli-Fiume G. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284:17370–17382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gierasch WW, et al. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206 [DOI] [PubMed] [Google Scholar]
- 18. Gilbert M, et al. 2011. Using centralized laboratory data to monitor trends in herpes simplex virus type 1 and 2 infection in British Columbia and the changing etiology of genital herpes. Can. J. Public Health 102:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirt B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365–369 [DOI] [PubMed] [Google Scholar]
- 20. Jurak I, et al. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 84:4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kinzler ER, Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kopp SJ, et al. 2009. Infection of neurons and encephalitis after intracranial inoculation of herpes simplex virus requires the entry receptor nectin-1. Proc. Natl. Acad. Sci. U. S. A. 106:17916–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krause PR, et al. 1995. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J. Exp. Med. 181:297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krummenacher C, et al. 2004. Comparative usage of herpesvirus entry mediator A and nectin-1 by laboratory strains and clinical isolates of herpes simplex virus. Virology 322:286–299 [DOI] [PubMed] [Google Scholar]
- 25. Lazear E, et al. 2008. Engineered disulfide bonds in herpes simplex virus type 1 gD separate receptor binding from fusion initiation and viral entry. J. Virol. 82:700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leone P. 2005. Reducing the risk of transmitting genital herpes: advances in understanding and therapy. Curr. Med. Res. Opin. 21:1577–1582 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Wang S, Zhu H, Zheng C. 2011. Cloning of the herpes simplex virus type 1 genome as a novel luciferase-tagged infectious bacterial artificial chromosome. Arch. Virol. 156:2267–2272 [DOI] [PubMed] [Google Scholar]
- 28. Looker KJ, Garnett GP, Schmid GP. 2008. An estimate of the global prevalence and incidence of herpes simplex virus type 2 infection. Bull. World Health Organ. 86:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a. Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. 2012. Genome sequence of herpes simplex virus 1 strain KOS. J.Virol. 86:6371–6372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malkin JE. 2004. Epidemiology of genital herpes simplex virus infection in developed countries. Herpes 11(Suppl 1):2A–23A [PubMed] [Google Scholar]
- 30. Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. 2004. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc. Natl. Acad. Sci. U. S. A. 101:12414–12421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Margolis TP, Imai Y, Yang L, Vallas V, Krause PR. 2007. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J. Virol. 81:1872–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGeoch DJ, et al. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531–1574 [DOI] [PubMed] [Google Scholar]
- 33. McGeoch DJ, Dolan A, Donald S, Rixon FJ. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1–13 [DOI] [PubMed] [Google Scholar]
- 34. Meseda CA, Schmeisser F, Pedersen R, Woerner A, Weir JP. 2004. DNA immunization with a herpes simplex virus 2 bacterial artificial chromosome. Virology 318:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miller CG, Krummenacher C, Eisenberg RJ, Cohen GH, Fraser NW. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160–168 [DOI] [PubMed] [Google Scholar]
- 36. Milne RS, et al. 2003. Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion. J. Virol. 77:8962–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mitchell WJ, Deshmane SL, Dolan A, McGeoch DJ, Fraser NW. 1990. Characterization of herpes simplex virus type 2 transcription during latent infection of mouse trigeminal ganglia. J. Virol. 64:5342–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Montgomery RI, Warner MS, Lum BJ, Spear PG. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427–436 [DOI] [PubMed] [Google Scholar]
- 39. Nagaike K, et al. 2004. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 22:4069–4074 [DOI] [PubMed] [Google Scholar]
- 40. Perry LJ, McGeoch DJ. 1988. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:2831–2846 [DOI] [PubMed] [Google Scholar]
- 41. Schmeisser F, Weir JP. 2006. Cloning of replication-incompetent herpes simplex viruses as bacterial artificial chromosomes to facilitate development of vectors for gene delivery into differentiated neurons. Hum. Gene Ther. 17:93–104 [DOI] [PubMed] [Google Scholar]
- 42. Sengupta A, Banerjee D, Chandra S, Banerjee S. 2006. Gene therapy for BCR-ABL+ human CML with dual phosphorylation resistant p27Kip1 and stable RNA interference using an EBV vector. J. Gene Med. 8:1251–1261 [DOI] [PubMed] [Google Scholar]
- 43. Shizuya H, et al. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. U. S. A. 89:8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spear PG. 2004. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 6:401–410 [DOI] [PubMed] [Google Scholar]
- 45. Spear PG, et al. 2006. Different receptors binding to distinct interfaces on herpes simplex virus gD can trigger events leading to cell fusion and viral entry. Virology 344:17–24 [DOI] [PubMed] [Google Scholar]
- 46. Stiles KM, Milne RS, Cohen GH, Eisenberg RJ, Krummenacher C. 2008. The herpes simplex virus receptor nectin-1 is down-regulated after trans-interaction with glycoprotein D. Virology 373:98–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J. Virol. 84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tang H, et al. 2010. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology 407:360–367 [DOI] [PubMed] [Google Scholar]
- 50. Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. 2011. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J. Virol. 85:4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang S, et al. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U. S. A. 105:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral miRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83:1433–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tischer BK, et al. 2007. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J. Virol. 81:13200–13208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turner A, Bruun B, Minson T, Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uchida H, et al. 2009. Generation of herpesvirus entry mediator (HVEM)-restricted herpes simplex virus type 1 mutant viruses: resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J. Virol. 83:2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Umbach JL, et al. 2010. Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J. Virol. 84:1189–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wagner M, Jonjic S, Koszinowski UH, Messerle M. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang K, Pesnicak L, Guancial E, Krause PR, Straus SE. 2001. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J. Virol. 75:8166–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang K, Pesnicak L, Straus SE. 1997. Mutations in the 5′ end of the herpes simplex virus type 2 latency-associated transcript (LAT) promoter affect LAT expression in vivo but not the rate of spontaneous reactivation of genital herpes. J. Virol. 71:7903–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36 doi:10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Weiss H. 2004. Epidemiology of herpes simplex virus type 2 infection in the developing world. Herpes 11(Suppl 1):24A–35A [PubMed] [Google Scholar]
- 62. Workowski KA, Berman S. 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recommd. Rep. 59(RR-12):1–110 [PubMed] [Google Scholar]
- 63. Yoshii H, Somboonthum P, Takahashi M, Yamanishi K, Mori Y. 2007. Cloning of full-length genome of varicella-zoster virus vaccine strain into a bacterial artificial chromosome and reconstitution of infectious virus. Vaccine 25:5006–5012 [DOI] [PubMed] [Google Scholar]
- 64. Zhang Z, et al. 2007. Genetic analysis of varicella-zoster virus ORF0 to ORF4 by use of a novel luciferase bacterial artificial chromosome system. J. Virol. 81:9024–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou FC, et al. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]