Abstract
Cu,Zn SOD1 (superoxide dismutase 1) is implicated in FALS (familial amyotrophic lateral sclerosis) through the accumulation of misfolded proteins that are toxic to neuronal cells. Loop VI (residues 102–115) of the protein is at the dimer interface and could play a critical role in stability. The free cysteine residue, Cys111 in the loop, is readily oxidized and alkylated. We have found that modification of this Cys111 with 2-ME (2-mercaptoethanol; 2-ME-SOD1) stabilizes the protein and the mechanism may provide insights into destabilization and the formation of aggregated proteins. Here, we determined the crystal structure of 2-ME-SOD1 and find that the 2-ME moieties in both subunits interact asymmetrically at the dimer interface and that there is an asymmetric configuration of segment Gly108 to Cys111 in loop VI. One loop VI of the dimer forms a 310-helix (Gly108 to His110) within a unique β-bridge stabilized by a hydrogen bond between Ser105-NH and His110-CO, while the other forms a β-turn without the H-bond. The H-bond (H-type) and H-bond free (F-type) configurations are also seen in some wild-type and mutant human SOD1s in the Protein Data Bank suggesting that they are interconvertible and an intrinsic property of SOD1s. The two structures serve as a basis for classification of these proteins and hopefully a guide to their stability and role in pathophysiology.
Keywords: superoxide dismutase 1 (SOD1), crystal structure, amyotrophic lateral sclerosis (ALS), asymmetric configuration
Abbreviations: ALS, amyotrophic lateral sclerosis; FALS, familial amyotrophic lateral sclerosis; 2-ME, 2-mercaptoethanol; PDB, protein data bank; SOD1, superoxide dismutase 1; WT, wild-type
INTRODUCTION
Cu,Zn SOD1 (superoxide dismutase 1) is a homodimer and contains one copper ion and one zinc ion per 16 kDa subunit. The enzyme catalyses the dismutation of the toxic superoxide anion radical (O2−) to produce O2 and H2O2, thereby protecting cells against oxidative stress. Further, mutations in SOD1 have been found in patients with FALS (familial amyotrophic lateral sclerosis), which is a neurological disease affecting motor neurons of the spinal cord [1,2]. Transgenic mice that express the mutant human SOD1 linked to ALS (amyotrophic lateral sclerosis) develop progressive neurodegeneration and a phenotype that clearly resembles human FALS [3,4]. Accumulating evidence suggests that the FALS-linked mutations cause misfolding/aggregation of SOD1 proteins, which is thought to be toxic to motor neurons [5,6]. Recent studies have suggested that WT-SOD1 (wild-type SOD1) shares an aberrant conformation with ALS mutant SOD1s, which may underlie the protein's toxity in sporadic ALS [7–10]. FALS-mutant SOD1s apparently give rise to distinct structures of loop VI (residues 102–115 in human SOD1 connecting β6/β7 and designated as a Greek key loop) because three monoclonal antibodies having epitopes in loop VI bind with differing affinities to different FALS mutants, even though the mutations exist in locations other than loop VI [11]. Clarification of the structure of loop VI and its permutations may lead to a better understanding of the molecular mechanism of the pathogenesity of the SOD1 protein.
The tertiary structure of SOD1 is characterized by the presence of a Greek key β-barrel containing an internal disulfide bond between Cys57 and Cys146 [12,13], both of which contribute to the protein's high stability. Although this disulfide bond is highly conserved in SOD1s from almost all organisms, free cysteine residues are not. Only the SOD1s from humans and great apes (chimpanzees and orangutans) among mammals have the two free cysteine residues, Cys6 and Cys111 [14]. Yeast and most plants have no free cysteine residues, and residue 6 is an alanine and residue 111 is a serine in these organisms [15]. More highly evolved organisms, such as frogs and mammals, including bovines and the Japanese monkey, have only one free cysteine residue, Cys6. Free cysteine residues are generally reactive and WT human SOD1 is less thermo-stable than the stable mutant SOD1s, C111S and C6A [16]. Cys111 located in loop VI at the surface of the human SOD1 molecule is particularly reactive. In fact, we previously demonstrated that the sulfhydryl group of Cys111 (Cys111-SH) is selectively peroxidized to a sulfinic acid (Cys111-SO2H) and further to sulfonic acid (Cys111-SO3H) even by air oxidation [17]. The oxidized SOD1 including Cys111-SO3H tends to misfold and aggregate, a characteristic relevant to the pathology of ALS [8,17]. The peroxidized monomers are readily distinguished from the natural reduced monomers having Cys111-SH as an upper shifted band on SDS/PAGE, and a half-oxidized SOD1 consisting of a reduced subunit and an oxidized subunit can be purified by anion-exchange chromatography [17]. Furthermore, modification of Cys111 with 4-nitrobenzoatothioate or 5,5′-dithiobis(2-nitrobenzoic acid) is not complete (never exceeding approximately 50%) under relatively mild conditions [18,19]. The half modification may be due to incomplete reaction [18] or steric hindrance by the first modified Cys111 [19]. In contrast, recombinant human SOD1 that has been modified by reaction with 2-ME (2-mercaptoethanol) only at Cys111 in a procedure developed by Ube Industries Ltd (2-ME-SOD1; Cys111-S-S-CH2CH2OH) is strongly resistant to oxidation and more stable than unmodified SOD1 [17].
In the present study, to explain the enhanced stability resulting from modification with 2-ME, the crystal structure of 2-ME-SOD1 was determined. Although 2-ME-SOD1 formed the typical homodimer of SOD1, we unexpectedly found that only loop VI, which includes the 2-ME-modified Cys111 residue, is completely asymmetric between the subunits. Asymmetrical interaction between the 2-ME moieties at the dimer interface appears to stabilize 2-ME-SOD1. The asymmetry of loop VI is composed of two types of structure, which can be defined by the presence or absence of a hydrogen bond between Ser105-NH and His110-CO on the main chain. The structures of loop VI can therefore be designated as either hydrogen bond type (H-type) or a hydrogen bond free type (F-type) and the distinction allows a classification of WT, chemically modified, and mutant human SOD1s, and can be extended to SOD1 structures from a variety of organisms.
EXPERIMENTAL
Proteins
Recombinant SOD1 (residues 1–153) chemically modified by reaction with 2-ME at Cys111 (Cys111-S-S-CH2CH2OH) was generously donated by Ube Industries Ltd. The fact that 2-ME only modified the side chain of Cys111, not Cys6, was verified by MALDI–TOF MS (matrix-assisted laser-desorption ionization–time-of-flight MS) analysis [17].
Oxidation and alkylation
The formation of WT-SOD1 by removal of 2-ME from 2-ME-SOD1 and the purification of half-oxidized SOD1 consisting of a reduced subunit with Cys111-SH and an oxidized subunit with Cys111-SO3H by anion-exchange chromatography on a Mono Q column were performed as previously reported [17]. For alkylation, 2-ME-SOD1 and WT-SOD1 were incubated with various concentrations of Maleimide-PEG5000 (Mol-PEG, ME-050MA, NOF Corporation) at 37°C.
Crystallography
Conditions for the crystallization of 2-ME-SOD1 were screened using a crystallizing robot [20]. Although crystals were obtained under many conditions, only two conditions have successfully yielded crystals suitable for X-ray analysis so far. One condition gave crystals within a minute, but the diffractions of these crystals were limited to approximately 3 Å (1 Å=0.1 nm) (results not shown). In the crystal structure of these samples, 2-ME was not visible on Cys111 for some unknown reason, possibly due to inadequate resolution. In contrast, the other condition gave crystals that diffracted better at beamline PF BL-5A, and were used in this report. Briefly, crystals of 2-ME-SOD1 were produced from a mixture of 100 mg/ml protein solution and an equal amount of a reservoir solution containing 15% (w/v) PEG3350/300 mM (NH4)2SO4 at pH 6.0 after being equilibrated against the reservoir solution by hanging drop vapour diffusion at a temperature of 16°C. As a cryo protectant, 20% (v/v) ethylene glycol was added to the reservoir solution. Diffraction data were collected at Photon Factory (Tsukuba, Japan). The diffraction data were processed with HKL2000 [21]. Molecular replacement by MOLREP [22] in the CCP4 suite [23] gave an initial model using the coordinates for SOD1 (PDB ID; 1P1V), followed by iterative auto and manual model building by COOT [24] and O [25] respectively with the aid of wARP [26]. Crystallographic refinement was performed by REFMAC5 [27]. Structural figures were prepared by the PyMOL Molecular Graphics System, RASMOL [28], MOLSCRIPT [29] and RASTER3D [30]. The stereochemical quality of the protein structures was checked using CCP4 validation tools [23], and no main-chain torsion angles were located in the disallowed region of the Ramachandran plot.
Comparison of various loop VIs
The loop VI region in each chain of the available SOD1 crystal structures in the PDB was compared with the chains A and B in 2-ME-SOD1 by the SSM superpose in COOT [24].
RESULTS AND DISCUSSION
Half-oxidized SOD1 is minimally alkylated
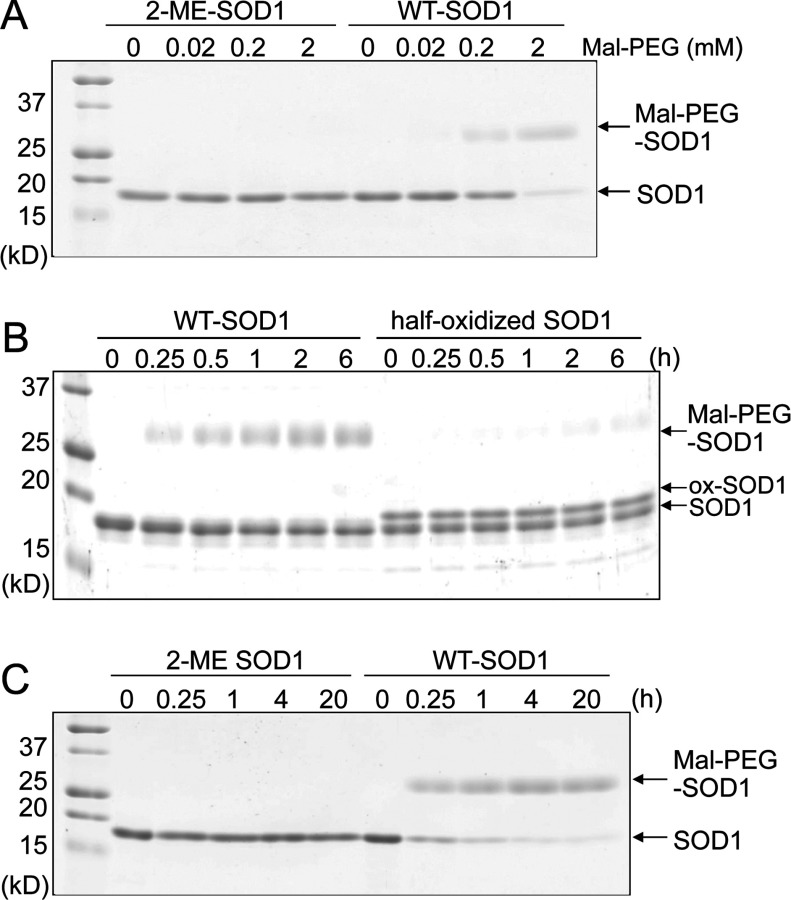
As reported previously, the side chain of Cys111 in human SOD1 is susceptible to oxidation and alkylation [17–19]. Indeed, WT-SOD1 but not 2-ME-SOD1 is easily alkylated when incubated with Maleimide-PEG5000 (Mal-PEG), which is a strong alkylating agent conjugated with polyethylene glycol 5000 in order to visualize the alkylated protein bands by SDS/PAGE (Figure 1A). However, the alkylation of Cys111 is not complete even by incubation with 100-fold equivalent of Mal-PEG over SOD1 dimer for 1 h. We thought that the modification of one of Cys111 residues of the dimer might block modification of the other. Thus, a half-oxidized SOD1 purified by anion-exchange chromatography [17] was incubated with 0.2 mM Mal-PEG. As shown in Figure 1(B), the half-oxidized SOD1 was hardly alkylated even though 50% of the subunits were present as the reduced Cys111-SH. In contrast, a long incubation with high concentrations of Mal-PEG (20 h with 2 mM Mal-PEG) led to the nearly complete alkylation of both subunits of WT-SOD1 (Figure 1C). These results suggest that the first modified Cys111 in one subunit sterically blocks a second modification in the other subunit.
Figure 1. Comparison of sensitivity to alkylation between 2-ME-SOD1 and half-oxidized SOD1.
(A) 2-ME-SOD1 (10 μM) and WT-SOD1 (10 μM) were incubated with 0, 0.02, 0.2 and 2.0 mM Maleimide-PEG5000 (Mal-PEG) at 37°C for 1 h, and then subjected to reducing SDS/PAGE. (B) Purified half-oxidized SOD1 (10 μM) and WT-SOD1 (10 μM) were incubated with 0.2 mM Mal-PEG for various hours, and then subjected to reducing SDS/PAGE. (C) 2-ME-SOD1 (10 μM) and WT-SOD1 (10 μM) were incubated with 2 mM Mal-PEG at 37°C for various times, and then subjected to reducing SDS/PAGE.
Asymmetric interactions between the 2-ME moieties are observed in the 2-ME-SOD1 dimer
To test whether complete modification of Cys111 causes structure distortions, the crystal structure of 2-ME-SOD1 was determined at a resolution of 1.8 Å in space group P212121 with cell constants, a=74.1 Å, b=163.3 Å, c=173.6 Å. The crystallographic and refinement data for 2-ME-SOD1 are summarized in Table 1. The structure of 2-ME-SOD1 is essentially identical with that for typical homodimeric SOD1, except for the modification with 2-ME and loop VI (Figure 2A). The asymmetric unit of the crystal includes six 2-ME-SOD1 dimers (chains A/B, D/E, F/G, H/I, J/K and L/M), consisting of 12 monomers. In fact, the crystal of 2-ME-SOD1, like many crystal structures of SOD1, also has unique crystallographic cell parameters and crystal packing. The overall averaged B-factor of all atoms in the crystal is 18.8 Å2, which is comparable with the Wilson B-value of 19.7 Å2, although most of chain M has poor electron density that is reflected in the high-averaged B-factor value of 40.4 Å2 for this chain. Among the 12 chains, the B-factors for chains A and B are 7.6 and 7.3 Å2 respectively, which indicates that chains A and B are the most stable dimeric pair out of the six dimers in the asymmetric unit. Therefore, we mainly used this dimer as a representative 2-ME-SOD1 in this study.
Table 1. Crystallographic data and refinement statistics.
R.m.s., root mean square. Values in parentheses are for the highest resolution bin.
| Parameter | |
|---|---|
| Crystallographic data | |
| Crystal | 2-ME-SOD1 |
| Beamline | PF BL5A |
| Wavelength (Å) | 1.0000 |
| Cell constants (Å) | a=74.1, b=163.3, c=173.6 |
| Cell constants (°) | α=β=γ=90.0 |
| Space group | P212121 |
| Solvent content (%) | 56 |
| Processed resolution (Å) | 200.0–1.8 (1.86–1.80) |
| Rsym (%)* | 11.5 (88.2) |
| Completeness (%) | 94.9 (92.4) |
| Averaged redundancy | 5.0 (4.8) |
| Averaged I/s(I) | 12.0 (1.6) |
| Mosaicity (°) | 0.24 |
| Refinement | |
| Resolution (Å) | 118.7–1.8 (1.85–1.80) |
| Rwork (%)† | 20.0 (27.6) |
| Rfree (%)‡ | 24.1 (32.3) |
| R.m.s bond lengths (Å) | 0.011 |
| R.m.s bond angles (°) | 1.27 |
| Wilson B value (Å2) | 19.7 |
| Overall B value (Å2) | 18.8 |
| TLS group (n) | 12 |
| Non-hydrogen atoms (n) | 15198 |
| Amino acid residues (n) | 1836 (1–153×12 chains) |
| Cu+ (n) | 12 |
| Zn2+ (n) | 12 |
| SO42− (n) | 12 |
| Water molecules (n) | 1746 |
| Ramachandran plot (%)§ | 95.6, 4.4, 0.0 |
*Rsym=100×S|I−〈I〉|/SI, where 〈I〉 is the mean intensity of symmetry-related reflections.
†Rwork=100×S|Fo−Fc|/SFo, where Fo and Fc are observed and calculated refelections.
‡A total of 5% of randomly chosen reflections were used for the calculation of Rfree.
§Percentage of preferred, allowed regions and outliers respectively.
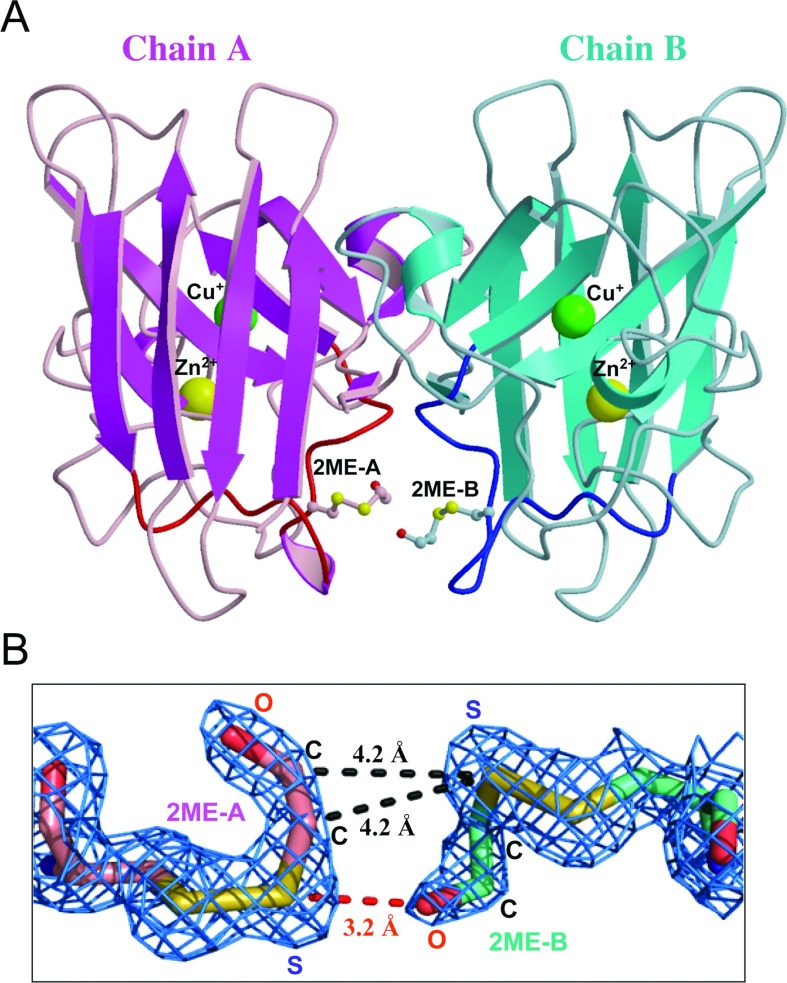
Figure 2. Structure of 2-ME-SOD1 dimer.
(A) A dimer of 2-ME-SOD1 is presented as a ribbon model based on the secondary structures. Only one dimer, chains A (magenta and pink) and B (cyan and light blue), out of six dimers in the crystallographic asymmetric unit, is shown as a representative structure. Loop VI of chains A and B are highlighted in red and blue respectively. The side chains of 2-ME-Cys111 are drawn as sticks with small spheres to indicate carbon (pink for chain A and cyan for chain B), sulfur (yellow), and oxygen (red) atoms. Large spheres indicate Cu+ (green) or Zn2+ (yellow). (B) 2mFo – DFc electron density map around 2-ME-Cys111 of chains A and B contoured at 1.5 σ (blue mesh) by the PyMOL Molecular Graphics System, Version 1.5.0.2, Schrödinger, LLC. Bonds are shown as sticks coloured to show carbon (pink for chain A and cyan for chain B), sulfur (yellow), and oxygen (red) atoms. Some distances between two 2-ME moieties are shown.
The electron density of 2-ME was clearly observed in 10 out of 12 monomers (densities for the two 2-ME moieties in chains G and I were not clear; results not shown). These results suggest that the 2-ME moiety is somewhat flexible; the B-factors for 2-ME of chains A and B are 17.9 and 19.0 Å2 respectively, which appeared to be higher than the average values for each individual chain. As shown in Figure 2(B), the electron density map shows that one 2-ME moiety is asymmetrically contacting the other on the adjacent Cys111 residue across the dimer interface. The sulfur atom of 2-ME, which is disulfide-bonded with the sulfur atom of Cys111 in the A chain, and the hydrogen atom of the OH group of 2-ME attached to the B chain participate in hydrogen bonding, because the distance between the S and the O is 3.2 Å. In contrast, the sulfur atom of 2-ME in the B chain is not involved in hydrogen bonding. The sulfur atom of 2-ME in the B chain and the two carbon atoms of the 2-ME group in the A chain seem to be involved in van der Waals contacts, because both the distances between the S and the C are approximately 4.2 Å. These asymmetric interactions between the 2-ME moieties likely stabilize the 2-ME-SOD1. Indeed, chemical cross-linking between Cys111 residues across the dimer interface stabilizes ALS-linked SOD1 proteins [31].
Binding of copper and zinc ions in each subunit was confirmed by the anomalous difference Fourier map calculated from the data collected at the wavelength of the copper and zinc absorption edges respectively (results not shown). The structure lacks the typical copper-coordinated water and the characteristic His63 bridge between zinc and copper in 10 out of the 12 monomers in the asymmetric unit of 2-ME-SOD1 crystal. This suggests a trigonal-planar configuration and caused us to model the copper ion as Cu+ (reduced CuI state) [32]. We also observed that the crystals of 2-ME-SOD1 were colourless again suggesting Cu+ (2-ME-SOD1 in water is blue green as is typical of Cu2+ solutions). Indeed, our NMR study of 2-ME-SOD1 in solution indicates that the copper ion state is largely Cu2+ (S. Hanashima, N. Fujiwara, K. Matsumoto, N. Iwasaki, G.-Q. Zheng, H. Torigoe, K. Suzuki, N. Taniguchi and Y. Yamaguchi, unpublished work). This discrepancy may be explained by the findings of Ogihara et al. [33], where EPR experiments indicate that some crystallization conditions with WT yeast SOD1 reduce the Cu2+ to Cu+.
A portion of loop VI of human SOD1 is also asymmetric
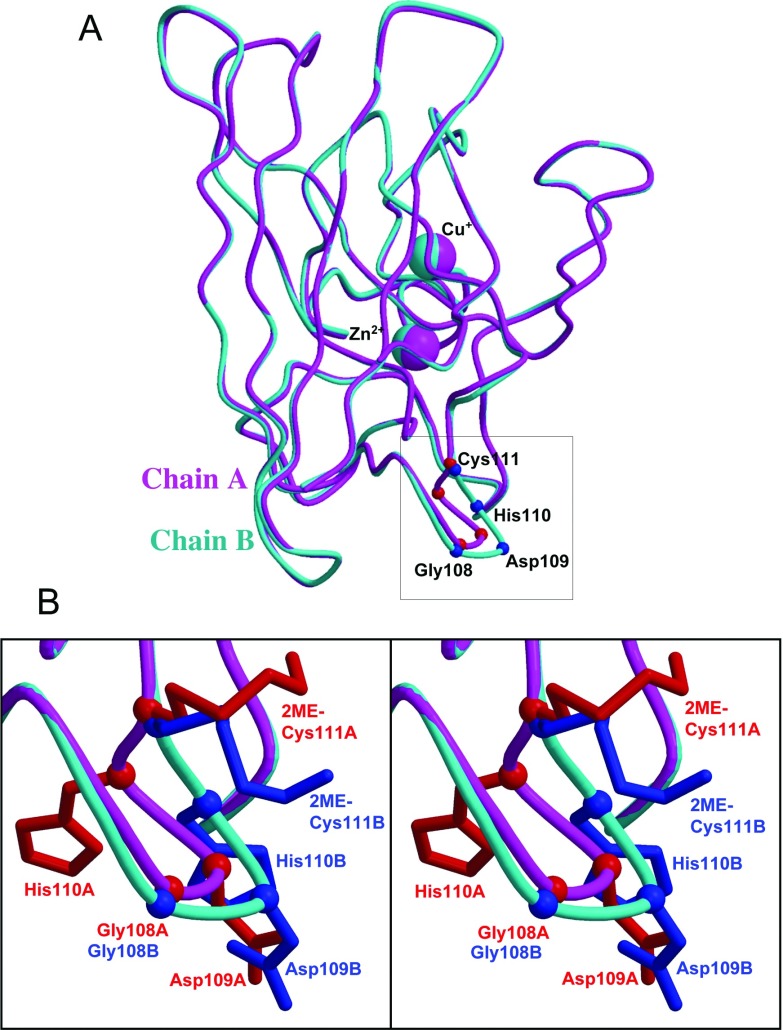
The asymmetric interaction between the two 2-ME moieties (Figure 2B) suggests that the A and B chains in the SOD1 dimer are not symmetrical. This prompted a detailed structural comparison between the A and B chains. As shown in Figure 3(A), the chains A and B superimpose well, except for portions of loop VI. The distances for the Cα atoms of Gly108, Asp109, His110, and Cys111 between the A and B chains in the superimposed structures are 0.97, 2.57, 2.61 and 0.48 Å respectively (see Supplementary Table S1 at http://www.bioscirep.org/bsr/032/bsr0320539add.htm). By contrast, the distances for the Cα atoms of all of the remaining regions between the A and B chains are less than 0.62 Å and the rmsd (root mean square deviation) of the 153 Cα atoms of all of the amino acids is 0.37 Å. Evidently, the asymmetry starts with Gly108, has the largest difference at His110, and ends at Cys111. Moreover, each side chain of Asp109, His110, and the 2-ME attached to Cys111 of the A and B chains points in different directions (Figure 3B). The largest distance between the A and B chains is 5.9 Å at CE1 in the His110 imidazole moiety.
Figure 3. Structural difference between chains A and B.
(A) Chains A (magenta) and B (cyan) are superposed using all 153 Cα atoms. Cα atoms of Gly108, Asp109, His110 and 2-ME-Cys111 are indicated as small balls in red for chain A and blue for chain B. In this superposition, only Asp109 and His110 are quite different. (B) Close-up view of the squared region in (A) is shown in stereo. Side chains of the four residues are drawn with sticks coloured as Cα. Note that the rotamers of 2-ME-Cys111 side chains are different despite the Cα positions being similar and the 2-ME moieties being far from symmetry. The side chains of Asp109 and His110 also point in different directions.
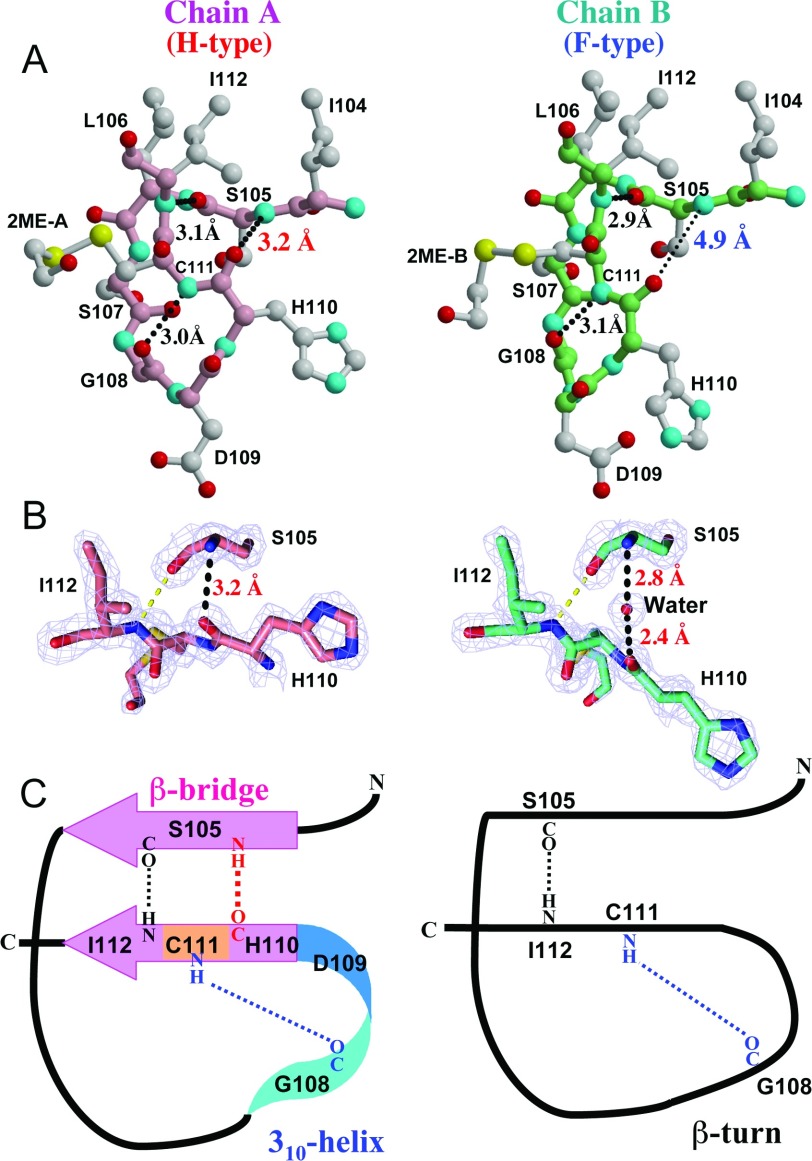
The structural difference between chains A and B may be explained as follows. As shown in Figure 4(A), three hydrogen bonds are found in loop VI of chain A: (i) between the main-chain oxygen of Ser105 (Ser105-O) and the main-chain nitrogen of Ile112 (Ile112-N) (3.1 Å), (ii) between Gly108-O and Cys111-N (3.0 Å) and (iii) Ser105-N and His110-O (3.2 Å). In contrast, loop VI of the B chain lacks the third hydrogen bond, here the distance between Ser105-N and His110-O is 4.9 Å. Instead there is a water molecule between Ser105 and His110. The distance from the water oxygen to the Ser105-N is 2.8 Å and to His110-O is 2.4 Å in the B chain (Figure 4B), suggesting strong hydrogen bonds. Therefore, the structural difference for the Gly108-Cys111 region between each dimer is exemplified by the existence or otherwise of the third hydrogen bond between Ser105 and His110 in the main chain. Hence, we designated the loop VI structure of chains A and B as H-type (hydrogen bond type) and F-type (hydrogen bond free type, albeit with hydrogen bonding to a water molecule) respectively (Figure 4B, and Supplementary Table S1). The third hydrogen bond causes two unique secondary structures, a β-helix-like isolated β-bridge and a 310-helix-like structure, in the H-type loop VI (Figure 4C, left). In contrast, only one β-turn is observed in the F-type loop VI (Figure 4C, right).
Figure 4. Structural determinants of loop VI.
(A) The residues Ile104–Ile112 in loop VI of chains A and B are shown in ball-and-stick models. Main-chain bonds and carbon atoms are coloured in pink and light green for chains A and B respectively. Side chain bonds and carbon atoms are coloured in grey. Colours of sulfur (yellow), nitrogen (cyan) and oxygen (red) atoms are common in both main and side chains. Key hydrogen bonds that maintain the loop VI structure are shown as black dots. In the B chain, the hydrogen bond between Ser105 and His110 does not exist, and the existence of this bond can be used to classify the loop VI structure into an H- or an F- type. (B) 2mFo – DFc electron density map around Ser105 and His110-Ile112 in chains A and B contoured at 2.0 σ (blue mesh) by the PyMOL Molecular Graphics System, Version 1.5.0.2, Schrödinger, LLC. Bonds are shown as sticks coloured to show carbon (pink for chain A and cyan for chain B), nitrogen (blue) and oxygen (red) atoms. (C) Two types of structures are drawn schematically. In the H-type, three hydrogen bonds maintain two secondary structures, a very short β-sheet with β-helix-like configuration and a 310-helix, while in the F-type there is a continuous loop stabilized by the two remaining hydrogen bonds.
A comparison of the remaining chains, D/E, F/G, H/I, J/K and L/M, with chain A or B showed that chains D, F, H, J and L fit well with chain A and chains E, G, I, K and M fit well with chain B (Supplementary Table S1 and Supplementary Figure S1A at http://www.bioscirep.org/bsr/032/bsr0320539add.htm). From the existence of a hydrogen bond between Ser105-NH and His110-CO in loop VI, the A-like chains, D, F, H, J and L are classified as H-type, and from its absence and the existence of one water molecule, the B-like chains, E, G, I, K and M are classified as F-type. These results indicate that the region from Gly108 to Cys111 in each 2-ME-SOD1 dimer (A/B, D/E, F/G, H/I, J/K and L/M) is an asymmetric HF-type heterodimer. Moreover, the six dimers in one asymmetric unit of the crystal can be arranged as shown in Supplementary Figure S1(B). In this configuration, all the H-type subunits lie inside a trimeric non-crystallographic symmetry, and all the F-type subunits surround the H-type subunits by forming dimer contacts.
Some human SOD1s are heterodimers having two forms of loop VI
We examined to what extent loop VI is either chain A-like or chain B-like in other human SOD1 structures is not modified with 2-ME (Supplementary Tables S2 and S3 available at http://www.bioscirep.org/bsr/032/bsr0320539add.htm). Each chain of available crystal structures in the PDB (protein data bank) was classified as being A-like or B-like by comparison with chains A and B in 2-ME-SOD1. In addition, when the distance between Ser105-N and His110-O in each chain is less than 3.6 Å (indicating a hydrogen bond), we classified it as H-type. As listed in Supplementary Table S3, hydrogen bonding between Ser105 and His110 was restricted to almost all of the A-like loop VI structures, indicating that an apparent A- or B-like structure is consistent with H- or F- type respectively. Many of the F-type chains in the various structures have water molecule accompanied with two hydrogen bonds to Ser105 and to His110 instead of the hydrogen bond between Ser105-NH and His110-CO, as shown in Figure 4(B). The hydrogen bond between Ser105-NH and His110-CO observed in H-type chains is weak because the average distances in various human SOD1s including our structure are 3.2 Å (the means and standard deviation of our structure: 3.22±0.90 Å; and other human SOD1: 3.23±0.16 Å). Due to this weakness, it is likely that the hydrogen bond is broken and instead Ser105-NH and His110-CO interact with the water molecule. H- and F-type conformations are potentially interconvertible by forming and breaking the weak hydrogen bond.
Of the 53 crystal structures of human SOD1 dimers including ours, 13 contain HF-type dimers having an asymmetric loop VI in their asymmetric units (Supplementary Table S2). Therefore, the asymmetry in loop VI is not caused by the 2-ME modification, but is an intrinsic property of human SOD1. However, unlike our structure that has complete asymmetry for all six dimers, the asymmetry is partial in many of the other structures. Evidently the interactions of the 2-ME moieties in both subunits stabilize the asymmetry of loop VI in the crystal structure of 2-ME-SOD1. Recently, the crystal structure of H80R SOD1 (PDB ID; 3QQD) that has Cys111-SO3H in subunit A and Cys111-SOH in subunit B was determined [34]. As Cys111-SOH can be reduced to Cys111-SH by a reducing agent, the SOD1 in the crystal may be half-oxidized. This is a good example of an asymmetrical modification even though the SOD1 dimer is HH-type.
Loop VI in other species can also be classified into two types
We further examined whether the HF rule specifying that the A-like or B-like loop VI is determined by the presence or absence of a third hydrogen bond can be applied to the SOD1s of other species (Supplementary Tables S4 and S5 available at http://www.bioscirep.org/bsr/032/bsr0320539add.htm). There are 23 and 11 crystal structures of bovine SOD1 and yeast SOD1 respectively, and they provide a good test of our classification criteria. The residues corresponding to Asp109-His110-Cys111 in loop VI of human SOD1 are Glu107-Tyr108-Ser109 in bovine SOD1 and Pro109-Tyr110-Ser111 in yeast.
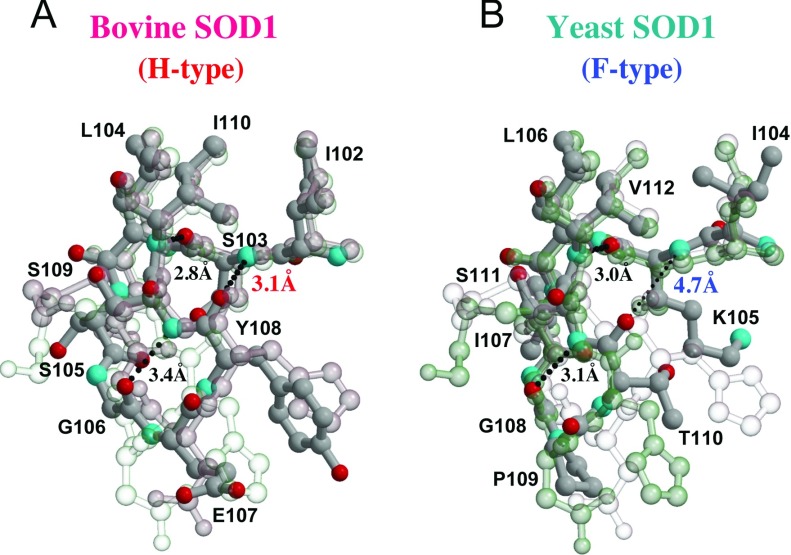
As shown in Figure 5(A), both chains A and B of bovine SOD1 (PDB; 1COB) contain three hydrogen bonds, namely between Ser103-O and Ile110-N (distance: 2.8 and 3.0 Å), Gly106-O and Ser109-N (3.4 and 2.8 Å), and Ser103-N and Tyr108-O (3.1 and 3.1 Å), which indicates that the loop VI of this bovine SOD1 can be classified as an HH-type dimer. The side chains of Glu107 and Tyr108 fit well with those of Asp109 and His110 in chain A of 2-ME-SOD1. Nineteen of the 23 structures of bovine SOD1 are HH-type dimers and the average distances between Ser103-N and Tyr108-O observed in the H-type chains are 3.26±0.17 Å (Supplementary Tables S4 and S5). It is noteworthy that in the remaining four structures of bovine SOD1 (PDB: 2Z7U, 2Z7W, 2Z7Y and 2Z7Z), the proteins had been treated with H2O2, and are classified as HF-type. As listed in Supplementary Table S5, a water molecule can be clearly observed between Ser103 and Tyr108 in chains A of 2Z7U, 2Z7Y and 2Z7Z, indicating that in these cases the A chains are F-type. These data suggest that the oxidation of bovine SOD1 with H2O2 may induce an H-type to F-type conformational change in loop VI in one subunit. Note that this conformational change is not related to oxidation of a cysteine residue because residue 111 of bovine SOD1 is a serine.
Figure 5. Loop VI in bovine and yeast SOD1.
Loop VI of bovine (A) and yeast (B) SOD1 (PDB code 1COB and 1B4L respectively) are superposed with chains A (transparent pink) and B (transparent cyan) of 2-ME-SOD1. Bonds and carbon atoms are shown as dark grey sticks and small spheres respectively, with small spheres of nitrogen (cyan) and oxygen (red) atoms. The transparency of chains A or B of 2-ME-SOD1 is less when the superposed structures are similar. Loop VI of bovine and yeast SOD1 can be also classified into H- and F-types by the existence of the hydrogen bond between 105th and 110th residues as in human SOD1. Moreover, the entire structures of loop VI of bovine and yeast SOD1 are quite similar to H- and F-types respectively.
In contrast, as shown in Figure 5(B), yeast SOD1 (PDB: 1B4L), which is shown as a monomer in the crystal but forms a dimer by crystallographic symmetry mate, has two hydrogen bonds between Lys105-O and Val112-N (distance: 3.0 Å), and Gly108-O and Ser111-N (3.1 Å), but not between Lys105-N and Thr110-O (4.7 Å), which indicates that the loop VI region of this yeast SOD1 can be classified as an FF-type dimer. The side chains of Pro109 and Tyr110 of yeast SOD1 fit well with the side chains of Asp109 and His110 in chain B of 2-ME-SOD1 (F-type) respectively. All of the 11 yeast SOD1 structures including FALS equivalent mutants are FF-type dimers (Supplementary Tables S4 and S5).
A classification based on the HF rule was also applied to SOD1s of other eukaryotic species and the results are listed in Supplementary Tables S4 and S5. Among SOD1 structures of prokaryotic species, only Bacillus subtilis SOD1 could be classified, because the amino acid residues 108–110 are absent in other bacterial SOD1s. B. subtilis SOD1 has a Lys-Gly-Ser-Lys-Leu insertion into a typical bacterial sequence (Supplementary Table S6 available at http://www.bioscirep.org/bsr/032/bsr0320539add.htm). The last two residues of the insertion plus an asparagine residue, Lys-Leu-Asn, structurally correspond to human SOD1 Asp109-His110-Cys111. All of the loop VI structures of B. subtilis SOD1s (PDB: 1S4I, 1XTL and 1XTM) appear to be H-type monomers.
The 105th amino acid residue may dictate the loop VI type
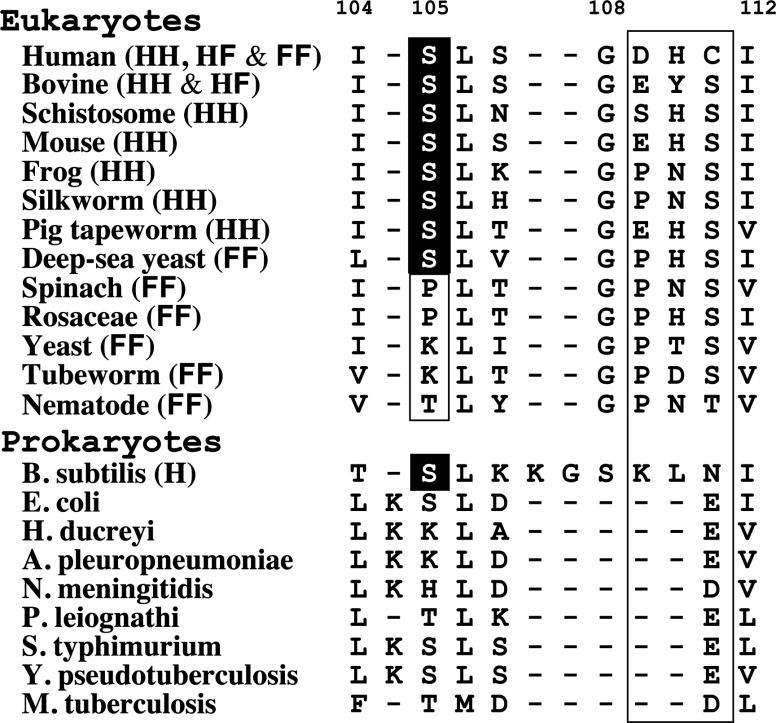
Application of the HF rule across all species indicates that the two types of loop VI structure of SOD1 are conserved during evolution. However, the triplet amino acid sequence corresponding to human SOD1 Asp109-His110-Cys111 does not appear to be a determinant of loop VI types. Figure 6 shows a summary of the loop VI type and amino acid sequence of SOD1 from various species corresponding to the human SOD1 region from Ile104 to Ile112. We found a relationship between the loop VI type and the 105th amino acid residue. When this residue is serine, an H-type loop VI predominates, although both types of loop VI structures can be formed. In contrast, when the 105th amino acid residue is proline, lysine and threonine, loop VI is F-type. The third hydrogen bond in the H-type loop VI requires that the main-chain amide of the 105th amino acid residue is a proton donor. However, Pro105 cannot function as a proton donor for the hydrogen bond because the prolyl residue lacks a hydrogen at the main-chain amide, due to the circular structure of the main chain. Therefore, it is quite reasonable that spinach and rosaceae SOD1s, which contain a Pro105, can exist as FF-type dimers. In addition, all yeast SOD1s and the tubeworm SOD1 having Lys105 are FF-type dimers (Figure 6). The side chain of a lysine residue is larger than that of a serine residue and cannot be accommodated in an H-type loop VI due to a clash with the side chain of the 110th amino acid residue. Therefore, we conclude that SOD1 molecules with Pro105 or Lys105 residues are likely to be FF-type dimers.
Figure 6. Amino acid alignments in loop VI corresponding to human SOD1 Ile104–Ile112.
The loop VI type and amino acid sequence of SOD1 from various species corresponding to the human SOD1 region from Ile104–Ile112 are summarized. The 105th, and 109th–111th amino acids are focused on to explain the relationship between loop VI type and amino acid sequence. The 105th amino acid residue but not 109th–111th appears to determine the loop VI type.
Is loop VI structure relevant to FALS mutant SOD1s?
To date, more than 150 mutations that induce FALS have been identified in the SOD1 subunit composed of 153 amino acids. The reasons why so many mutations can cause FALS remain unknown. FALS-mutated SOD1s have certain characteristics including weakened metal binding, unstable structure and a tendency to undergo oligomerization or aggregation [5]. However, the crystal structures of A4V, G37R, H43R and I113T [35–37], which are metal-filled structures, reveal only slight structural perturbations relative to the WT-SOD1. On the other hand, the structures of the metal-binding mutants H46R, D125H and S134N [38–40] reveal conformational disorder in the electrostatic and zinc loop elements [5,6]. Clear structural changes in loop VI by a FALS mutation have been reported only in one paper in which an A4V mutation results in the movement of the side chains of Leu106 and Ile113 which in turn, affects the loop VI structure [41]. Based on this, loop VI seems to contribute to the stability of the entire SOD1 structure. Leu106 and Ile113 may stabilize the β-barrel through hydrophobic interactions [42]. Leu106 is well conserved in SOD1s from bacteria to humans (Figure 6). As shown in Supplementary Table S2, applying the HF-rule shows that there are 29 HH-types and 5 HF-types among the 34 structures of FALS mutants, whereas, as we have seen, WT-SOD1s have more HF types (8 among 19 structures). In particular, the asymmetry of loop VI found in 2-ME-SOD1, stably mutated SOD1s such as C6A/C111S seems to contribute to the structural stabilization of SOD1 proteins.
We previously reported that three monoclonal antibodies having epitopes in loop VI showed different binding abilities to different FALS mutants although the mutations exist in locations outside of loop VI [11]. This observation indicates that the structure of loop VI depends on the nature of the mutation. Thus, the locations of each amino acid in loop VI in FALS mutants were compared with those in the H- or F-types of 2-ME-SOD1 (WT). However, the differences between FALS mutants and WT-SOD1 are much less than the difference between H-type and F-type structures (results not shown). A molecular mechanics simulation using demetallated human A4V SOD1 suggested that there is a conformational change in loop VI. The isolated β-bridge in loop VI of A4V SOD1 is destroyed to make a stable π-helix, which does not appear in WT-SOD1 [43]. This conformational change of loop VI from β-bridge to π-helix should correspond to a configurational switch from H-type to F-type. Flexible switching between H- and F-types as occurs in WT human SOD1 may be inhibited by ALS-mutations. Furukawa et al. demonstrated that amino acid residues in the 90–120 region in human SOD1, including loop VI, are one of three core structures of fibrillar aggregates that are resistant to the action of a protease [44]. These findings suggest that the small conformational changes of loop VI in FALS-mutant SOD1s detected by the monoclonal antibodies may occur in solution and lead to the oligomerization and/or aggregation of SOD1 molecules.
Another interesting finding is that all loop VI structures of the monomeric SOD1 mutants (PDB: 1MFM, 2XJK, 2XJL and 3HFF) are F-type (Supplementary Table S2). Moreover, disulfide-bond-free mutant (C6A/C111A/C57A/C146A; PDB: 2GBU and 2GBV) also contains some FF dimers and HF dimers in the crystallographic asymmetric unit (Supplementary Table S3). This mutant, when metal ions are absent, will be a monomer, because elimination of the disulfide bond between Cys57 and Cys146 by mutation or reduction leads to monomerization of apo-SOD1 [45,46]. Therefore, the SOD1 monomer may tend to F-type. Monomeric SOD1 of WT and FALS-mutant SOD1 is a common misfolding intermediate [47] and the most facile state for formation of amyloid-like fibrillar aggregates [48], which are involved in ALS pathology. The monomeric SOD1 intermediate may induce misfolding of its native counterpart in a template-directed reaction, thereby forming a seed of aggregated protein for transmission in a prion-like mechanism [49]. Whether or not the intermediate monomer with H- or F-type is involved in seed formation of aggregated protein is an interesting idea deserving further research in ALS.
In this report, we determined the crystal structure of human SOD1 modified with 2-ME at Cys111 and found that asymmetric interactions of the 2-ME moieties in both subunits and other interactions in loop VI stabilize the asymmetry at loop VI and likely contribute to the stability of the entire protein. Furthermore, we show that the Gly108-Cys111 segment in loop VI of several human SOD1s is of the asymmetric HF-type and that the existence or otherwise of the hydrogen bond between Ser105-NH and His110-CO switches the type of loop VI. Asymmetric loop structures in some dimeric enzymes have been reported to play a part role in the catalytic reaction [50,51]. Although the role of loop VI asymmetry in some human SOD1s in the catalysis remains unclear, the different affinity of substrate, superoxide anion radical, against Cys111 in human SOD1 might affect catalytic reaction. Loop VI structures in SOD1s from other species could also be limited to the two types, H- and F-type. The surprising structural match of loop VI across species, its restriction to two interconvertible conformations and their link to protein stability could be key to our understanding of the structure, function and pathophysiology of SOD1.
Online data
AUTHOR CONTRIBUTION
Kentaro Ihara performed the crystallization experiments, refined and analysed the structural data, and wrote the paper Hidetaka Torigoe performed the biochemical experiments and commented on the paper. Yoshiki Yamaguchi and Naoyuki Taniguchi analysed the results and helped to write the paper. Soichi Wakatsuki and Keiichiro Suzuki provided scientific guidance and commented on the paper. Noriko Fujiwara designed the research, analysed the data and wrote the paper. All authors contributed to editing the paper, and approved the final version of the paper.
ACKNOWLEDGEMENTS
We are grateful to Ube Industries for kindly providing the 2-ME-SOD1. We thank staff of the Photon Factory (Tsukuba, Japan) for discussion on the work as well as assistance in data collection.
FUNDING
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan [grant numbers 19500313, 23591259] and in part by the Ministry of Health, Labor and Welfare of Japan via a grant for the Research Group on Development of Novel Therapeutics for ALS.
References
- 1.Deng H. X., Hentati A., Tainer J. A., Zafar I., Cayabyab A., Hung W. Y., Getzoff E. D., Hu P., Herzfeldt B., Roos R. P., et al. Amyotrophic lateral sclerosis and structural defects in Cu,Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 2.Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., Deng H. X., et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 4.Ripps M. E., Huntley G. W., Hof P. R., Morrison J. H., Gordon J. W. Transgenic mice expressing an altered murine superoxide dismutase gene provide an animal model of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 1995;92:689–693. doi: 10.1073/pnas.92.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentine J. S., Doucette P. A., Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu. Rev. Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 6.Seetharaman S. V., Prudencio M., Karch C., Holloway S. P., Borchelt D. R., Hart P. Immature copper–zinc superoxide dismutase and familial amyotrophic lateral sclerosis. J. Exp. Biol. Med. 2009;234:1140–1154. doi: 10.3181/0903-MR-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haidet-Phillips A. M., Hester M. E., Miranda C. J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M. J., Foust K. D., et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruzman A., Wood W. L., Alpert E., Prasad M. D., Miller R. G., Rothstein J. D., Bowser R., Hamilton R., Wood T. D., Cleveland D. W., et al. Common molecular signature in SOD1 for both sporadic and familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12524–12529. doi: 10.1073/pnas.0705044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zetterström P., Graffmo K. S., Andersen P. M., Brännström T., Marklund S. L. Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic amyotrophic lateral sclerosis (ALS) model mice. J. Biol. Chem. 2011;286:20130–20136. doi: 10.1074/jbc.M111.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara N., Miyamoto Y., Ogasahara K., Takahashi M., Ikegami T., Takamiya R., Suzuki K., Taniguchi N. Different immunoreactivity against monoclonal antibodies between wild-type and mutant copper/zinc superoxide dismutase linked to amyotrophic lateral sclerosis. J. Biol. Chem. 2005;280:5061–5070. doi: 10.1074/jbc.M406106200. [DOI] [PubMed] [Google Scholar]
- 12.Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., Richardson D. C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J. Mol. Biol. 1982;160:181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- 13.Parge H. E., Hallewell R. A., Tainer J. A. Atomic structures of wild-type and thermostable mutant recombinant human Cu,Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6109–6113. doi: 10.1073/pnas.89.13.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuhara R., Tezuka T., Kageyama T. Structure, molecular evolution, and gene expression of primate superoxide dismutases. Gene. 2002;296:99–109. doi: 10.1016/s0378-1119(02)00837-5. [DOI] [PubMed] [Google Scholar]
- 15.Fink R. C., Scandaliios J. G. Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 2002;399:19–36. doi: 10.1006/abbi.2001.2739. [DOI] [PubMed] [Google Scholar]
- 16.Repock J. R., Frey H. E., Hallewell R. A. Contribution of conformational stability and reversibility of unfolding to the increased thermostability of human and bovine superoxide dismutase mutated at free cysteines. J. Biol. Chem. 1990;265:21612–21618. [PubMed] [Google Scholar]
- 17.Fujiwara N., Nakano M., Kato S., Yoshihara D., Ookawara T., Eguchi H., Taniguchi N., Suzuki K. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper-zinc superoxide dismutase. J. Biol. Chem. 2007;282:35933–35944. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- 18.Okado-Matsumoto A., Guan Z., Fridovich I. Modification of cysteine 111 in human Cu,Zn-superoxide dismutase. Free Radical Biol. Med. 2006;41:1837–1846. doi: 10.1016/j.freeradbiomed.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Liu H., Zhu H., Eggers D. K., Nersissian A. M., Faull K. F., Goto J. J., Ai J., Sanders-Loehr J., Gralla E. B., Valentine J. S. Copper(2+) binding to the surface residue cysteine 111 of His46Arg human copper-zinc superoxide dismutase, a familial amyotrophic lateral sclerosis mutant. Biochemistry. 2000;39:8125–8132. doi: 10.1021/bi000846f. [DOI] [PubMed] [Google Scholar]
- 20.Hiraki M., Kato R., Nagai M., Satoh T., Hirano S., Ihara K. Development of an automated large-scale protein-crystallization and monitoring system for high-throughput protein-structure analyses. Acta Crystallogr. D: Biol. Crystallogr. 2006;62:1058–1065. doi: 10.1107/S0907444906023821. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 23.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:A110–A119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov G. N., Vagin A. A., Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Sayle R. A., Milner-White E. J. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 30.Merritt E. A., Murphy M. E. P. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. Sect D Biol. Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 31.Auclair J. R., Boggio K. J., Petsko G. A., Ringe D., Agar J. N. Strategies for stabilizing superoxide dismutase (SOD1), the protein destabilized in the most common form of familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21394–21399. doi: 10.1073/pnas.1015463107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blackburn N. J., Hasnain S. S., Binsted N., Diakun G. P., Garner C. D., Knowles P. F. An extended-X-ray-absorption-finestructure study of bovine erythrocyte superoxide dismutase in aqueous solution. Direct evidence for three-co-ordinate Cu(I) in reduced enzyme. Biochem. J. 1984;219:985–990. doi: 10.1042/bj2190985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogihara N. L., Parge H. E., Hart P. J., Weiss M. S., Goto J. J., Crane B. R., Tsang J., Slater K., Roe J. A., Valentine J. S., et al. Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase. Biochemistry. 1996;35:2316–2321. doi: 10.1021/bi951930b. [DOI] [PubMed] [Google Scholar]
- 34.Seetharaman S. V., Winkler D. D., Taylor A. B., Cao X., Whitson L. J., Doucette P. A., Valentine J. S., Sfiurf V., Demeler B., Carroll M. C., et al. Disrupted zinc-binding sites in structures of pathogenic SOD1 variants D124V and H80R. Biochemistry. 2010;49:5714–5725. doi: 10.1021/bi100314n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hough M. A., Grossmann J. G., Antonyuk S. V, Strange R. W., Doucette P. A., Rodriguez J. A., Whitson L. J., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banci L., Bertini I., D'Amelio N., Libralesso E., Turano P., Valentine J. S. Metalation of the amyotrophic lateral sclerosis mutant glycine 37 to arginine superoxide dismutase (SOD1) apoprotein restores its structural and dynamical properties in solution to those of metalated wild-type SOD1. Biochemistry. 2007;46:9953–9962. doi: 10.1021/bi700620r. [DOI] [PubMed] [Google Scholar]
- 37.DiDonato M., Craig L., Huff M. E., Thayer M. M., Cardoso R. M., Kassmann C. J., Lo T. P., Bruns C. K., Powers E. T., Kelly J. W., Getzoff E. D., Tainer J. A. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 38.Elam J. S., Taylor A. B., Strange R., Antonyuk A., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., Hart P. J. Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 2003;10:461–467. doi: 10.1038/nsb935. [DOI] [PubMed] [Google Scholar]
- 39.Antonyuk S., Elam J. S., Hough M. A., Strange R. W., Doucette P. A., Rodriguez J. A., Hayward L. J., Valentine J. S., Hart P. J., Hasnain S. S. Structural consequences of the familial amyotrophic lateral sclerosis SOD1 mutant His46Arg. Protein Sci. 2005;14:1201–1213. doi: 10.1110/ps.041256705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X., Antonyuk S. V., Seetharaman S. V., Whitson L. J., Taylor A. B., Holloway S. P., Strange R. W., Doucette P. A., Valentine J. S., Tiwari A., et al. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:16169–16177. doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardoso R. M., Thayer M. M., DiDonato M., Lo T. P., Bruns C. K., Getzoff E. D., Tainer J. A. Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase. J. Mol. Biol. 2002;324:247–256. doi: 10.1016/s0022-2836(02)01090-2. [DOI] [PubMed] [Google Scholar]
- 42.Getzoff E. D., Tainer J. A., Stempien M. M., Bell G. I., Hallewell R. A. Evolution of CuZn superoxide dismutase and the Greek key beta-barrel structural motif. Proteins. 1989;5:322–336. doi: 10.1002/prot.340050408. [DOI] [PubMed] [Google Scholar]
- 43.Schmidlin T., Kennedy B. K., Daggett V. Structural changes to monomeric CuZn superoxide dismutase caused by the familial amyotrophic lateral sclerosis-associated mutation A4V. Biophys. J. 2009;97:104–111. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Furukawa Y., Kaneko K., Yamanaka K., Nukina N. Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2010;285:22221–22231. doi: 10.1074/jbc.M110.113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., O'Hallorann T. V. The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 2004;279:47998–48003. doi: 10.1074/jbc.M406021200. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi S., Fujiwara N., Ido A., Ono M., Takeuchi Y., Tateno M., Suzuki K., Takahashi R., Tooyama I., Taniguchi N., et al. Induction of protective immunity by vaccination with wild-type apo superoxide dismutase 1 in mutant SOD1 transgenic mice. J. Neuropathol. Exp. Neurol. 2010;69:1044–1056. doi: 10.1097/NEN.0b013e3181f4a90a. [DOI] [PubMed] [Google Scholar]
- 47.Rakhit R., Crow J. P., Lepock J. R., Kondejewski L. H., Cashman N. R., Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 48.Furukawa Y., Kaneko K., Yamanaka K., O'Halloran T. V., Nukina N. Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:24167–24176. doi: 10.1074/jbc.M802083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polymenidou M., Cleveland D. W. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henriksen A., Aghajari N., Jensen K. F., Gajhede M. A flexible loop at the dimer interface is a part of the active site of the adjacent monomer of Escherichia coli orotate phosphoribosyltransferase. Biochemistry. 1996;35:3803–3809. doi: 10.1021/bi952226y. [DOI] [PubMed] [Google Scholar]
- 51.Lee M., Maher M. J., Christopherson R. I., Guss J. M. Kinetic and structural analysis of mutant Escherichia coli dihydroorotases: a flexible loop stabilizes the transition state. Biochemistry. 2007;37:10538–10550. doi: 10.1021/bi701098e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.