Abstract
The X-ray crystal structure of the Head module, one-third of the Mediator of transcriptional regulation, has been determined as a complex with the C-terminal domain (CTD) of RNA polymerase II. The structure reveals multiple points of interaction with an extended conformation of the CTD; it suggests a basis for regulation by phosphorylation of the CTD. Biochemical studies show a requirement for Mediator–CTD interaction for transcription.
Keywords: transcriptional initiation, X-ray crystallography, cross-linking, yeast
Mediator, a megaDalton multiprotein complex, enables the regulation of transcription (reviewed in refs 1 and 2); it bridges between gene activator proteins at enhancers and RNA polymerase II (pol II) at promoters. Mediator makes multiple contacts with pol II, including with the carboxyl-terminal domain (CTD) of the Rpb1 subunit and with the Rpb3 subunit (3). The importance of contact with the CTD is suggested by the consequences of CTD truncation. The CTD comprises 27 repeats of a heptapeptide in yeast and 52 repeats of the same sequence in human cells. Truncation of the yeast CTD to 10–12 repeats causes conditional growth phenotypes, and truncation to fewer than 10 repeats results in loss of viability (4). CTD truncation also correlates with a dampened transcriptional response, and CTD lengthening rescues activator protein mutations (5–7). Suppressors of CTD truncation phenotypes identified the first genes for Mediator subunits (8), later shown to be components of the purified Mediator complex (9).
Phosphorylation of the CTD plays multiple roles in transcription. Pol II, with an unmodified CTD, enters a preinitiation complex and is phosphorylated on Ser-5 of the heptapeptide repeat by TFIIH (10–12). Phosphorylation disrupts a Mediator–pol II complex in vitro (13), and phosphorylated pol II is devoid of Mediator in vivo (14). Phosphorylation of Ser-5 creates a binding site for the mRNA capping apparatus (15), which acts on nascent transcripts about 25 residues in length, whereupon the transition-to-transcription elongation takes place. Phosphorylation may be obligatory for the transition to elongation, as inhibition of phosphorylation increases stalling of pol II at promoters in vitro (16) and decreases the occupancy of elongating pol II on yeast ORFs in vivo (17). Elongating pol II is dephosphorylated at Ser-5 and rephosphorylated at Ser-2, leading to the recruitment of RNA splicing and cleavage/polyadenylation factors. Recently, the number of physiologically significant CTD modifications has been expanded with Thr-4 phosphorylation, found to function in histone mRNA 3′ processing (18) and mammalian RNA pol II elongation, and Tyr-1 phosphorylation, which appears to affect the association of the CTD with elongation and termination factors (19).
Electron microscopy has suggested a division of Mediator in three modules, termed Head, Middle, and Tail (20). Evidence for discrete modules comes from the separate isolation of Head and Middle modules, and from the creation of Tail-less Mediator by deletion of genes for Tail subunits. The entire Mediator and isolated Head and Middle modules bind to immobilized CTD peptides (21, 22).
Despite biochemical and genetic evidence for Mediator–CTD interaction, the molecular details of the complex are largely unknown. A failure of genetic experiments to identify mutations in individual Mediator genes causing disruption of the complex suggests an extensive interaction surface involving multiple Mediator components, but the subunit identity and the basis of molecular specificity remain to be determined. Furthermore, the importance of Mediator–CTD interaction in the regulatory role of CTD phosphorylation has not been addressed.
Here we investigate structural aspects of Mediator–CTD interaction, defining the path of the CTD across a highly conserved surface of the Mediator Head module and suggesting a basis for binding, which involves several important interactions with CTD Tyr-1. We also report on the role of CTD phosphorylation in Mediator-dependent transcription in vitro.
Results and Discussion
X-ray crystallography of recombinant Head module, made up of seven Mediator subunits, with a mass of 220 kDa, revealed about 60% of the primary structure (23). Here we report on the X-ray crystallography of native Head module, with improved experimental phases, revealing about 80% of the structure (Fig. S1). Addition of a CTD peptide gave rise to difference electron density showing the Mediator–CTD interaction.
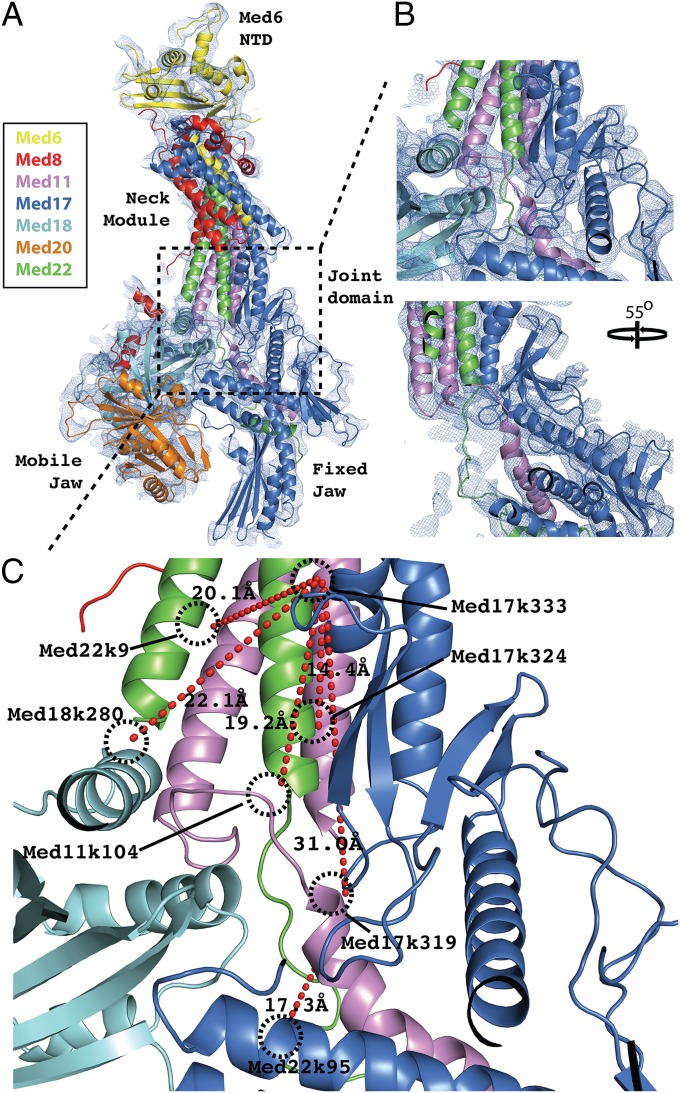
We produced a native Saccharomyces cerevisae Head module by genetic manipulation to reduce interaction with Middle module and by affinity chromatography (see Materials and Methods). Diffraction was phased by multiple isomorphous replacement with anomalous scattering (MIRAS) from Ta6Br12 and Au derivatives. Molecular replacement with the previous recombinant head structure [Protein Database Identification (PID): 3RJ1] was unsuccessful because of a difference in orientation of the Fixed Jaw domain (see domain labels in Fig. 1). Initial substructure solutions were calculated from anomalous difference maps, aided by phases from partial molecular replacement solutions that were calculated by searching independently for the Neck and Mobile Jaw domains. Substructure refinement with the use of MIRAS phases resulted in electron density maps revealing many features not seen previously, such as domain connectivity in the central “Joint” region (Fig. 1) and large sections of β-sheet within both the Med6 N-terminal domain (112–164) and Med17 C-terminal domain (322–410 and 457–479) (Fig. S2). Distance constraints from MS/MS-coupled cross-linking analyses guided and validated new model build (Fig. 1) (24) and supported revisions of previous secondary structure assignments in the Med11 N-terminal domain (Fig. S3) and in the Med17 C-terminal domain (Fig. S4). Selenomethionine anomalous difference maps were also used to guide sequence assignments during the building of revised models.
Fig. 1.
Resolving interdomain connectivity within the Joint region. (A) Secondary structure representation of the Apo-Mediator Head structure with each chain individually colored: Med6 (yellow), Med8 (red), Med11 (violet), Med17 (blue), Med18 (cyan), Med20 (orange), Med22 (green). The model is overlaid with the map from experimental MIRAS phasing contoured at 1.0 σ (blue mesh). The dashed box bounds the Joint region within which a significant portion of new interdomain connectivity has been resolved and new model built. (B) Two close-up views of the Joint region, related by a 55° rotation around the vertical axis. For clarity, the Med18 model was omitted from the second panel. (C) Modeling of the Joint region is validated with MS/MS-cross-linking data. Mediator Head module cross-linked with 8 mM BS3 was digested fully with trypsin before LC-coupled MS/MS analysis. Analysis of the MS spectra from parent ions with Z ≥ 3 identified a number of cross-links between residues in the Joint region (red dashed lines). The residues involved and the modeled Cβ cross-link distance are indicated.
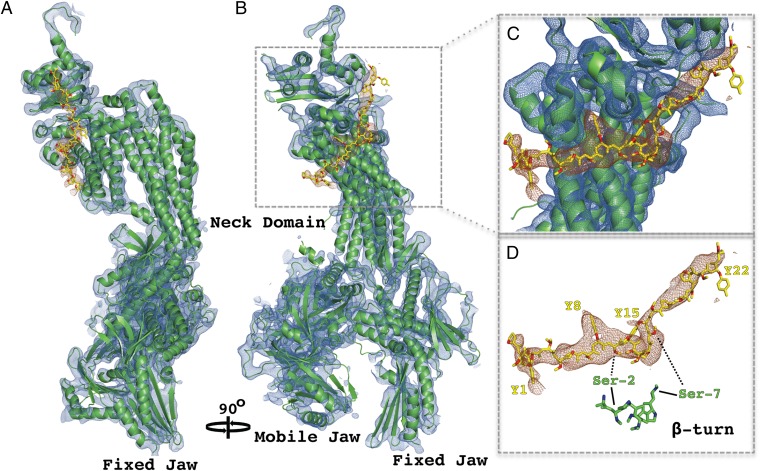
Crystals soaked with a 35-residue peptide containing five CTD heptapeptide repeats gave rise to difference electron density for the CTD (Fig. 2), whereas crystals soaked with a peptide containing only two repeats did not. Despite the moderate resolution of the CTD difference maps (4.5 Å), clear structural features allow the sequence register and directionality of the CTD to be modeled with confidence. Bulky side-chain density attributable to tyrosine residues was clearly visible in some places (Fig. 2C). A kinked central section of the difference density can be accounted for by a β-turn motif (2SPTSPS7), stabilized by an intramolecular hydrogen-bond network (25), seen in previous X-ray structures of CTD peptides (Fig. 2D). This placement of the β-turn motif was supported by the occurrence of bulky side-chain density for tyrosine residues located on either side (Fig. 2D). A CTD model was built by extending both ends of the β-turn motif, with sequence register constraints provided by bulky side-chain densities, and with all-trans peptide bond geometry imposed, in keeping with structures of CTD peptides in complexes with RNA processing proteins (26, 27). The entire CTD model comprises almost four heptapeptide repeats (25 residues), beginning with P-1 and ending with S23, where residue numbering relates to the first tyrosine Y1 (Fig. 3A). The CTD adopts an almost entirely extended conformation, interacting with the Head module over a distance of 73 Å. CTD interaction with RNA processing proteins so far observed is more limited, spanning at most 17 residues (27).
Fig. 2.
Structure of Mediator Head module–CTD complex. (A) Secondary structure representation of the Head module (green) superimposed on weighted electron-density map of the Head alone (2FO-FC; blue mesh), contoured at 1.25 σ. Difference map showing electron density for the CTD (brown mesh), calculated with Fsoak-Fnative amplitudes and MIRAS phases, contoured at 2.5 σ. CTD model encompassing almost four heptapeptide repeats is shown in stick representation (yellow). (B) Same as A rotated 90° around the vertical axis. (C) Close-up view of CTD interaction region bounded by dashed box in B. (D) CTD model and density from C shown with an aligned section of unmodified CTD peptide (β-turn) from high-resolution crystal structure (PID: 3D9O).
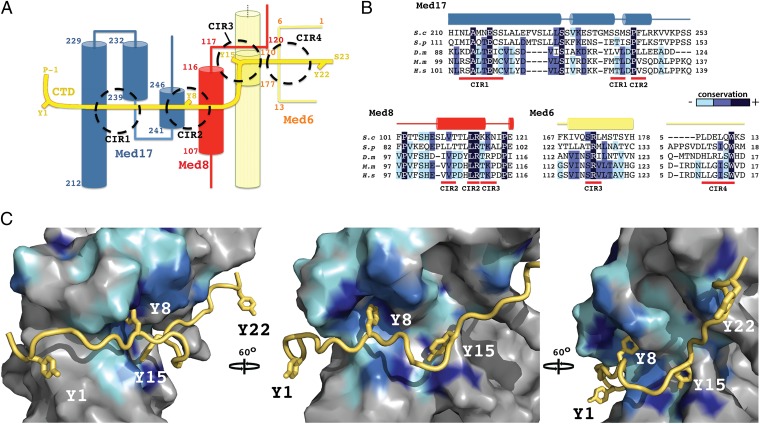
Fig. 3.
CTD-interacting regions of Mediator Head module. (A) Schematic representation of the path of the CTD (yellow) across the Head module. CTD tyrosines (Y1–Y22) and terminal residues are highlighted, as well as major interacting regions (CIR1-4; dashed circles). Secondary structure elements of the interacting Mediator polypeptides, Med17 (blue), Med8 (red), and Med6 (straw) are shown with corresponding residue numbering. (B) Sequence alignments covering the CTD-interacting regions of Med17, Med8, and Med6 from S cerevisiae (S.c), Shizosaccharomyces pombe (S.p), Drosophila melanogaster (D.m), Mus Musculus (M.m) and Homo sapiens (H.s). Conserved residues are shaded in dark blue, marine, and cyan, in order of decreasing conservation. Secondary structure elements are shown above the sequence. (C) Three views of a surface representation of the CTD (yellow)–Head module (gray) interaction, related by 60° rotations. Mediator sequence conservation in represented with surface coloring according to B. The CTD backbone is shown as a ribbon with the four tyrosines depicted as sticks.
The CTD interacts with three Mediator subunits, Med6, Med8, and Med17 (Fig. 3A). The interaction surface can be divided into four CTD-interacting regions (CIR), termed CIR1 to -4 (Fig. 3 A and B). CIR1, which interacts with CTD residues 2–5, comprises two Med17 helices (Fig. 3A, CIR1). The longer of these helices (197–228) forms the outer surface of the smallest helical bundle in the Neck domain. The CTD abuts a section of this helix containing residues that are highly conserved across eukaryotes, including invariant Ala214 and Glu217 residues. The interacting surface at the base of the second Med17 helix (232–239) includes two serines (240 and 242), the location of which, with respect to CTD residue S5, suggests the formation of a stabilizing hydrogen-bonding network. Such a network would likely be sensitive to the addition of a phosphate group to S5, with a resultant decrease in CTD binding affinity.
CIR2 buries CTD tyrosine Y8 within a deep pocket, created by the juxtaposition of short Med17 (232–239 and 241–246) and Med8 (107–116) helices (Fig. 3 A and C, Y8). The interacting surfaces of these helices are composed of very highly conserved residues, including invariant Med8 Leu114 and Arg115 and Med17 Pro243 residues (Fig. 3B). The interior of the binding pocket is rich in hydrophobic residues, providing a favorable environment for the tyrosine side chain. The high degree of sequence conservation identifies the pocket as a core element of Mediator–CTD interaction, in keeping with a report that substitution of all CTD tyrosines with phenylalanine is lethal in S. cerevisiae (28).
Immediately C-terminal to the buried Y8 of CIR2 is the putative β-turn section of the CTD, which appears not to interact with Head directly but rather to position Y15 within the heart of CIR3. Side-chain density attributable to Y15 abuts Med8 loop density (117–119) and projects toward the surface of the Med6 helix, located within the small helical bundle of the Neck domain (Fig. 3 A and C). The section of helix within range of interaction with Y15 contains a patch of highly conserved residues, including invariant Arg173 (Fig. 3B), which is well positioned for hydrogen bonding with the tyrosine hydroxyl group. CIR4, consisting of a Med6 loop region close to the extreme N terminus (4–11), interacts with CTD residues 17–20. Although the sequence conservation in this area is lower and molecular interactions are less apparent, close proximity (5 Å Cα spacing) of Med6 density to CTD residue S19 (S5 equivalent) suggests that phosphorylation at this position could interfere with interaction. In general, the high degree of sequence conservation in Mediator surface residues concentrated at the major CTD interaction points provides clear evidence for the biological significance of the observed complex.
We pursued the effects of phosphorylation on Mediator–CTD interaction in biochemical studies. We found by surface plasmon resonance measurements that the affinity constant of the Mediator Head module for the 35-residue CTD peptide was significantly diminished by phosphorylation of all Ser-5 residues, from 4 × 107 M−1 to 5.5 × 106 M−1. We tested the effect of CTD phosphorylation on transcription with the use of TFIIH bearing a mutation in the kinase subunit Kin28 (Kin28-AS) that confers sensitivity to the inhibitor NA-PP1 (17). It was reported that transcription of the HIS4 promoter with nuclear extract from this mutant was reduced by about 40% by the inhibitor (17). We found that transcription with highly purified Mediator, mutant TFIIH, and transcription factors, was reduced by about 35% by the inhibitor (Fig. S5). In the absence of Mediator the inhibitor had no effect on transcription (Fig. S5A), despite producing a marked reduction in kinase activity (Fig. S5B). The correlation of transcription with level of CTD phosphorylation was entirely Mediator-dependent. The importance of CTD phosphorylation only in the presence of Mediator points to a role of Mediator–CTD interaction in transcription.
We were surprised to find that Mediator was slightly inhibitory to transcription (Fig. S5, compare lanes 1 and 3), whereas Mediator has been reported to stimulate the “basal” reaction in the past. To test whether our purified Mediator was inactive or contained an inhibitory contaminant, we assayed for transcriptional regulation in yeast extract. The response to a transcriptional activator was abolished by depletion of Mediator from the extract. Addition of our purified Mediator restored the transcriptional response to a level greater than that obtained with the undepleted extract (Fig. S6). We suspect that the discrepancy with previous results may reflect a greater purity of all of our purified transcription protein preparations, which may have contained inhibitors in the past whose effects were reversed by Mediator.
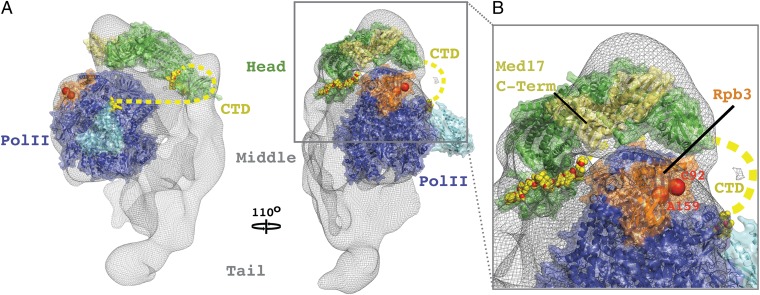
The significance of our findings is several-fold. First, the structure of the Head module–CTD complex reveals directly the Mediator–CTD interaction that was inferred from biochemical and genetic studies in the past. The Head module is seen to contact four CTD heptapeptide repeats, each in a different way. Second, the structure of the complex forms a basis for understanding the effects of CTD-phosphorylation, shown here to influence transcription in a Mediator-dependent manner. Serine residues of the CTD appear to make contacts with Mediator that would be disrupted by phosphorylation. Details of these contacts await extension of the analysis to higher resolution. Finally, the structure of the Head module–CTD complex provides a starting point for modeling a complete Mediator-pol II complex (Fig. 4). Such a model places constraints on the path of the entire CTD and on the possibilities for additional Mediator–pol II interaction. Ultimately it may give insight into the communication of regulatory information through Mediator to pol II.
Fig. 4.
Docking of X-ray structure of Head module–CTD complex to electron microscope structure of Mediator–pol II complex. (A) Side and front views (related by 110° rotation) showing the Mediator Head module (green) and 12-subunit pol II (blue; PDB ID: 1WCM) models docked to the electron density map of the Mediator–pol II complex from electron microscopy (gray mesh). CTD residues are depicted as yellow sticks and a putative path of the entire CTD is represented by a dashed yellow line. The Rpb4 and Rpb7 subunits are cyan. Locations of mutations in Rpb3 that disrupt Mediator–pol II interaction (C92R and A159G) are shown as red spheres. The C-terminal region of Med17 (pale-green) contains residues whose mutations supress the Rpb3 mutant phenotype. (B) Close-up view of Mediator–pol II interaction surfaces.
Materials and Methods
Yeast Strains.
To isolate the native Mediator Head module a C-terminal TAP tag was first introduced into the endogenous MED8 gene in S. cerevisiae strain CB010 (Matα pep4::HIS3 prb1::LEU2 prc1::HISG can1 ade2 trp1 ura3 his3 leu2–3 leu2–112) with the TRP1 gene for selection as described previously (29). Next, the Mediator complex was destabilized by the replacement of the nonessential Mediator gene SIN4 (30) with URA3 for selection. Holo-TFIIH containing analog sensitive Kin28 (L83G) was generated from the strain SHY532 (gift from S. Hahn, Fred Hutchinson Cancer Research Center, Seattle, WA) containing a TFB3-FLAG1-TAP fusion described previously (17), except for one major alteration. To stabilize the mutant Holo-TFIIH the TFB6 ORF was replaced with the KANMX gene for selection. Wild-type Holo-TFIIH was prepared from a CB010 strain carrying TFB3-TAP::TRP1 and TFB6Δ::KANMX. RNA polymerase II and Mediator were prepared from a single CB010 strain. First, the LEU2 gene at the prb1 endogenous locus was replaced with a loxP-flanked KANMX gene. Next, an N-terminal TAP tag was introduced into the Mediator Head subunit MED17, as previously described (31). Cre recombinase expression from pSH47 led to the removal of the KANMX and TRP1 (TAP tag) marker genes through LoxP recombination. Finally, an Rpb1 C-terminal protein G-tagging cassette (RPB1-3C-PROTEING::KANMX) was introduced to tag RNA polymerase II. TFIIF was prepared from a strain of CB010 harboring TFG2-TAP::TRP1. Successful transformants were verified by PCR amplification.
Protein Purification.
TAP-tagged Mediator Head module was isolated from S. cerevisiae lysates with IgG affinity chromatography followed by additional chromatography steps to separate subcomplexes and improve sample homogeneity (see SI Materials and Methods for details). In vitro transcription assays were performed with pure preparations of each of the S. cerevisiae general transcription factors (32, 33): TBP, TFIIA, -B, -E, -H, RNA polymerase II-TFIIF (PolII-IIF), and Mediator. A full description of the purification protocols for each assay component is provided in SI Materials and Methods.
Mediator Head Crystal Growth, Derivatization, Peptide Soaking, and Cryoprotection.
The first poorly-diffracting crystals of the native S. cerevisiae Mediator Head complex were obtained by vapor diffusion at 18.0 °C in hanging drops comprising 0.5 μL 10 mg/mL protein and 0.5 μL 1.1 M ammonium tartrate dibasic pH 7.0. Significant crystal improvement was achieved by microseeding these crystals into 2.0-μL hanging drops containing 1.0 μL 7 mg/mL protein and 1.0 μL 275 mM ammonium citrate tribasic pH 7.0, 10% wt/vol PEG 3350, 2 mM DTT that had been preincubated at 18.0 °C for 3 d before seed introduction. Cryoprotectant [25% (vol/vol) glycerol, 275 mM ammonium citrate pH 7.0, 10% (wt/vol) PEG 3350, 2 mM DTT] was introduced gradually in steps of 5% glycerol over a period of 24 h before plunge-freezing in liquid nitrogen. For MIRAS phasing experiments, crystals were soaked for roughly 2 wk with either Au68 (2 mM) or Ta6Br12 (2 mM) clusters in a final cryoprotectant buffer in which 2 mM DTT had been substituted with 1 mM TCEP. For CTD peptide soaks, crystals pre-equilibrated in 25% glycerol cryoprotectant buffer were soaked with either 2× CTD or 5× CTD peptide for ∼36 h at a concentration of ∼6 mg/mL made up in 25% glycerol cryo buffer. To provide sequence markers for tracing novel stretches of polypeptide chain in the MIRAS-phased maps, Mediator Head module containing selenomethione substitutions was prepared essentially as described previously (34) (see SI Materials and Methods and Tables S1–S4 for full details).
In Vitro Assays.
In vitro transcription and promoter-specific CTD kinase assays were performed to investigate the Mediator-dependent effects of CTD kinase inhibition and the stimulatory effects of full-length and Mediator Head module complexes in wild-type and depleted yeast whole-cell extracts using similar methods to those described previously (35, 36). Minor modifications are described in detail in SI Materials and Methods.
MS Analysis of Cross-Linked Head Module.
Mediator Head module was cross-linked with a labeled BS3 reagent (D0:D12) mix at 1:1 molar ratio and fully digested with trypsin. A portion of the digested peptides were fractionated before MS analysis using high pH C18 chromatography. Fractionated and unfractionated samples were analyzed by LC-MS/MS with MS1 measured from 300 to 2,000 m/z and HCD product ion MS2 spectra collected for peptides with Z ≥ 3. Cross-links were identified using the Protein Prospector suite as described previously (24, 37) (see full description in SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Dr. Maia Azubel for Au68 clusters; Drs. Kenji Murakami and Xin Liu for purified transcription factors; and Dr. Ralph Davis for Mediator-depleted yeast whole-cell extract. This research was supported by National Institutes of Health (NIH) Grants GM49985, GM36659, and AI21144 (to R.D.K.); mass spectrometry was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at the University of California at San Francisco supported by NIH National Institute of General Medical Sciences Grant 8P41GM103481 and the Howard Hughes Medical Institute; crystallographic data were collected at the Stanford Synchrotron Radiation Lightsource beamlines 12-2 and 11-1, which has support from the Department of Energy and NIH (Grants P41GM103393 and P41RR001209) and the Advanced Photon Source beamline 23-ID-D (GM/CA CAT), supported by the National Cancer Institute (Grant Y1-CO-1020) and the National Institute of General Medical Sciences (Grant Y1-GM-1104).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics, www.rcsb.org (RCSB ID codes rcsb074727 and rcsb074728) and Protein Data Bank, www.pdb.org (PDB ID codes 4GWP and 4GWQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215241109/-/DCSupplemental.
References
- 1.Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 4.Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 5.Allison LA, Ingles CJ. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci USA. 1989;86:2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scafe C, et al. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 7.Liao SM, Taylor IC, Kingston RE, Young RA. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991;5(12B):2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- 8.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 10.Laybourn PJ, Dahmus ME. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J Biol Chem. 1990;265:13165–13173. [PubMed] [Google Scholar]
- 11.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feaver WJ, Gileadi O, Li Y, Kornberg RD. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 13.Søgaard TM, Svejstrup JQ, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 14.Svejstrup JQ, et al. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh A, Shuman S, Lima CD. Structural insights to how mammalian capping enzyme reads the CTD code. Mol Cell. 2011;43:299–310. doi: 10.1016/j.molcel.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Yan M, Gralla JD. A three-step pathway of transcription initiation leading to promoter clearance at an activation RNA polymerase II promoter. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol Cell Biol. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336:1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 20.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 21.Myers LC, et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang JS, et al. The structural and functional organization of the yeast mediator complex. J Biol Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 23.Imasaki T, et al. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding chimeras: A bioinformatics strategy for identification of cross-linked peptides. Mol Cell Proteomics. 2010;9:25–31. doi: 10.1074/mcp.M800555-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker R, Loll B, Meinhart A. Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2008;283:22659–22669. doi: 10.1074/jbc.M803540200. [DOI] [PubMed] [Google Scholar]
- 26.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 27.Fabrega C, Shen V, Shuman S, Lima CD. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 28.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 30.Zhang F, Sumibcay L, Hinnebusch AG, Swanson MJ. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol Cell Biol. 2004;24:6871–6886. doi: 10.1128/MCB.24.15.6871-6886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 32.Ranish JA, Lane WS, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bushnell DA, Cramer P, Kornberg RD. Selenomethionine incorporation in Saccharomyces cerevisiae RNA polymerase II. Structure. 2001;9:R11–R14. doi: 10.1016/s0969-2126(00)00554-2. [DOI] [PubMed] [Google Scholar]
- 35.Sayre MH, Tschochner H, Kornberg RD. Reconstitution of transcription with five purified initiation factors and RNA polymerase II from Saccharomyces cerevisiae. J Biol Chem. 1992;267:23376–23382. [PubMed] [Google Scholar]
- 36.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 37.Trnka MJ, Burlingame AL. Topographic studies of the GroEL-GroES chaperonin complex by chemical cross-linking using diformyl ethynylbenzene: The power of high resolution electron transfer dissociation for determination of both peptide sequences and their attachment sites. Mol Cell Proteomics. 2010;9:2306–2317. doi: 10.1074/mcp.M110.003764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.