Abstract
GABAA receptors are pentameric ligand-gated ion channels involved in fast inhibitory neurotransmission and are allosterically modulated by the anxiolytic, anticonvulsant, and sedative-hypnotic benzodiazepines. Here we show that the prokaryotic homolog ELIC also is activated by GABA and is modulated by benzodiazepines with effects comparable to those at GABAA receptors. Crystal structures reveal important features of GABA recognition and indicate that benzodiazepines, depending on their concentration, occupy two possible sites in ELIC. An intrasubunit site is adjacent to the GABA-recognition site but faces the channel vestibule. A second intersubunit site partially overlaps with the GABA site and likely corresponds to a low-affinity benzodiazepine-binding site in GABAA receptors that mediates inhibitory effects of the benzodiazepine flurazepam. Our study offers a structural view how GABA and benzodiazepines are recognized at a GABA-activated ion channel.
GABA (γ-aminobutyric acid) is one of the most important inhibitory neurotransmitters in the central nervous system, and its effects are mediated mostly through ionotropic GABAA receptors. These proteins belong to the family of pentameric ligand-gated ion channels (pLGICs), which includes glycine, nicotinic acetylcholine, and 5-HT3 receptors. GABAA receptors are heteropentamers, with a majority of receptors composed of two α, two β, and one γ2 subunit. They contain two binding sites for GABA, which have been localized to two β/α interfaces formed by the principal (+) face of the β-subunit and the complementary (−) face of the α-subunit. GABAA receptors are modulated by a variety of clinically used drugs, including benzodiazepines, barbiturates, neuroactive steroids, and anesthetics (1). Benzodiazepines, such as diazepam and flurazepam, modulate GABAA receptors through binding at an extracellular intersubunit site formed between the principal (+) face of the α-subunit and the complementary (−) face of the γ2-subunit (2). Benzodiazepines are among the most widely prescribed psychoactive drugs worldwide because of their anxiolytic, anticonvulsant, muscle relaxant, and sedative-hypnotic effects. More recently developed nonbenzodiazepine hypnotics, such as zopiclone and zolpidem, differ in chemical structure from the classical benzodiazepines but also act as positive allosteric modulators at GABAA receptors through the benzodiazepine-binding site. Amino acids that form the recognition sites for both GABA and benzodiazepines are located on six noncontiguous regions historically designated as loops A, B, and C on the principal (+) face of the binding site and D, E, and F on the complementary (−) face of the binding site (Fig. 1A and SI Appendix, Fig. S1).
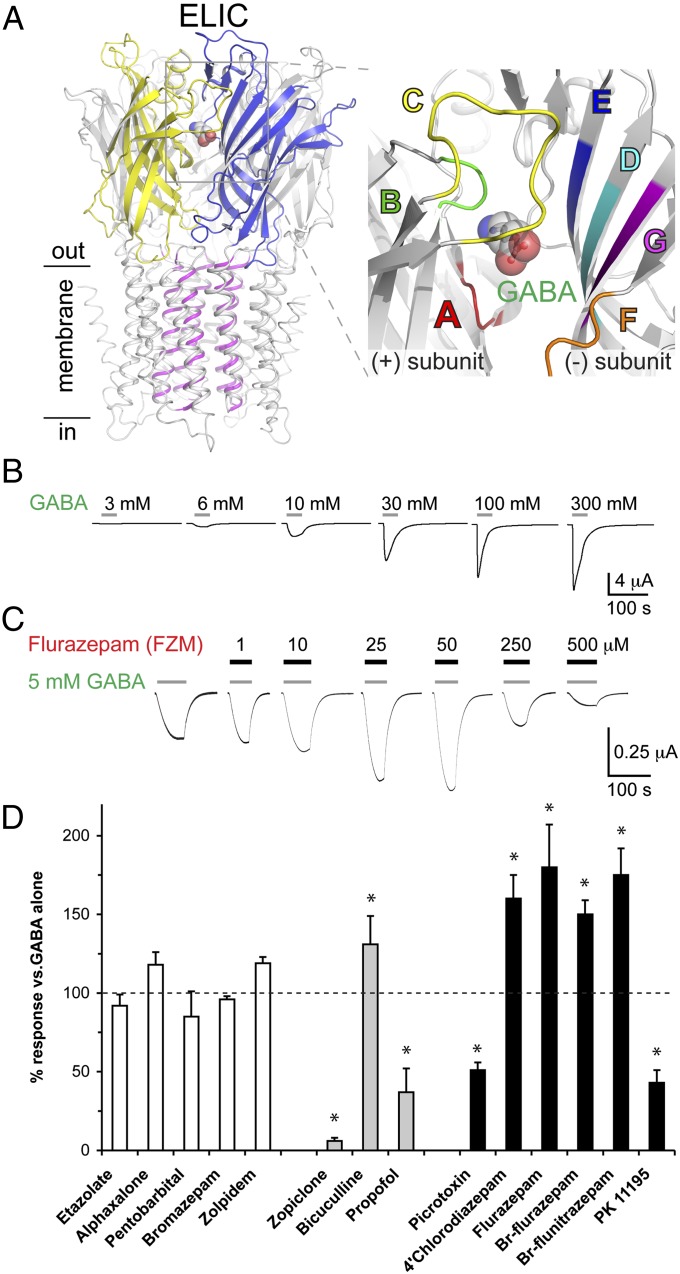
Fig. 1.
ELIC is a bacterial GABA-gated channel, which is modulated by benzodiazepines. (A) (Left) X-ray crystal structure of ELIC in complex with GABA showing the location of the GABA-binding site between the principal (yellow) and complementary (blue) subunits. GABA is shown in spherical representation. (Right) Close-up view showing the different loops (A–G) that contribute to the ligand-binding site. (B) Typical traces of concentration-dependent GABA-induced current recorded from oocytes injected with ELIC. Horizontal bars indicate duration of agonist application. (C) Flurazepam (black bars) biphasically modulates the GABA-induced current. (D) Application of a range of GABAA-receptor modulators [10 μM each, except propofol (100 μM), etazolate (30 μM), alphaxalone (100 μM), pentobarbital (1 mM), flurazepam (50 μM), and PK11195 (100 μM)] to 3–5 mM GABA-induced ELIC currents shows that the effects of some GABAA-receptor modulators are conserved in ELIC. No modulators had effects when applied alone. Data are mean ± SEM, n = 3–5. *P < 0.05.
Binding-site residues contributing to recognition of both GABA and benzodiazepines previously have been studied extensively by mutagenesis. Both natural (3, 4) and unnatural amino acid mutagenesis (5, 6) have demonstrated that interactions between the γ-amino nitrogen of GABA and aromatic side chains of loop A in the GABAA receptor and loop B in the GABAC receptor are crucial for recognition of GABA (summarized in SI Appendix, Table S1). In addition, it has been suggested that the GABA carboxylate group is stabilized through electrostatic interactions with Arg residues on the principal and complementary faces of the binding site (4, 7–9). For benzodiazepines, the individual contributions of residues in loops A–F of the high-affinity binding site have been investigated extensively (for a recent review, see ref. 10. or references given in SI Appendix, Table S2). In brief, several studies have revealed the importance of a histidine residue in loop A (11, 12) and of several aromatic residues on the principal and complementary faces of the benzodiazepine-binding site (13–18).
Despite these insights, detailed understanding of the nature of these interactions is currently lacking because of the absence of X-ray crystal structures for eukaryote GABAA receptors. Recently, however, the crystal structures of two prokaryotic pLGICs, namely ELIC (19) and GLIC (20, 21), and the glutamate-gated chloride channel from Caenorhabditis elegans, GluCl (22), have revealed the molecular architecture of closed and open pLGICs.
In this study, we show that ELIC is activated by GABA and modulated by benzodiazepines. Crystal structures of ELIC in complex with GABA and different benzodiazepines reveal the structural architecture of ligand recognition and provide a structural framework to understand both the enhancement and the inhibition of GABA-elicited responses by benzodiazepines.
Results and Discussion
ELIC Is Activated by GABA and Modulated by Benzodiazepines.
Using high-throughput electrophysiological techniques, we screened a library of compounds composed of natural amino acids, photosynthesis intermediates, neurotransmitters, agonists, and known modulators of ligand-gated ion channels. We discovered that application of GABA to Xenopus oocytes expressing ELIC produced concentration-dependent, rapidly activating inward currents, which desensitized slowly (Fig. 1B). From plotting current amplitude against a series of GABA concentrations, we calculated an EC50 of 21 mM [pEC50 (−log value of EC50): 1.69 ± 0.03, n = 4] on ELIC, a value that is ∼400-fold greater than at α1β3γ2 GABAA receptors (EC50 = 50 μM). When calcium was omitted from the extracellular recording solution (SI Appendix, Fig. S2), we observed a decrease in the rate of activation and desensitization and an increase in sensitivity to GABA (EC50 = 7.3 mM; pEC50: 2.14 ± 0.02, n = 5). This effect was not ablated by 1,2-bis(o-aminophenoxy)ethane- N,N,N',N'-tetraacetic acid (BAPTA) or compounds that block endogenous Ca2+-activated Cl− channels in oocytes (SI Appendix, Fig. S2), suggesting that calcium modulates GABA sensitivity and channel function in ELIC directly. Other known agonists for GABAA receptors, including muscimol and THIP (4,5,6,7-Tetrahydroisoxazolo[5,4-c]pyridin-3-ol hydrochloride), had no effect on ELIC (1–10 mM, n = 4). Zimmermann and Dutzler (23) reported that ELIC could be activated by a series of primary amines, including GABA. These authors also demonstrated that ELIC forms cation-selective channels with a large single-channel conductance, making it a suitable model channel to study the molecular mechanisms underlying ligand recognition and coupling of ligand binding to channel opening in the family of pLGICs. Very recently, Pan et al. (24) reported a crystal structure of ELIC in complex with acetylcholine, which acts as an antagonist at this channel. However, high-resolution acetylcholine-bound structures already have been determined in complex with acetylcholine-binding proteins from eukaryotes (25, 26), which are better homologs of the nicotinic acetylcholine receptor than the distant, prokaryotic ELIC structure. In contrast, no structural data are currently available that provide detailed insight into the recognition of GABA and allosteric modulators such as benzodiazepines.
Next, we investigated the effect on ELIC of a series of GABAA-receptor modulators, including benzodiazepines, barbiturates, neuroactive steroids, and nonbenzodiazepine hypnotics. The effect of these compounds was characterized by coapplication with an EC3 or EC10 concentration of GABA. Some compounds, such as alphaxalone and pentobarbital, had no significant effect on ELIC (white bars in Fig. 1D), suggesting that the binding sites for these modulators have not yet been formed at this stage of evolution or that these compounds bind to ELIC but have no effect on channel function. Others had an effect opposite that at GABAA receptors (gray bars in Fig. 1D): Propofol and zopiclone have inhibitory effects on ELIC, although both compounds potentiate GABA responses in human GABAA receptors. Bicuculline, which acts as an antagonist at most GABAA receptors, potentiates ELIC responses, an effect also seen at homomeric β3 GABAA receptors (27). Some compounds, however, affect ELIC similarly to GABAA receptors (black bars in Fig. 1D). For example, flurazepam potentiates ELIC responses up to 180 ± 27% (n = 6, P < 0.05) at 50 μM, but at higher concentrations it inhibits GABA-induced currents (Fig. 1C). These effects are similar to the biphasic effect of flurazepam on GABAA receptors, which has been attributed to an interaction of flurazepam with a high- and a low-affinity binding site at these receptors (28, 29). Bromo-analogs of flurazepam and flunitrazepam (here termed “Br-flurazepam” and “Br-flunitrazepam,” respectively) (SI Appendix, Fig. S3), which facilitate structural studies, also potentiated the GABA response to 150 ± 9% (n = 4, P < 0.05) and 175 ± 17% (n = 4, P < 0.05), respectively (black bars in Fig. 1D).
Flurazepam potentiates the GABA response with an EC50 of 12.5 μM (pEC50: 4.90 ± 0.26, n = 4), which is ∼30-fold higher than that at GABAA receptors (EC50 of 0.39 μM; pEC50: 6.41 ± 0.14, n = 6). Potencies of drugs usually are lower in bacterial proteins than in mammalian proteins. For example, the bacterial neurotransmitter sodium symporter LeuT displays affinities for tricyclic antidepressants, such as clomipramine and imipramine, that are more than 200-fold lower than for their binding sites in human symporters (30, 31). Nevertheless, binding mechanisms and molecular determinants for drug actions often are conserved throughout evolution.
Together, these results demonstrate that ELIC is a potentially useful model to study GABA recognition and channel modulation by known allosteric modulators of the GABAA receptor, including the benzodiazepine flurazepam and the nonbenzodiazepine hypnotic zopiclone. In addition, combined with previous data showing GABA modulation of benzodiazepine-binding sites in insects (32, 33), these results establish a clear evolutionary link between bacterial and eukaryote pLGICs. Previous studies have shown that GABA plays a role in quorum sensing (34) in certain bacteria, including Erwinia species (35), indicating that specialized receptors involved in GABA signaling possibly arose during the early evolution of proteobacteria.
Molecular Recognition of GABA.
To understand the structural basis for molecular recognition of GABA and benzodiazepines, we determined three separate cocrystal structures of ELIC with GABA and flurazepam, Br-flurazepam, and the nonbenzodiazepine modulator zopiclone. Depending on the relative amounts of compounds used during cocrystallization conditions, we found that benzodiazepines can localize to two different binding sites in ELIC (see below).
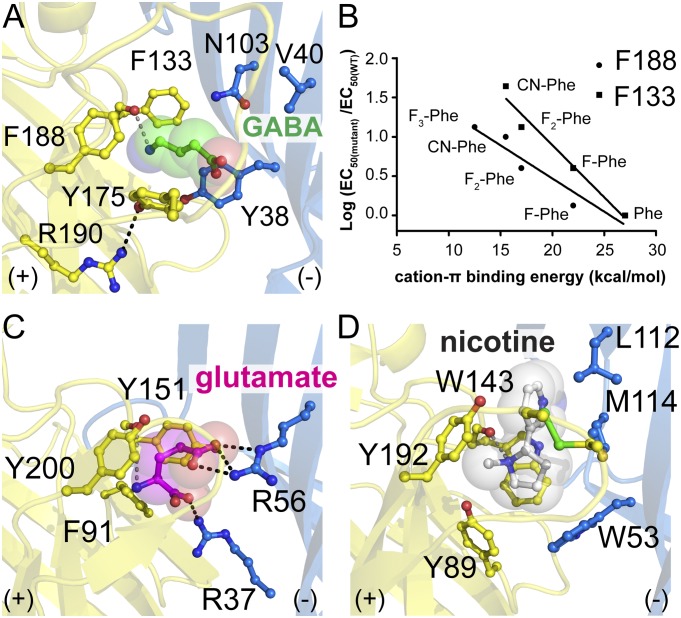
First, we obtained 3.8-Å diffraction data from ELIC crystals grown in the presence of excess GABA versus flurazepam (SI Appendix, Table S3). Although the resolution of these data is relatively low, we observe clear peaks at 3σ in unaveraged Fo-Fc omit electron density maps in two extracellular ligand-binding sites at the subunit interface (SI Appendix, Fig. S4). This sausage-shaped density, which we do not observe in crystals grown in the absence of GABA (SI Appendix, Fig. S4), is sufficiently clear to assign a likely binding pose for a GABA molecule. In agreement with previous mutagenesis experiments of GABAA receptor residues involved in ligand contacts with GABA (3, 6), we find that the γ-amino nitrogen of GABA interacts with the carbonyl oxygen of F133 in loop B (Fig. 2A), which mimics the interaction of the pyrrolidine N+-H of nicotine with the carbonyl oxygen of W143 in loop B of acetylcholine-binding protein (AChBP) from Lymnaea stagnalis (Fig. 2D) (25, 36). Also similar to nicotine in AChBP, we find that the amino-moiety of GABA is caged by aromatic side chains of F133 (loop B), Y175 and F188 (loop C), and Y38 (loop D) and forms a cation-π interaction with F133 and F188 (Fig. 2A). In Lymnaea AChBP (Fig. 2D) (37), these residues correspond to W143 (loop B), Y192 (loop C), and W53 (loop D). Residue Y89, which forms loop A in AChBP, also contributes an aromatic side chain to the binding pocket, but this residue is not conserved in ELIC (I79). We observe that R190 in loop C of ELIC is involved in a hydrogen bond with the hydroxyl group of Y175 (loop C), consistent with mutagenesis and modeling data showing that the homologous residue in GABAC receptors (R249 in ρ1) contributes to the structure of the binding pocket through a salt bridge and/or hydrogen bond (9), although the equivalent residue in GABAA receptors (R207 in β1) was proposed to contact GABA directly (38). The observed GABA-binding pose also is consistent with a low-resolution structure of ELIC in complex with bromopropylamine (23) and the recently determined crystal structure of GluCl in complex with glutamate (22) (Fig. 2C). We find that the α-amino nitrogen of glutamate occupies a position that is almost identical (<1.0 Å) to the γ-amino nitrogen of GABA and is involved in similar interactions, including cation-π interactions with Y151 and Y200 (homologous to F133 and F188 in ELIC). Arginine residues of loop D, which may stabilize the carboxyl tail of GABA in vertebrate receptors (9, 39), are not conserved in ELIC (R67 in α1, R104 in ρ1, R56 in GluCl, and V40 in ELIC). This difference likely explains the different binding pose of the γ-carboxylate group of glutamate and carboxyl tail of GABA (Fig. 2 A and C).
Fig. 2.
Locating GABA in the ELIC-binding pocket. (A) GABA is encompassed by four aromatic residues (Y38, F133, Y175, and F188) in the binding pocket and is in close proximity to N103 and V40. The amino-moiety of GABA forms a hydrogen bond with the carbonyl oxygen of F133 in loop B, which mimics the interaction of the pyrrolidine N+-H of nicotine with the carbonyl oxygen of W143 in loop B of AChBP from L. stagnalis (25, 36) shown in D. Similar interactions also are observed in the glutamate-bound structure of GluCl (22) as shown in C. The γ-amino nitrogen of GABA also forms cation-π interactions with F133 and F188, as corroborated by the plots shown in B. Here the aromatic residues surrounding GABA were replaced with a range of fluorinated Phe or cyano-Phe, and relative EC50s were determined; a linear relationship between the log ratio of EC50-values for wild type and mutant and the cation-π binding energy, as seen for F133 and F188, is indicative of a cation-π interaction.
Because the resolution of the ELIC structure in complex with GABA is relatively low, and GABA is a reasonably small and symmetric molecule, we further investigated possible cation-π interactions between the amino-moiety of GABA and the aromatic side chains of the intersubunit binding site by unnatural amino acid mutagenesis. To facilitate this study, we took advantage of L240S-containing ELIC receptors, which have improved GABA potency. Consistent with the described effects of similar mutations at the 9′ position of the pore-lining M2 helix in the nicotinic acetylcholine receptor (nAChR) (36, 40), we find that the L240S mutation in ELIC decreases the EC50 for GABA to 4.2 mM (pEC50: 2.37 ± 0.03, n = 5). In the background of L240S ELIC, we probed residues Y38 (loop D), F133 (loop B), Y175 (loop C), and F188 (loop C) by incorporating a series of fluorinated phenylalanine derivatives. The data reveal a serial decrease in agonist potency with increasing fluorination, indicative of a cation-π interaction, at positions 133 and 188 (Fig. 2B) but not at positions 38 and 175 (SI Appendix, Fig. S5). These data make a strong argument for a cation-π interaction between the γ-amino group of GABA and both the loop B and C aromatics, similar to our recent observation in an insect GABA receptor (41). These data also are consistent with the assigned binding pose for GABA in the crystal structure, which places the amino moiety almost equidistant between the loop B and C aromatic residues in the binding pocket.
Intrasubunit Benzodiazepine Site Facing the Channel Vestibule in ELIC.
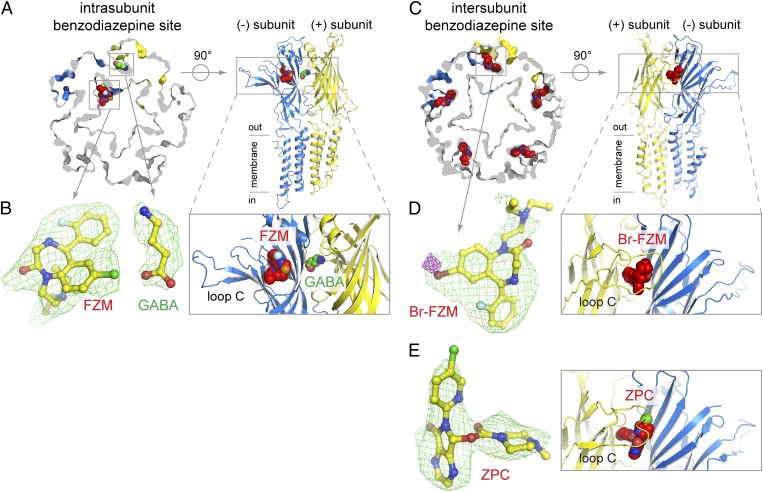
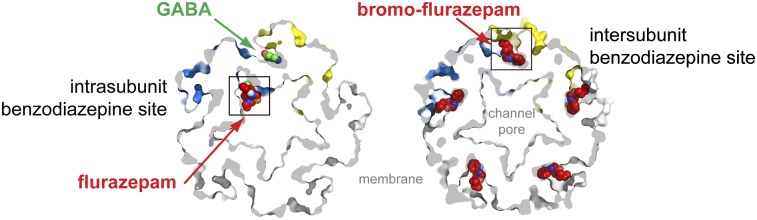
The unaveraged Fo-Fc omit density map for the GABA-bound ELIC structure shows strong density at 3.5σ (Fig. 3B and SI Appendix, Fig. S6), which we interpreted as flurazepam bound in one subunit because it was present at relatively low amounts in the crystallization solution. The difference density shows a protrusion for the chlorine atom and the curvature for the benzazepine moiety, which allowed us to assign a likely binding pose for flurazepam. Unexpectedly, this density is localized at an intrasubunit cavity facing the vestibule of the extracellular domain (Fig. 3A). This intrasubunit benzodiazepine-binding site is localized at the same height as the GABA intersubunit-binding site but lies opposite the inner walls formed by loop B (+ face) and loop D (− face) of the neighboring GABA-recognition site (Fig. 3A). This benzodiazepine site is, in consequence, ideally positioned to modulate GABA function allosterically in ELIC. Interestingly, this density could be observed in only one subunit. The slightly lower cavity volume of the other four subunits, which might have resulted from a local asymmetry imposed by the crystal lattice or the binding of one flurazepam to ELIC, might have prevented more flurazepam molecules from binding.
Fig. 3.
Two different benzodiazepine-binding sites in ELIC. (A) Cross-section through a surface representation of the ELIC structure obtained from crystals grown in the presence of excess GABA (10 mM) and 50 μM flurazepam (FZM). One GABA molecule is shown as green spheres at the intersubunit binding site composed of the principal (+) face (yellow) and the complementary (−) face (blue). Flurazepam is shown as red spheres and is localized at an intrasubunit benzodiazepine site that faces the channel vestibule in ELIC. The view, perpendicular to the membrane, is a cartoon representation and shows only two subunits of the pentamer. (B) (Left) A detailed view of GABA and flurazepam, shown in sticks, and Fourier Fo-Fc density (shown as green mesh, contoured at 3σ). (Right) A detailed view of the opposing locations of the GABA- (green spheres) and flurazepam- (red spheres) binding sites. GABA and flurazepam are localized at the same height but on opposing binding sites. (C–E) Same views as in A and B but for the ELIC structure obtained from crystals grown in the presence of equal amounts (10 mM each) of GABA and Br-flurazepam (Br-FZM) or of GABA and zopiclone (ZPC). At these concentrations, Br-flurazepam and zopiclone displace GABA and occupy the intersubunit benzodiazepine site. The green mesh is Fourier Fo-Fc density, contoured at 3σ, and the magenta mesh in D is anomalous density for the bromine atom, contoured at 4.5 σ.
To probe the contribution of the intrasubunit benzodiazepine-binding site, we performed cysteine-scanning mutagenesis on a selection of residues lining the intrasubunit benzodiazepine site in ELIC. We found that potentiation of GABA responses by 50 μM flurazepam was abolished in N60C (100 ± 5%, n = 4) and I63C (100 ± 7%, n = 4) mutants (SI Appendix, Fig. S7 A and B). These data indicate that the potentiating effects of flurazepam on ELIC arise from an interaction with the intrasubunit benzodiazepine site. An intrasubunit pocket exactly matching the intrasubunit benzodiazepine site in ELIC has been identified in the crystal structure of the muscle α1 nAChR subunit (42) and was used in a structure-based drug-design approach for nAChR modulators (43), indicating that the intrasubunit site in ELIC also is conserved in eukaryote pLGICs. The possible existence of a similar intrasubunit site in eukaryote GABAA receptors is supported by a striking sequence similarity of residues lining the intrasubunit binding site (see alignment in SI Appendix, Fig. S1). Mutagenesis studies in eukaryote GABAA receptors demonstrated a large change in the flurazepam EC50 for γ2-I76C (17), which is a residue equivalent to G37 in the intrasubunit benzodiazepine site in ELIC. However, further studies are warranted to investigate a possible functional role of the intrasubunit site in eukaryote receptors.
Intersubunit Benzodiazepine Site in ELIC.
We obtained two additional crystal structures at 3.6-Å and 3.3-Å resolution from crystals grown in the presence of equal amounts of Br-flurazepam and GABA or zopiclone and GABA, respectively (Fig. 3 C–E). No crystals could be obtained in the absence of GABA or at less than 1 mM Br-flurazepam. The anomalous signal for the bromine atom in Br-flurazepam and the higher-resolution data for zopiclone allowed us to assign ligand-binding poses with certainty. Stereo figures of electron densities are shown in SI Appendix, Figs. S8 and S9. For Br-flurazepam (Fig. 3D) and zopiclone (Fig. 3E), we observe clear Fourier Fo-Fc density (3σ) and anomalous density (4σ) for the bromine atom in Br-flurazepam (magenta mesh in Fig. 3D) at five binding sites, which we termed the “intersubunit benzodiazepine site” and which partially overlaps the recognition site for GABA in crystal structure for the low-flurazepam concentration (Fig. 3A). No GABA could be identified in the Br-flurazepam and zopiclone density maps, likely because benzodiazepines and zopiclone compete with higher affinity (micromolars) than GABA (millimolars) for a partially overlapping binding site. Considerable overlap exists between the GABA and the intersubunit benzodiazepine site, but the structural data allowed us to pinpoint a residue, F19, which is involved in an interaction with benzodiazepines, but not GABA, because it lies at the periphery of the intersubunit site. To investigate whether the intersubunit benzodiazepine site mediates inhibitory effects of flurazepam on ELIC, we characterized the functional effects of 50 μM and 500 μM flurazepam on F19A mutants of ELIC (SI Appendix, Fig. S7 A and C). In agreement with the structural data, we find that the F19A mutant still is activated by GABA but no longer is inhibited by 500 μM flurazepam. In contrast, lower concentrations of flurazepam (50 μM), still potentiate the GABA response. Together, these data provide clear evidence that the inhibitory effects on ELIC of flurazepam at >100 μM arise from an interaction at the intersubunit benzodiazepine site.
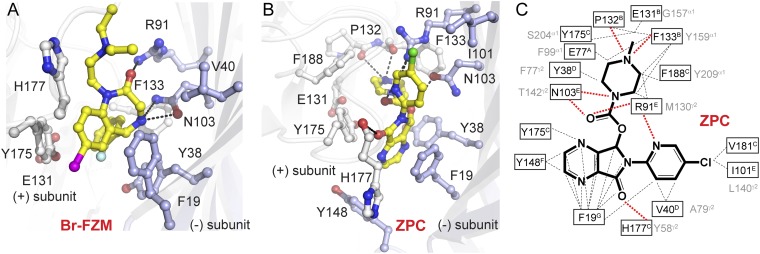
Detailed analysis of molecular interactions for Br-flurazepam (Fig. 4A) shows contributions of conserved aromatic residues previously shown to be important for benzodiazepine actions on GABAA receptors. These residues include F133 (loop B, homologous to Y159 in α1), F188 (loop C, homologous to Y209 in α1), and Y38 (loop D, homologous to F77 in γ2). A notable π-stacking of the bromophenyl moiety between Y175 (loop C) and F19 can be observed, as can a π–π interaction between the fluorophenyl moiety and Y38 (loop D, F77 in the γ2-subunit). In GABAA receptors, F19 corresponds to Y58 in the γ2-subunit, which is located at the periphery of the intersubunit benzodiazepine site and was identified previously as an important determinant of benzodiazepine action termed “loop G” (2, 18). Hydrogen bonds are formed with R91 and N103 in loop E, which correspond to M130 and T142 in the γ2-subunit. Although Br-flurazepam occupies a pocket that resembles the location of the α/γ interface in GABAA receptors, the bromine substituent on position 7 of the phenyl moiety in flurazepam points out of the benzodiazepine pocket in ELIC. In contrast, previous photolabeling studies with flunitrazepam (44) or experiments with isothiocyanate substitutions at position 7 of diazepam (45, 46) position this moiety near α1H101 of loop A in GABAA receptors, a location deep inside the pocket (I79 in ELIC). Presumably, the divergent amino acid composition between ELIC and the high-affinity α/γ benzodiazepine site in GABAA receptors causes differences in the pocket architecture and results in different benzodiazepine-binding poses in ELIC. Nevertheless, the observed binding interactions of Br-flurazepam in ELIC may assist in clarifying previously described inhibitory effects at >100 μM concentration of benzodiazepines, which could displace GABA from the β/α interface in GABAA receptors (47).
Fig. 4.
Molecular determinants of Br-flurazepam and zopiclone recognition in ELIC and GABAA receptors. (A and B) Detailed view of ligand interactions of Br-flurazepam (Br-FZM) and zopiclone (ZPC) at the intersubunit benzodiazepine site in ELIC. Ligands are shown in yellow ball-and-stick representation. Residues involved in ligand interactions are shown as white sticks on the principal (+) face and light-blue sticks on the complementary (−) face. Dashed lines represent hydrogen bonds. (C) Chemical structure of zopiclone, with the surrounding ligand contacts in ELIC. Black dashed lines represent hydrophobic interactions; red dashed lines represent hydrogen bonds. Superscript letters indicate the binding-site loop (A–G) at which each residue is located. Important residues previously identified by mutagenesis at the high affinity α/γ benzodiazepine site in GABAA receptors are indicated in gray next to homologous residues in ELIC. Superscript letters indicate whether the residue was identified on the principal (+) face of the α1-subunit or the complementary (−) face of the γ2-subunit.
Interestingly, we observed a preferential fit in electron density of the S-enantiomer of zopiclone even though the racemic mixture was used for cocrystallization with ELIC (Fig. 3E and SI Appendix, Fig. S9). This enantioselectivity corresponds to what is known from S-zopiclone binding to the high-affinity benzodiazepine-binding site in GABAA receptors (48). Zopiclone features extensive interactions (Fig. 4 B and C) with binding-site residues deep inside the principal (+) face, including cation-π interactions with F133 and F188 and hydrogen bonds with the carbonyl oxygen molecules of P132 and F133, which are interactions not observed in the Br-flurazepam structure (Fig. 4A). The hydrogen bond between the piperazine nitrogen of zopiclone and the carbonyl oxygen of the conserved aromatic residue of loop B (F133, which corresponds to Y159 in the α1-subunit) is strikingly similar to the interaction between tertiary and quaternary amines and the loop B aromat W145 in AChBP cocrystal structures (49). On the complementary (−) face, zopiclone is involved in aromatic stacking with F19, which is homologous to Y58 in the γ2-subunit. Similarly, V40 in ELIC corresponds to A79 in the γ2-subunit, which is a crucial residue in zopiclone affinity (48). Taken together, zopiclone is involved in key interactions with conserved aromatic residues, namely F19 (Y58 in γ2-subunit, loop G), Y38 (F77 in the γ2-subunit, loop D), F133 (Y159 in α1-subunit, loop B), and F188 (Y209 in α1-subunit, loop C), suggesting that zopiclone adopts a binding pose that possibly mimics the ligand orientation in the α/γ interface of the GABAA receptors.
Benzodiazepines Induce Conformational Changes in the Binding Pocket of ELIC.
Upon binding of Br-flurazepam, loop C undergoes a significant conformational movement (SI Appendix, Figs. S10 and S11), similar to the outward extension of loop C observed upon binding of certain partial agonists and antagonists in AChBP (49). We quantified the outward movement of loop C by measuring the distance between the F133 carbonyl oxygen and Cα atom of L178, which is 17.1 ± 0.1 Å in the Br-flurazepam structure, 13.9 ± 0.1 Å in the zopiclone structure, 11.4 ± 0.1 Å in GABA-bound ELIC, and 9.8 ± 0.3 Å in a closed-channel state of ELIC (here termed “apo-ELIC”). Except for the conformational change of loop C and local side-chain reorientations in the binding pocket, ELIC in complex with GABA and modulators shows an overall conformation that strongly resembles the published ELIC structure (19), which is thought to represent apo-ELIC. The GABA-bound ELIC structure can be superposed onto apo-ELIC with a rmsd value of 0.59 Å (1,514 residues aligned for pentamer 1) and 0.61 Å (1,512 residues aligned for pentamer 2). No conformational changes can be observed that resemble those in the GLIC (20, 21) and GluCl structures (22), which are thought to resemble open-channel states. There are two possible explanations for the lack of conformational changes in the GABA-bound structure of ELIC. First, crystal lattice contacts may stabilize ELIC in a closed, unactivated conformational state. Alternatively, because our electrophysiological recordings show that ELIC desensitizes over a time course of several minutes during exposure to GABA, the GABA-bound structure may represent a desensitized state, that resembles the published apo-ELIC structure.
Concluding Remarks
GABAA receptors are critical for fast synaptic inhibition in the mammalian brain and are the target of many therapeutic agents; understanding their mechanism of action therefore has long been an important goal. Our study, which reveals the critical elements of both GABA and benzodiazepine recognition in ELIC, brings this goal a step closer. We find that GABA binds at an intersubunit binding site using a range of interactions, including a cation-π interaction as observed in GABAA and GABAC receptors. Benzodiazepines can occupy two different sites in ELIC: an intrasubunit site responsible for potentiating effects, and an intersubunit site responsible for inhibitory effects, which partially overlaps with the GABA-recognition site. In eukaryote GABAA receptors there are at least three, but possibly more, different low-affinity binding sites for benzodiazepine-site ligands in addition to the high-affinity benzodiazepine-binding site at the α/γ interface. One of these sites is the low-affinity diazepam-binding site described by Walters et al. (29). A second site is a low-affinity flurazepam-binding site that likely localizes to the α/β interface (28). A third site is a low-affinity binding site that accounts for the low-potency inhibition of the GABA current in α/β receptors (47). This latter site corresponds to the intersubunit benzodiazepine site in ELIC, which mediates inhibitory effects of flurazepam. The location of the intrasubunit benzodiazepines site in ELIC was unexpected, but if a similar site exists in GABAA receptors, it could provide a framework for understanding previously unexplained actions of benzodiazepines on GABAA receptors.
Our study offers a structural view how GABA and allosteric ligands are recognized, which is a key question in understanding modulation of ligand-gated ion channels. Overall, this work paves the way for structural elucidation of eukaryote GABA receptors in the presence of different allosteric modulators.
Methods
Protein Production and Crystallization.
ELIC was expressed and purified as previously described with minor modifications. The ELIC sequence (GenBank accession number ADN00343.1) was purchased from GenScript as a synthetic gene with optimized codon use for expression in Escherichia coli. In the published sequence of ELIC [Protein Data Bank (PDB) ID code 2VL0] a Gly residue is missing after R163. Consequently, all residues after G164 are renumbered +1 in our ELIC sequence. The synthetic gene corresponding to mature ELIC with an N-terminal truncation (residue numbers 8–322) was subcloned in pET-11a (Novagen) and N-terminally fused to maltose-binding protein (MBP) with a 3CV protease cleavage site for removal of MBP. The MBP-ELIC fusion protein was expressed in the C43 E. coli strain, which was grown in LB medium supplemented with carbenicilline to an absorbance A600 = 1.8. Upon addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), the temperature was lowered to 20 °C for overnight incubation. E. coli membranes were isolated by ultracentrifugation at 125,000 × g at 4 °C for 1 h after cell lysis with an Emulsiflex C-5 (Avestin) in 50 mM Na-phosphate (pH 8.0), 150 mM NaCl (buffer A), 1 mg/mL lysozyme, 20 μg/mL DNase, 5 mM MgCl2, and the addition of protease inhibitors [either complete tablets (Roche) with the addition of 1 mM PMSF or 1 μg/mL leupeptin, μg/mL pepstatin, and 1 mM PMSF]. Membrane proteins were solubilized with 2% (wt/vol) anagrade n-undecyl-β-d-maltoside (Anatrace) at 4 °C overnight, and the solubilized fraction was cleared by ultracentrifugation at 30,000 × g at 4 °C for 1 h. Solubilized proteins were batch purified by affinity chromatography on amylose resin (New England Biolabs). After incubation with beads for 1–2 h at 4 °C, the column was washed with three column volumes buffer A + 0.15% n-undecyl-β-d-maltoside. The MBP-ELIC fusion protein was cleaved overnight at 4 °C by incubation of the beads with 3CV protease in the presence of 1 mM EDTA and 1 mM DTT. Cleaved ELIC and 3CV protease were passed through the column and concentrated to less than 1 mL on a Vivaspin concentrating column with a cutoff of 100 kDa (Sartorius). ELIC was purified further on a Superdex 200 10/300 GL (GE Healthcare) column equilibrated with buffer containing 10 mM Na-phosphate (pH 8.0), 150 mM NaCl, and 0.15% n-undecyl-β-d-maltoside. Peak fractions corresponding to cleaved pentameric ELIC were pooled, concentrated to 10 mg/mL, and supplemented with 0.5 mg/mL E. coli lipids (Avanti Polar Lipids).
Crystallization of ELIC was carried out at 4 °C by vapor diffusion of sitting drops that were set up with a Mosquito crystallization robot in MRC plates (Molecular Dimensions). The crystallization buffer was composed of 200 mM ammonium sulfate, 50 mM ADA (N-(2-acetamido)iminodiacetic acid) (pH 6.5), and 9–12% (vol/vol) PEG4000. During the course of this study more than 1,000 crystals for different ELIC complexes were screened, and datasets were collected when diffraction was better than 4.0 Å. Crystals for ELIC in complex with GABA and allosteric modulators were obtained by screening in two dimensions with a range of GABA concentrations from 100 μM to 100 mM and a range of benzodiazepine (flurazepam, flunitrazepam, or bromazepam) concentrations from 10 μM to 15 mM. The crystal structure of ELIC in complex with GABA and flurazepam was obtained from crystals grown in the presence of 10 mM GABA and 50 μM flurazepam. The crystal structures for ELIC in complex with Br-flurazepam and zopiclone were obtained by screening with 10 mM GABA and a range of Br-flurazepam or zopiclone concentrations from 1–15 mM Br-flurazepam or 0.05–10 mM zopiclone. Anomalous diffraction data were collected from crystals grown in the presence of 10 mM GABA and 10 mM Br-flurazepam. No crystals were obtained at Br-flurazepam concentrations lower than 1 mM. No crystals were obtained for Br-flunitrazepam. Stock solutions for GABA and flurazepam were made in milliQ water. The stock solutions for Br-flurazepam, Br-flunitrazepam, flunitrazepam, bromazepam, and zopiclone were made in DMSO. Crystals were harvested after 1 wk by adding 30% (vol/vol) glycerol as a cryoprotectant to the mother liquor. Crystals were cryo-cooled by immersion in liquid nitrogen. The synthesis of Br-flurazepam and Br-flunitrazepam is described in SI Appendix, Fig. S12.
Structure Determination and Refinement.
All crystals obtained in this study belonged to the same spacegroup as the ELIC structure published by Hilf and Dutzler (PDB ID code 2VL0). The dataset for GABA+flurazepam was obtained by merging partial datasets obtained by exposing a single crystal at three different positions. For GABA+Br-flurazepam the best quality in electron density maps was obtained after merging five partial datasets obtained by exposure on different positions of two isomorphous crystals. Data integration was done in XDS and scaling either in XSCALE (GABA+Br-flurazepam, GABA+zopiclone) or SCALA (GABA+flurazepam). Structure solutions were obtained after molecular replacement with PHASER and refinement with REFMAC. Final model building and refinement for the GABA+flurazepam structure was carried out by iterative cycles of manual rebuilding in COOT and refinement in PHENIX. The loops preceding and following the β8-strand in the extracellular domain were built incorrectly in previously published ELIC structures (PDB ID codes 2VL0, 2YKS, 3RQU, 3RQW, 3UQ4, 3UQ5, and 3UQ7). In our structures, we have built residues 137–139 of the loop preceding β8 in a different conformation, resulting in a shift of the β8-strand register by +2 residues. After residue 153 in the loop following the β8-strand, our structures follow the same register as the previously published structures. Ligand restraints for GABA, flurazepam, Br-flurazepam, and zopiclone were generated using ELBOW in the PHENIX suite or with PRODRG. In PHENIX we used autoNCS to detect automatically noncrystallographic symmetry (NCS) restraints, which were maintained during refinement for an entire subunit per NCS group. One TLS (Translation/Libration/Screw) body was assigned per subunit. Model validation was done using MOLPROBITY and all figures were prepared using PYMOL (19).
Two-Electrode Voltage-Clamp Recordings.
For electrophysiological recordings from Xenopus oocytes, the mature sequence of ELIC (residue numbers 8–322) was cloned into pGEMHE with the signal sequence of the human α7 nAChR (MRCSPGGVWLALAASLLHVSLQ). mRNA was transcribed in vitro using the mMESSAGE mMACHINE T7 transcription kit (Ambion). Mutants were engineered using a QuikChange strategy (Stratagene) and verified by sequencing.
For oocyte recordings stage V–VI Xenopus laevis oocytes were stored and incubated at 18 °C in modified Barth’s medium [88 mM NaCl, 10 mM Hepes-NaOH (pH 7.4), 2.4 mM NaHCO3, 1 mM KCl, 0.82 mM MgSO4, 0.41 mM CaCl2, 0.34 mM Ca(NO3)2] supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin. Collagenase treatment (type IA, Sigma) and mechanical defolliculation of the oocytes were performed as described (47). Oocytes were injected with 10 ng RNA, and recordings were made 18–36 h postinjection.
Supplementary Material
Acknowledgments
We thank Jessy Winters for assisting with protein preparations, Estelle Neveu for technical assistance with oocyte recordings, and Jan Tytgat for mass spectrometry. For model refinement we used the infrastructure of the Flemish Supercomputer Center funded by the Hercules Foundation and the Flemish Government. This project was supported by Grant KULeuven OT/08/048 (to C.U.); by the European Union Seventh Framework Programme under Grant Agreement HEALTH-F2-2007-202088 (Structure, function and disease of Cys-loop receptors, “NeuroCypres” project) (to W.S., I.J.P.d.E., S.C.R.L., D.B., P.-J.C., and C.U.); by the Wellcome Trust (S.C.R.L. and K.P.); by Austrian Science Fund Grant P19653 (to M.E.); and by National Institutes of Health Grant NS 34407 (to D.A.D.). S.C.R.L. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The structures of ELIC in complex with GABA and flurazepam, zopiclone, and Br-flurazepam have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4A96, 4A97, and 4A98, respectively).
See Author Summary on page 17752 (volume 109, number 44).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208208109/-/DCSupplemental.
References
- 1.Sieghart W. Structure and pharmacology of gamma-aminobutyric acidA receptor subtypes. Pharmacol Rev. 1995;47:181–234. [PubMed] [Google Scholar]
- 2.Ernst M, Brauchart D, Boresch S, Sieghart W. Comparative modeling of GABA(A) receptors: Limits, insights, future developments. Neuroscience. 2003;119:933–943. doi: 10.1016/s0306-4522(03)00288-4. [DOI] [PubMed] [Google Scholar]
- 3.Amin J, Weiss DS. GABAA receptor needs two homologous domains of the beta-subunit for activation by GABA but not by pentobarbital. Nature. 1993;366:565–569. doi: 10.1038/366565a0. [DOI] [PubMed] [Google Scholar]
- 4.Wagner DA, Czajkowski C. Structure and dynamics of the GABA binding pocket: A narrowing cleft that constricts during activation. J Neurosci. 2001;21:67–74. doi: 10.1523/JNEUROSCI.21-01-00067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lummis SC, L Beene D, Harrison NJ, Lester HA, Dougherty DA. A cation-pi binding interaction with a tyrosine in the binding site of the GABAC receptor. Chem Biol. 2005;12:993–997. doi: 10.1016/j.chembiol.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Padgett CL, Hanek AP, Lester HA, Dougherty DA, Lummis SC. Unnatural amino acid mutagenesis of the GABA(A) receptor binding site residues reveals a novel cation-π interaction between GABA and beta 2Tyr97. J Neurosci. 2007;27:886–892. doi: 10.1523/JNEUROSCI.4791-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boileau AJ, Evers AR, Davis AF, Czajkowski C. Mapping the agonist binding site of the GABAA receptor: Evidence for a beta-strand. J Neurosci. 1999;19:4847–4854. doi: 10.1523/JNEUROSCI.19-12-04847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westh-Hansen SE, et al. Arginine residue 120 of the human GABAA receptor alpha 1, subunit is essential for GABA binding and chloride ion current gating. Neuroreport. 1999;10:2417–2421. doi: 10.1097/00001756-199908020-00036. [DOI] [PubMed] [Google Scholar]
- 9.Harrison NJ, Lummis SC. Locating the carboxylate group of GABA in the homomeric rho GABA(A) receptor ligand-binding pocket. J Biol Chem. 2006;281:24455–24461. doi: 10.1074/jbc.M601775200. [DOI] [PubMed] [Google Scholar]
- 10.Sigel E, Lüscher BP. A closer look at the high affinity benzodiazepine binding site on GABAA receptors. Curr Top Med Chem. 2011;11:241–246. doi: 10.2174/156802611794863562. [DOI] [PubMed] [Google Scholar]
- 11.Dunn SM, Davies M, Muntoni AL, Lambert JJ. Mutagenesis of the rat alpha1 subunit of the gamma-aminobutyric acid(A) receptor reveals the importance of residue 101 in determining the allosteric effects of benzodiazepine site ligands. Mol Pharmacol. 1999;56:768–774. [PubMed] [Google Scholar]
- 12.Wieland HA, Lüddens H, Seeburg PH. A single histidine in GABAA receptors is essential for benzodiazepine agonist binding. J Biol Chem. 1992;267:1426–1429. [PubMed] [Google Scholar]
- 13.Amin J, Brooks-Kayal A, Weiss DS. Two tyrosine residues on the alpha subunit are crucial for benzodiazepine binding and allosteric modulation of gamma-aminobutyric acidA receptors. Mol Pharmacol. 1997;51:833–841. doi: 10.1124/mol.51.5.833. [DOI] [PubMed] [Google Scholar]
- 14.Buhr A, Baur R, Malherbe P, Sigel E. Point mutations of the alpha 1 beta 2 gamma 2 gamma-aminobutyric acid(A) receptor affecting modulation of the channel by ligands of the benzodiazepine binding site. Mol Pharmacol. 1996;49:1080–1084. [PubMed] [Google Scholar]
- 15.Buhr A, Schaerer MT, Baur R, Sigel E. Residues at positions 206 and 209 of the alpha1 subunit of gamma-aminobutyric AcidA receptors influence affinities for benzodiazepine binding site ligands. Mol Pharmacol. 1997;52:676–682. doi: 10.1124/mol.52.4.676. [DOI] [PubMed] [Google Scholar]
- 16.Morlock EV, Czajkowski C. Different residues in the GABAA receptor benzodiazepine binding pocket mediate benzodiazepine efficacy and binding. Mol Pharmacol. 2011;80:14–22. doi: 10.1124/mol.110.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teissére JA, Czajkowski C. A (beta)-strand in the (gamma)2 subunit lines the benzodiazepine binding site of the GABA A receptor: Structural rearrangements detected during channel gating. J Neurosci. 2001;21:4977–4986. doi: 10.1523/JNEUROSCI.21-14-04977.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucken AM, et al. Identification of benzodiazepine binding site residues in the gamma2 subunit of the gamma-aminobutyric acid(A) receptor. Mol Pharmacol. 2000;57:932–939. [PubMed] [Google Scholar]
- 19.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 20.Bocquet N, et al. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- 21.Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- 22.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmermann I, Dutzler R. Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS Biol. 2011;9:e1001101. doi: 10.1371/journal.pbio.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan J, et al. Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine. Nat Commun. 2012;3:714. doi: 10.1038/ncomms1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 26.Brams M, et al. Crystal structures of a cysteine-modified mutant in loop D of acetylcholine-binding protein. J Biol Chem. 2011;286:4420–4428. doi: 10.1074/jbc.M110.188730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wooltorton JR, Moss SJ, Smart TG. Pharmacological and physiological characterization of murine homomeric beta3 GABA(A) receptors. Eur J Neurosci. 1997;9:2225–2235. doi: 10.1111/j.1460-9568.1997.tb01641.x. [DOI] [PubMed] [Google Scholar]
- 28.Baur R, et al. Covalent modification of GABAA receptor isoforms by a diazepam analogue provides evidence for a novel benzodiazepine binding site that prevents modulation by these drugs. J Neurochem. 2008;106:2353–2363. doi: 10.1111/j.1471-4159.2008.05574.x. [DOI] [PubMed] [Google Scholar]
- 29.Walters RJ, Hadley SH, Morris KD, Amin J. Benzodiazepines act on GABAA receptors via two distinct and separable mechanisms. Nat Neurosci. 2000;3:1274–1281. doi: 10.1038/81800. [DOI] [PubMed] [Google Scholar]
- 30.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Z, et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson T, MacAllan D, Lunt G, Battersby M. gamma-Aminobutyric acid receptor complex of insect CNS: Characterization of a benzodiazepine binding site. J Neurochem. 1986;47:1955–1962. doi: 10.1111/j.1471-4159.1986.tb13114.x. [DOI] [PubMed] [Google Scholar]
- 33.Lummis SC, Sattelle DB. Binding sites for [(3)H]GABA, [(3)H]flunitrazepam and [(35)S]TBPS in insect CNS. Neurochem Int. 1986;9:287–293. doi: 10.1016/0197-0186(86)90065-3. [DOI] [PubMed] [Google Scholar]
- 34.Chevrot R, et al. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faure D, Dessaux Y. Quorum sensing as a target for developing control strategies for the plant pathogen Pectobacterium. (Translated from English) Eur J Plant Pathol. 2007;119:353–365. [Google Scholar]
- 36.Xiu X, Puskar NL, Shanata JA, Lester HA, Dougherty DA. Nicotine binding to brain receptors requires a strong cation-pi interaction. Nature. 2009;458:534–537. doi: 10.1038/nature07768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 38.Wagner DA, Czajkowski C, Jones MV. An arginine involved in GABA binding and unbinding but not gating of the GABA(A) receptor. J Neurosci. 2004;24:2733–2741. doi: 10.1523/JNEUROSCI.4316-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldschen-Ohm MP, Wagner DA, Jones MV. Three arginines in the GABAA receptor binding pocket have distinct roles in the formation and stability of agonist- versus antagonist-bound complexes. Mol Pharmacol. 2011;80:647–656. doi: 10.1124/mol.111.072033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revah F, et al. Mutations in the channel domain alter desensitization of a neuronal nicotinic receptor. Nature. 1991;353:846–849. doi: 10.1038/353846a0. [DOI] [PubMed] [Google Scholar]
- 41.Lummis SC, McGonigle I, Ashby JA, Dougherty DA. Two amino acid residues contribute to a cation-π binding interaction in the binding site of an insect GABA receptor. J Neurosci. 2011;31:12371–12376. doi: 10.1523/JNEUROSCI.1610-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Structural determinants for alpha-neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels (Austin) 2007;1:234–237. doi: 10.4161/chan.4909. [DOI] [PubMed] [Google Scholar]
- 43.Dey R, Chen L. In search of allosteric modulators of a7-nAChR by solvent density guided virtual screening. J Biomol Struct Dyn. 2011;28:695–715. doi: 10.1080/07391102.2011.10508600. [DOI] [PubMed] [Google Scholar]
- 44.Duncalfe LL, Carpenter MR, Smillie LB, Martin IL, Dunn SM. The major site of photoaffinity labeling of the gamma-aminobutyric acid type A receptor by [3H]flunitrazepam is histidine 102 of the alpha subunit. J Biol Chem. 1996;271:9209–9214. doi: 10.1074/jbc.271.16.9209. [DOI] [PubMed] [Google Scholar]
- 45.Berezhnoy D, et al. On the benzodiazepine binding pocket in GABAA receptors. J Biol Chem. 2004;279:3160–3168. doi: 10.1074/jbc.M311371200. [DOI] [PubMed] [Google Scholar]
- 46.Tan KR, Baur R, Charon S, Goeldner M, Sigel E. Relative positioning of diazepam in the benzodiazepine-binding-pocket of GABA receptors. J Neurochem. 2009;111:1264–1273. doi: 10.1111/j.1471-4159.2009.06419.x. [DOI] [PubMed] [Google Scholar]
- 47.Ramerstorfer J, et al. The GABAA receptor alpha+beta- interface: A novel target for subtype selective drugs. J Neurosci. 2011;31:870–877. doi: 10.1523/JNEUROSCI.5012-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanson SM, Morlock EV, Satyshur KA, Czajkowski C. Structural requirements for eszopiclone and zolpidem binding to the gamma-aminobutyric acid type-A (GABAA) receptor are different. J Med Chem. 2008;51:7243–7252. doi: 10.1021/jm800889m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brams M, et al. A structural and mutagenic blueprint for molecular recognition of strychnine and d-tubocurarine by different cys-loop receptors. PLoS Biol. 2011;9:e1001034. doi: 10.1371/journal.pbio.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]