Abstract
SNARE-dependent membrane fusion in eukaryotic cells requires that the heptad-repeat SNARE domains from R- and Q-SNAREs, anchored to apposed membranes, assemble into four-helix coiled-coil bundles. In addition to their SNARE and transmembrane domains, most SNAREs have N-terminal domains (N-domains), although their functions are unclear. The N-domain of the yeast vacuolar Qc-SNARE Vam7p is a binding partner for the homotypic fusion and vacuole protein sorting complex (a master regulator of vacuole fusion) and has Phox homology, providing a phosphatidylinositol 3-phosphate (PI3P)-specific membrane anchor. We now report that this Vam7p N-domain has yet another role, one that does not depend on its physical connection to the Vam7p SNARE domain. By attaching a transmembrane anchor to the C terminus of Vam7p to create Vam7tm, we bypass the requirement for the N-domain to anchor Vam7tm to reconstituted proteoliposomes. The N-domain of Vam7tm is indispensible for trans-SNARE complex assembly in SNARE-only reactions. Introducing Vam7(1-125)p as a separate recombinant protein suppresses the defect caused by N-domain deletion from Vam7tm, demonstrating that the function of this N-domain is not constrained to covalent attachment to Vam7p. The Vam7p N-domain catalyzes the docking of apposed membranes by promoting transinteractions between R- and Q-SNAREs. This function of the Vam7p N-domain depends on the presence of PI3P and its affinity for PI3P. Added N-domain can even promote SNARE complex assembly when Vam7 still bears its own N-domain.
Keywords: phosphoinositide, Phox homology domain
Membranes fuse by conserved mechanisms during endocytic and exocytic vesicular trafficking (1). The initial step of association between membranes, termed tethering, is mediated by a Rab-family GTPase and its associated tethering proteins (2). Selected proteins and lipids then become enriched in a fusion-competent microdomain. SNARE proteins anchored to apposed membranes form four-helix bundles, the trans-SNARE complex, in conjunction with SNARE-binding factors such as Sec1-Munc18 proteins and, at neuronal synapses, Munc13, complexin, and synaptotagmin (3, 4). Although SNARE proteins are central to the ensuing fusion, little is known of the roles of their N-domains.
We study fusion mechanisms with yeast vacuoles (lysosomes) (5). Vacuoles undergo constant homotypic fusion and fission in the cell in response to the osmolarity of the growth environment. When fusion is genetically impaired, continuing fission causes a fragmented vacuole morphology, the vam phenotype (6), which allowed identification of the genes for proteins that catalyze vacuole fusion. Their roles were confirmed and extended through studies of the fusion of the isolated organelle (7). Vacuole clustering (tethering) is mediated by the Rab GTPase Ypt7p through its direct binding to the hexameric homotypic fusion and vacuole protein sorting (HOPS) complex (8–11). As vacuole membranes are drawn together, the proteins and lipids needed for fusion become enriched in a ring-shaped microdomain which surrounds each pair of apposed membranes (12–14). In addition to Ypt7p and HOPS, the four SNARE proteins become highly enriched in this ring-shaped microdomain. Vacuole fusion requires Nyv1p, the R-SNARE (in reference to the arginyl residue at the center of its SNARE domain), and three Q-SNAREs (with central glutamyl residues), Vam3p, Vti1p, and Vam7p, members of the conserved Qa, Qb, and Qc families (15, 16), respectively. cis-SNARE complexes, with each SNARE anchored to the same membrane, are disassembled by the chaperones Sec18p and Sec17p, permitting the individual SNAREs to participate in the assembly of trans-SNARE complexes (17–21). The roles of the SNARE N-domains, whether for localization to the fusion ring microdomain or for SNARE complex assembly, are largely unknown.
Vam7p is unique among the vacuolar SNAREs in lacking an apolar membrane anchor domain. Instead, it has an N-domain with Phox homology (PX), which binds phosphatidylinositol 3-phosphate (PI3P) and thereby contributes to its membrane association (22). The N-domain of Vam7p may also contribute to its membrane binding by its direct affinity for HOPS (23). Does the Vam7p N-domain have any function other than binding PI3P and HOPS? We now exploit a minimal system of reconstituted proteoliposomes (RPLs) bearing the vacuolar R-SNARE or Q-SNAREs (24, 25) to show that the Vam7p N-domain, whether joined to Vam7p or bound to membranes separately, promotes functional trans-SNARE interactions.
Results
R-SNARE RPLs were prepared by dialysis from mixed micellar solutions of vacuolar lipids [with PI3P and PI(4,5)P2, termed PIPx, where indicated], octyl glucoside, nitrobenzoxadiazole (NBD)-PE and rhodamine-PE, and the purified recombinant R-SNARE Nyv1p (25). The NBD-PE fluorescence of these RPLs is quenched by the rhodamine-PE. 3Q-SNARE RPLs were also prepared without fluorescent lipids and with the three recombinant vacuolar Q-SNAREs instead of Nyv1p. When these two RPL preparations are mixed, the transinteractions of the R- and Q-SNAREs triggers lipidic changes (e.g., membrane fusion, lysis and reannealing, or other lipid phase changes), relieving the quenching of the NBD fluorescence (25). This dequenching is a basic measure of functional trans-SNARE interactions.
Requirement for Vam7p N-Domain in Reconstituted Reactions.
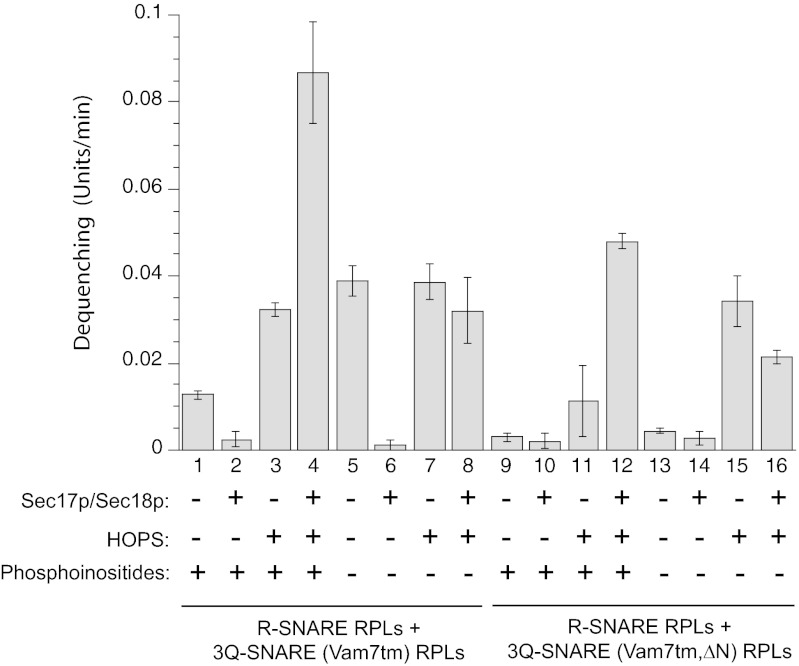
To assay additional functions of the N-domain of the Qc-SNARE Vam7p, we relieved this domain of its role of anchoring Vam7p to the vacuole by joining a transmembrane (tm) anchor domain from the Qb-SNARE Vti1p to the Vam7p C terminus, creating a fusion protein termed Vam7tm (Fig. 1A). This recombinant protein was expressed in Escherichia coli, purified, and substituted for Vam7p in the preparation of 3Q-SNARE RPLs in the presence or absence of phosphoinositides (Fig. 1B). R-SNARE proteoliposomes were mixed with these 3Q-SNARE RPLs and incubated at 27 °C, and the rate of dequenching of NBD fluorescence was monitored (Fig. 2). Vam7tm supports SNARE-dependent dequenching that is sensitive to SNARE disassembly chaperones Sec17p/Sec18p (Fig. 2, lanes 1 and 2 and lanes 5 and 6) unless HOPS is also present (Fig. 2, lanes 3 and 4 and lanes 7 and 8), in accord with published studies (25) with WT Vam7p.
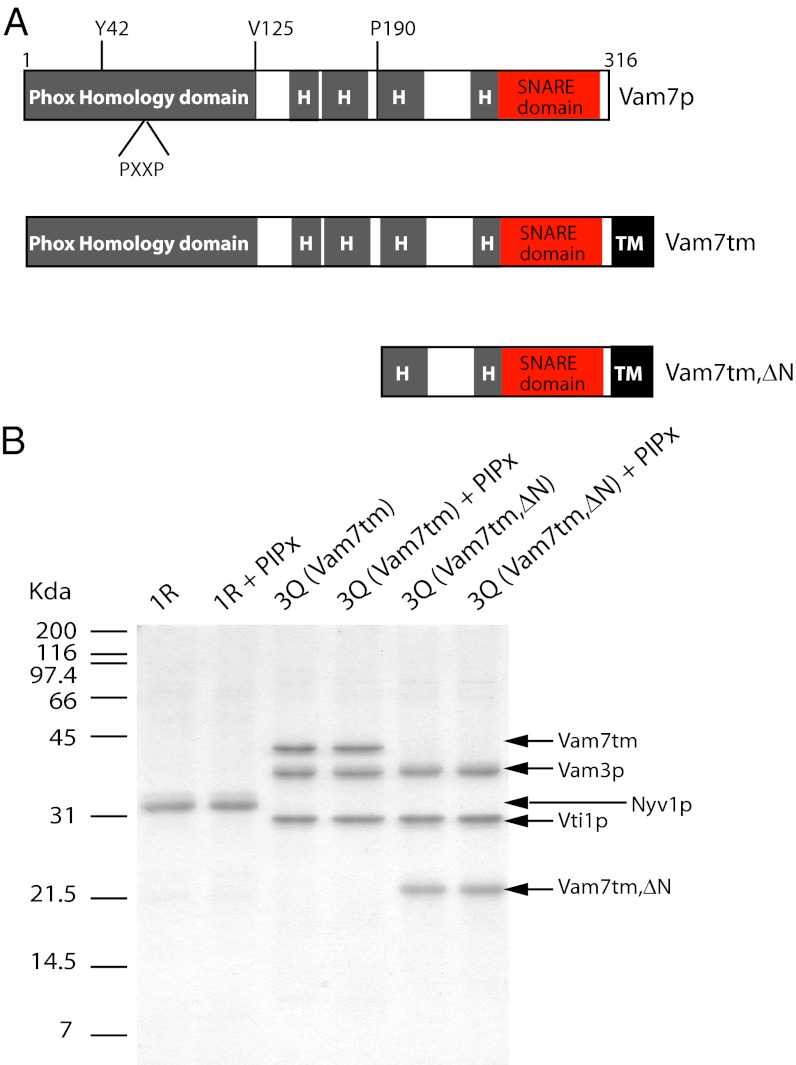
Fig. 1.
Qc-SNARE Vam7p derivatives and their reconstitution into proteoliposomes. (A) Vam7p has an N-terminal domain with PX and C-terminal SNARE domains. The linker region contains regions with predicted tendencies to adopt α-helical conformation (gray areas, labeled “H”). Residue Y42 is required for PI3P binding, and the PXXP motif is highly conserved in PX domains. The tm domain (TM) from the Qb SNARE Vti1p was attached to the C-terminal end of full-length Vam7p to generate Vam7tm (Middle). N-terminal truncation then generated Vam7tm,∆N (Bottom). Vam7tm and Vam7tm,∆N were used to make proteoliposomes with other Q-SNAREs. (B) Proteoliposomes were reconstituted with the R-SNARE Nyv1p or with 3 Q-SNAREs and evaluated by SDS/PAGE and Coomassie blue staining. PIPx denotes the inclusion of PI3P and PI(4,5)P2 in lipid mixtures. The positions of protein markers are indicated on the left.
Fig. 2.
Requirement for the Vam7p N-domain in reconstituted proteoliposomal reactions. R- and Q-SNARE proteoliposomes were incubated with HOPS (0.12 μM), Sec17p (0.68 μM)/Sec18p (0.24 μM), both, or neither in a fluorescence plate reader. Where indicated, phosphoinositides [PI3P and PI(45)P2] were also included during proteoliposome reconstitution. The NBD fluorescence signal was measured every minute. The maximal rate of signal increase in the initial phase of the reaction is displayed.
Phosphoinositides inhibit the SNARE-only reaction when Vam7tm is the Qc SNARE (Fig. 2, lanes 1 and 5). Earlier studies (26) with 3Q-SNARE proteoliposomes bearing WT Vam7p had suggested that the Vam7p PX domain has the flexibility to bind PI3P from the same membrane (Q-SNARE RPLs), or from the R-SNARE RPLs, with inhibitory or stimulatory effect on dequenching. The PX domain of Vam7tm may inhibit dequenching by binding PI3P from the same membrane (i.e., Q-SNARE RPLs). Phosphoinositides are required for optimal synergy between HOPS and Sec17p/Sec18p (Fig. 2, compare lanes 3 and 4 vs. lanes 7 and 8), as seen with WT Vam7p (25, 27).
The dequenching seen with proteoliposomes bearing Vam7tm in the absence of phosphoinositides, HOPS, or Sec17p/Sec18p is lost upon deletion of the N-domain of Vam7p (Fig. 2, compare lane 5 with Vam7tm vs. lane 13 with Vam7tm,∆N). Dequenching was restored to incubations with Vam7tm,∆N when the tethering complex HOPS was added (Fig. 2, lane 15), suggesting that the deletion of the N-domain of Vam7tm had removed an important function rather than completely denaturing the protein.
Dequenching with Vam7tm,∆N Is Reactivated by Recombinant Vam7 (1-125)p.
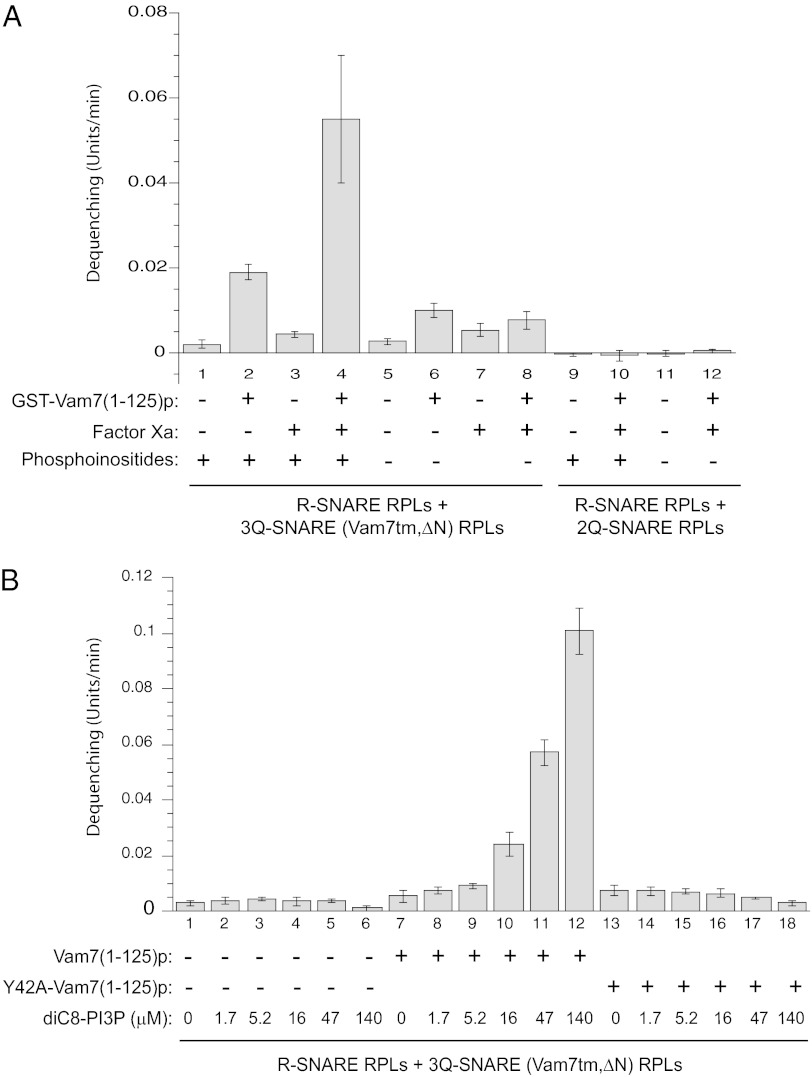
Because HOPS can restore dequenching in the absence of the Vam7p N-domain, we further tested whether the Vam7p N-domain itself has a defined function that can be most clearly observed in the absence of HOPS or other SNARE chaperones. The addition of GST-Vam7(1-125)p to incubations of mixed R-SNARE RPLs and 3Q-SNARE (Vam7tm,ΔN) RPLs gives a partial restoration of dequenching (Fig. 3A, lanes 1 and 2 and lanes 5 and 6), but the dimerization of the GST tag might have artificially promoted the docking of RPLs. We therefore introduced Factor Xa to cleave the GST tag (Materials and Methods). The tag-free Vam7(1-125)p is much more potent in stimulating dequenching (Fig. 3A, lanes 2 and 4). This stimulation requires phosphoinositides (Fig. 3A, lanes 4 and 8), and the presence of Vam7tm,∆N on the Q-SNARE RPLs (Fig. 3A, lane 4 vs. lane 10). Factor Xa alone does not stimulate (Fig. 3A, lane 1 vs. lane 3). Thus, the isolated recombinant Vam7p N-domain, separate from the Vam7p SNARE domain, is sufficient to restore the activity of the membrane-anchored Vam7p N-terminal truncation mutant.
Fig. 3.
Recombinant Vam7 N-domain restores activity to RPLs bearing Vam7tm,∆N. (A) R-SNARE RPLs and 3Q-SNARE RPLs bearing Vam7tm,∆N (lanes 1–8) were incubated in the fusion reaction with 1.1 μM GST-Vam7(1-125)p, factor Xa (10μg/mL), or GST-Vam7(1-125)p that had been preincubated with Factor Xa (lanes 4 and 10). 2Q-SNARE RPLs bearing Vam3p and Vti1p (lanes 9–12) were used as control. Proteoliposomes were prepared with phosphoinositides [PI3P and PI(4,5)P2] where specified. (B) To address the requirement for PI3P, proteoliposomes prepared in the absence of phosphoinositides were incubated with increasing concentrations of soluble diC8-PI3P. GST-Vam7(1-125)p or GST-Vam7(1-125,Y42A)p had been preincubated with Factor Xa (10 μg/mL).
To determine whether Vam7(1-125)p might promote dequenching by modulating the level of free PI3P on the RPLs to some low, optimal value, we added graded levels of diC8-PI3P [a soluble form of PI3P (27)] to preformed RPLs that are otherwise free of phosphoinositides (Fig. 3B). Di-C8 phosphoinositides have the useful property of partitioning into proteoliposomes and fulfilling their roles there in supporting fusion (27). This enables the very same proteoliposomes to be assayed with a curve of added phosphoinositide concentrations (Fig. 3B), eliminating any batch-to-batch RPL variability. DiC8-PI3P by itself does not activate dequenching with Vam7tm,∆N (Fig. 3B, lanes 1–6), whereas GST-free Vam7(1-125)p activates the dequenching supported by Vam7tm,∆N at increasing diC8-PI3P levels (Fig. 3B, lanes 7–12). Y42A, a point mutation in Vam7(1-125)p that interferes with its capacity to bind PI3P, renders it completely inactive (Fig. 3B, lanes 13–18). Therefore, the specific interaction between PI3P and the PX residues of Vam7(1-125)p is essential for it to act in concert with a separate C-terminal portion of Vam7p to promote lipid mixing. This interaction may anchor Vam7(1-125)p to the membrane, stabilize it in an active conformation, or both.
Isolated Vam7p N-Domain Stimulates Lipid Mixing in Presence of Full-Length Vam7p.
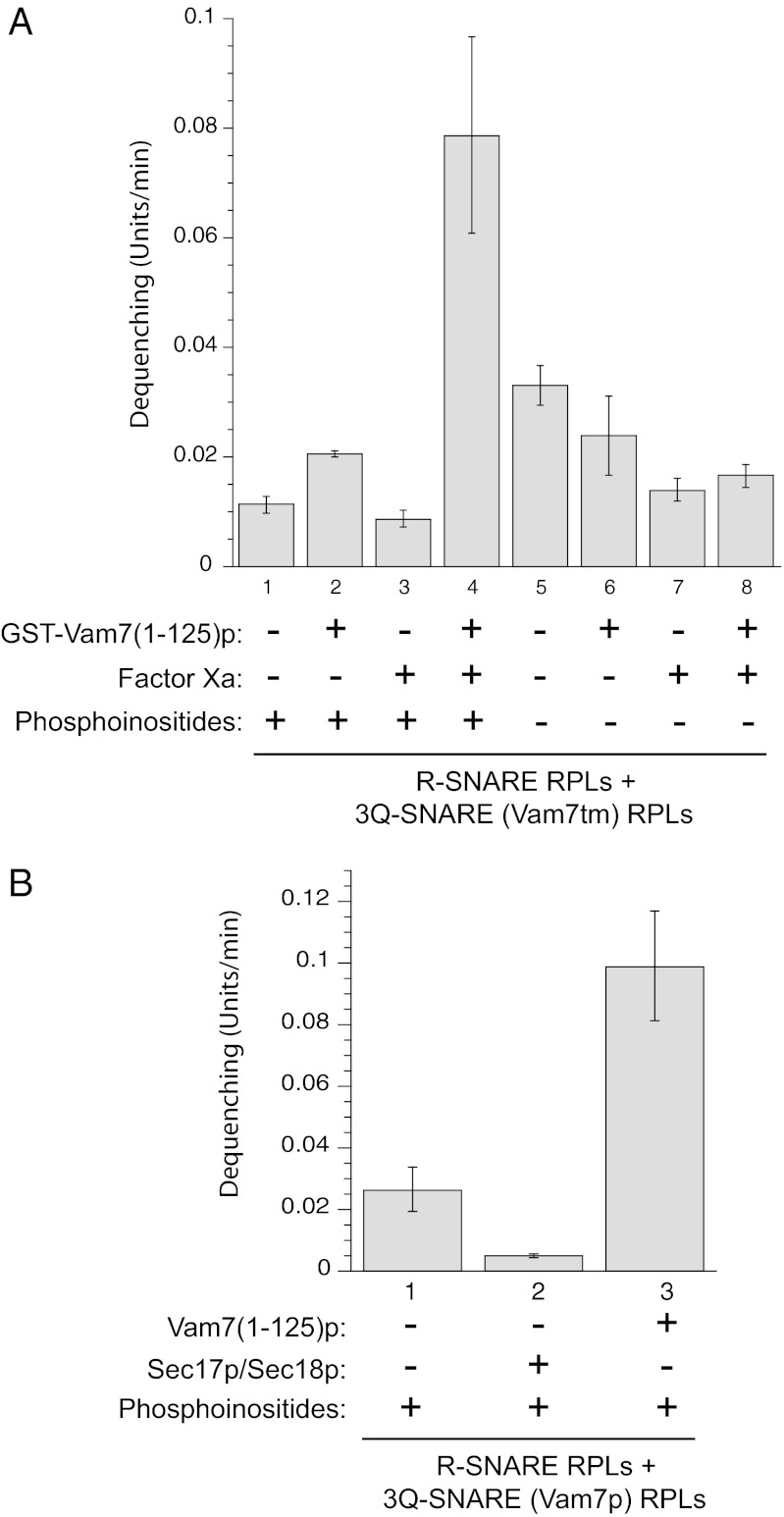
Although the Vam7p N-domain, in its new role, does not depend on covalent connection with the rest of the Vam7p, might it simply restore activity by spatially replacing much of the deleted N-domain? We therefore asked whether Vam7(1-125)p stimulates dequenching when the Vam7-tm in the Q-SNARE RPLs retains its normal N-domain. R-SNARE RPLs were mixed with 3Q-SNARE RPLs bearing Vam7-tm, with or without the addition of recombinant Vam7(1-125)p. As observed with Vam7-tm,∆N, the isolated Vam7 N-domain, in the presence of phosphoinositides, robustly stimulates lipid mixing even when the Vam7p from the Q-SNARE RPLs has its endogenous PX domain (Fig. 4A, lane 4). Stimulation by Vam7(1-125)p is not limited to incubations with Vam7tm, as it is also seen in reactions with WT Vam7p (Fig. 4B). Thus, this function of the Vam7p N-domain does not require it to be in a 1:1 stoichiometry with the Vam7p C-terminal SNARE domain.
Fig. 4.
The Vam7 N-domain stimulates SNARE-dependent dequenching in the presence of full-length Vam7p. (A) Incubations with Vam7tm. R-SNARE RPLs and 3Q-SNARE RPLs bearing Vam7tm were incubated with control buffer addition, 1.1 μM GST-Vam7(1-125)p, Factor Xa (10 μg/mL), or GST-Vam7(1-125)p that had been preincubated with Factor Xa. Reactions 1 to 4 used RPLs that bore phosphoinositides [PI3P and PI(4,5)P2]. (B) Incubations with WT Vam7p. R-SNARE RPLs and 3Q-SNARE RPLs bearing WT Vam7p were incubated with control buffer addition, with Sec17p (100 nM) and Sec18p (83 nM), or with 1.1 μM Vam7(1-125)p that had been preincubated with Factor Xa.
Vam7(1-125)p Promotes SNARE-Dependent Docking.
The fusion of two lipid bilayers is preceded by the apposition of these membranes and the formation of the trans-SNARE complex. To address which fusion subreaction is catalyzed by the Vam7p N-domain, R-SNARE and Q-SNARE RPLs bearing Vam7tm,∆N were incubated at 27 °C for 15 min to measure the kinetics of lipid mixing (Fig. 5A) before the membranes were solubilized with detergent. SNARE complex formation was gauged by the amount of R-SNARE (Nyv1p) that coimmunoprecipitated with the Qa-SNARE Vam3p from membrane extracts (Fig. 5B). Minimal Nyv1p and Vam3p association was detected on Western blotting when R-SNARE and Q-SNARE RPLs were incubated with diC8-PI3P or Vam7p N-domain alone. However, in the presence of both, a clear Nyv1p signal was detected by Western blotting. This Nyv1p-Vam3p association is not simply caused by protein interaction in detergent, because R-SNARE and Q-SNARE RPLs that were incubated separately before detergent solubilization and then mixed do not exhibit Nyv1p coimmunoprecipitation with Vam3p (Fig. 5B, control).
Fig. 5.
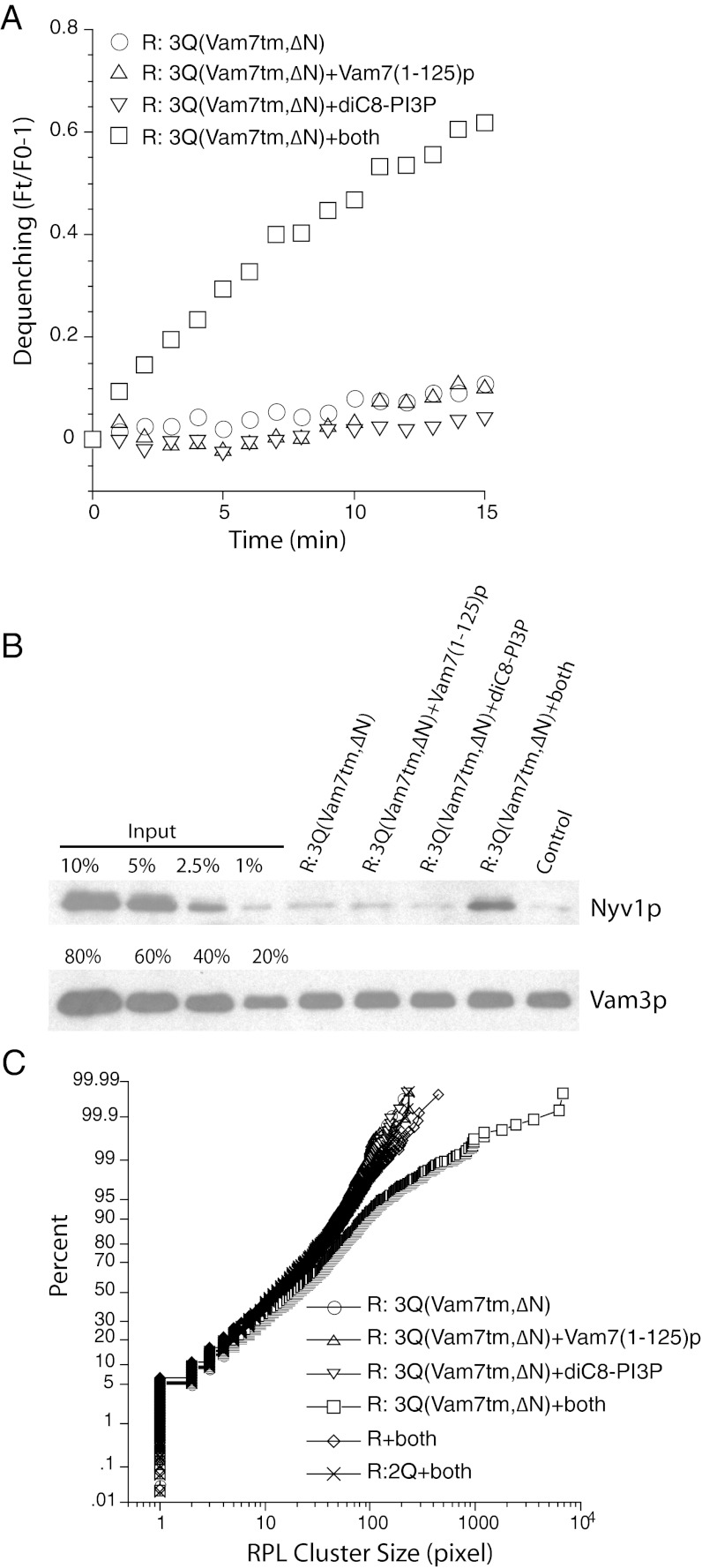
The Vam7 N-domain promotes trans-SNARE complex-dependent RPL docking. R-SNARE RPLs and 3Q-SNARE RPLs bearing Vam7tm,∆N were incubated at 27 °C for 15 min with Vam7(1-125)p from which the GST domain had been cleaved, diC8-PI3P (47 μM), or both. (A) NBD dequenching, expressed as its ratio over the fluorescent signal at t = 0. At the end of the 15-min incubation, samples were transferred to ice, solubilized in RIPA buffer, and subjected to immunoprecipitation using immobilized anti-Vam3p antibody (21). Western blotting of the immunoprecipitated Vam3p and the coprecipitated Nyv1p are shown in B. As control for protein interaction in membrane lysates, R-SNARE RPLs were incubated separately from 3Q-SNARE RPLs [which had Vam7(1-125)p and diC8-PI3P] and mixed immediately after the addition of RIPA buffer. (C) To measure the clustering of R-SNARE RPLs under various reaction conditions, 1 μL from each sample was diluted 40-fold in 20 mM Hepes-KOH, pH 7.5, 150 mM NaCl, and 10% glycerol. An aliquot (4 μL) of the diluted sample were transferred to a microscope plate and covered by a 22-mm2 coverslip. Samples were observed with an Olympus BX51 microscope with a 100-W mercury arc lamp, with images captured by a CCD camera. The size distribution was analyzed by ImageJ and plotted accordingly (26).
In vivo, tethering of membranes by a Rab-family GTPase and its effectors initiates the docking process. Docking is completed by trans-SNARE pairing. In our simplified proteoliposomal reaction, trans-SNARE pairing bypasses the requirement for the Rab GTPase and its effector. To determine whether docking is promoted by Vam7(1-125)p, we assayed the clustering of RPLs by quantitative fluorescence microscopy. After a 15-min incubation, an aliquot of the reaction mixture was subject to fluorescence light microscopy, with images captured by a CCD camera. The size distribution of proteoliposome clusters was analyzed by ImageJ and displayed as a cumulative distribution plot (Fig. 5C). In samples in which both diC8-PI3P and Vam7(1-125)p were incubated with R-SNARE and 3Q-SNARE liposomes that bore Vam7tm,ΔN, large clusters formed (Fig. 5C, open squares). R-SNARE liposomes alone (Fig. 5C, open diamonds) or R-SNARE liposomes incubated with 2Q-SNARE liposomes (only Qc SNARE missing; Fig. 5C, × symbol) do not increase in cluster size in response to diC8-PI3P and Vam7(1-125)p, showing that Vam7(1-125)p does not cluster membranes by itself. Clustering has been shown (11) to result from tethering or docking, with little effect of fusion on cluster size. Thus, the clustering we are seeing, and which requires Vam7(1-125)p and PI3P, only comes about by trans-SNARE complex assembly (assayed directly in Fig. 5B and shown to be functional for dequenching in Fig. 5A). We conclude that Vam7(1-125)p catalyzes the clustering of membranes by promoting the transinteractions between R- and 3Q-SNAREs.
Specificity of PI3P Binding Domains in Fusion Reaction.
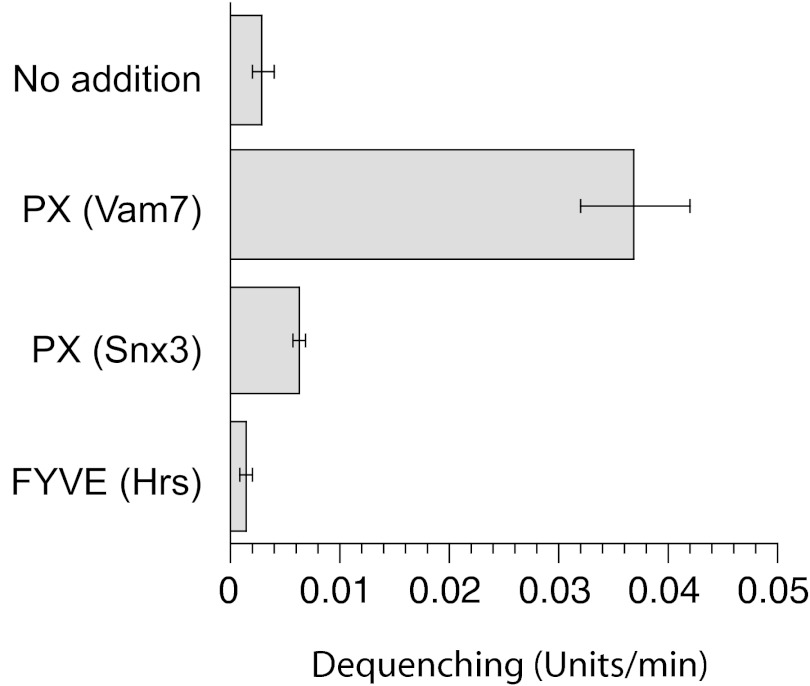
To determine whether PI3P binding modules from other proteins have similar effects on trans-SNARE complex formation, we isolated recombinant SNX3 PX domain and Hrs FYVE domain, each with similar affinity for PI3P as the Vam7p PX domain (28). The Vam7p PX provides robust stimulation (Fig. 6), whereas much less activity was detected for the others. We conclude that binding to PI3P is essential, but insufficient, to facilitate trans-SNARE complex formation.
Fig. 6.
Activities of PI3P binding domains. R-SNARE RPLs and 3Q-SNARE RPLs bearing Vam7tm,∆N were incubated with diC8-PI3P (47 μM) and control buffer or 1.1 μM of PX domain from Vam7p (Vam7(1-125)p), PX domain from SNX3, or FYVE domain from Hrs. The N-terminal GST tag of these PI3P binding domains had been precleaved with thrombin.
Discussion
A hydrophobic tm anchor was fused to the C terminus of Vam7p to bypass the need for the PX region of its N-domain to anchor the SNARE to the membrane. 3Q-SNARE RPLs bearing this recombinant Vam7tm are active in transinteractions with R-SNARE RPLs, as assayed by lipid fluorescence dequenching. Although dequenching is lost when the N-domain is deleted from Vam7tm, this does not reflect an overall denaturation of the protein, as dequenching is restored by addition of HOPS, Sec17p, and Sec18p, or by recombinant Vam7 N-domain. Although trans-SNARE interactions with the anchored Vam7tm do not require phosphoinositides, added Vam7p N-domain needs its membrane receptor PI3P to promote dequenching. The restoration of dequenching by Vam7p N-domain is specific, in that it is far more potent than PX domain from Snx3 or the FYVE domain from Hrs. The definition of just which parts of the Vam7p N-domain are required for this activity will require the preparation of a systematic collection of various regions of Vam7p and their use in the assays we present herein.
Strikingly, the stimulation by recombinant Vam7p N-domain is still observed when the Vam7tm from the 3Q-SNARE RPLs bears a WT N-domain. The function of the added recombinant N-domain is thus not limited to “filling in” where and when it had been deleted. Rather, the WT Vam7p has at least four functions: providing the Qc SNARE domain to four-helical trans-SNARE complexes, binding to PI3P for its own membrane association (22), binding to PI3P in trans as an auxiliary tethering mechanism (26), and as a vehicle to deliver its N-domain to the site of SNARE clustering where it facilitates trans-SNARE complex assembly. In a similar fashion, its has been shown that the N-peptide domain of syntaxin has a required role in Sec1-Munc18 protein recruitment, but can function well when released from syntaxin and moved to its own membrane anchor (29). The Vam7p N-domain, incubated separately with R-SNARE RPLs, does not promote clustering; N-domain promotes clustering only when PI3P and both R- and 3Q-SNARE RPLs are present, suggesting that it functions to directly promote trans-SNARE complex assembly. In accord with this, the direct physical association of the Q- and R-SNAREs is strongly enhanced by added Vam7p N-domain.
Other SNARE N-domain functions have been explored. The Nyv1p N-terminal longin domain is needed for its trafficking to the vacuole (30), although it may have additional functions as well. Several syntaxins have three-coiled coils as N-domains, but only in certain isoforms do they interact directly with the C-terminal SNARE domains (31). The syntaxin N-terminal residues also serve as a receptor for Munc18p (32, 33). On the contrary, the N-domain of Vam3p (34), the syntaxin homologue of yeast vacuoles, is not strictly required for the membrane association of HOPS (which bears the Sec1/Munc18 homologue subunit Vps33p) or for fusion (35). Other Qc SNAREs do not in general have PX, but further studies will be required to determine whether they promote trans-SNARE complex assembly. A more complete understanding of the roles of SNARE N-domains will require determination of the structure of the full SNARE complex with N-domains and systematic exploration of the effects of modifying the several N-domains within each SNARE complex, singly and in combination.
The molecular interactions between the N-domains and SNARE domains during the assembly of the vacuolar trans–4-SNARE complex are not known. Our working model is that the Qc-SNARE Vam7p N-domain binds to HOPS and to PI3P, which allows its SNARE domain to intertwine with those of the other Q-SNAREs, Vam3p and Vti1p, to assemble the 3Q-SNARE complex. Our current studies reveal an additional function of the N-domain of Vam7p, which is to catalyze the association in trans between the R-SNARE Nyv1p and the assembled 3Q-SNARE subcomplex. This function of the Vam7p N-domain is independent of HOPS, although HOPS may also continue to promote trans-SNARE complex assembly.
Materials and Methods
Plasmids.
cDNAs encoding Vam7tm and Vam7tm,ΔN were generated by PCR by using pET41H-Vam7 (36) and primers 1 and 3 and primers 2 and 3, respectively. The cDNAs were digested with NcoRI and EcoRI and ligated into the NcoRI/EcoRI sites of pMBD-Parallel1 (37), generating pMBP-TCS (TEV cleavage site)-Vam7tm and pMBP-TCS-Vam7tm,ΔN. The nucleotide sequence underlined in primer-3 encodes the entire tm domain of Vti1p. Both constructs were verified by DNA sequencing.
Primer 1: 5′-tgccATGGCAGCTAATTCTGTAGGG.
Primer 2: 5′-tgccatgGACCCAAGCACGGAAGTGACTC.
Primer 3: 5′-cggaattcTTATTTAAACTTTGAGAACAAAACTAGCAAAATCAATAATATAAGGACTGCGATAATGGCATAGCTTATGAAAGCACTGTTGTTAAAATGTC.
Proteins.
To purify Vam7tm or Vam7tm,ΔN, E. coli Rossetta2(DE3)pLys (Novagen) transformed with the respective plasmid was inoculated into 100 mL Luria-Bertani medium containing 100 μg/mL ampicillin and 25 μg/mL chloramphenicol. Following overnight growth at 37 °C, the culture was added to 1 L of Terrific Broth medium containing 100 μg/mL ampicillin and 25 μg/mL chloramphenicol, and shaken at 37 °C until an OD600 of 1.5 was achieved. Isopropyl β-d-1-thiogalactopyranoside was added to 1 mM. After 4 h growth at 37 °C, bacteria were harvested by centrifugation (3,000 × g, 5 min, room temperature; JLA 10.500 rotor in a Beckman centrifuge). The pellet was resuspended in 20 mL of 100 mM Hepes-KOH, pH 7.5, 0.5 M KCl, 10% (vol/vol) glycerol, 1 mM DTT, 0.5 mM benzamidine Cl, 1 mM PMSF, 0.62 μg/mL leupeptin, 4 μg/mL pepstatin A, and 24.4 μg/mL Pefabloc-SC, and subjected to three passes through a French press at 900 psi. One-tenth volume of 1 M n-octyl-β-d-glucoside was added and incubation was continued at 4 °C for 1 h with nutation. The detergent lysate was centrifuged at 177,000 × g for 1 h in a Beckman 60Ti rotor. The supernatant was added to 4 mL of amylose resin (NEB) preequilibrated with wash buffer (50 mM Hepes-HCl, pH 7.5, 0.3 M KCl, 10% glycerol, and 0.1 M n-octyl-β-d-glucoside). Following 1 h nutation at 4 °C, the resin was packed into a column in the cold room, washed with 40 mL of wash buffer, and eluted with 10 mM maltose in wash buffer. The N-terminal MBP was cleaved from these proteins during RPL formation by the TEV protease in the reconstitution mixture (25).
GST-Vam7(1-125)p (38), GST-Vam7p (1-125, Y42A) (38), GST-PX(SNX3)p (28), and GST-FYVE(Hrs) (28) were produced in Rossetta 2 strains, purified according to Boeddinghaus et al. (38), and dialyzed in 20 mM Pipes-KOH, pH 6.4, 125 mM KCl, 5 mM MgCl2. When specified, the GST tag was cleaved with Faxtor Xa or thrombin (Haematologic Technologies); GST-tagged recombinant proteins (5.5 μM) were incubated in 40 μL of 10 mM Pipes⋅KOH, pH 6.4, 62.5 mM KCl, 2.5 mM MgCl2 with 0.05 mg/mL Factor Xa or 0.3 mg/mL thrombin. After 1 h at 27 °C, 5 mM PMSF was added to inactivated thrombin. All other proteins and antibodies have been described previously (26).
RPLs and the Dequenching Assay.
R-SNARE and 3Q-SNARE proteoliposomes with palmitoyl, oleoyl (PO) phosphatidylcholine (POPC; 44% or 46% for donor or acceptor), PO phosphatidylethanolamine (POPE; 18%), soy phosphatidylinositol (PI; 18%), PO phosphatidylserine (POPS; 4.4%), PO phosphatidic acid (POPA; 2%), cardiolipin (CL; 1.6%), ergosterol (ERG; 8.0%), diacylglycerol (DAG; 1.0%), and fluorescent lipids (1.5% NBD-PE and 1.5% Rh-PE for donor; 1.0% dansyl-PE for acceptor) were prepared as described previously (25). Where specified, PI3P (1%) and PI(4,5)P2 (1%) were added, with corresponding reduction in the amount of POPC.
Dequenching assays with R-SNARE RPLs (50 μM lipids) and Q-SNARE RPLs (400 μM) were performed in 20 μL mixtures as described previously (21), but with 1 mM ATP and 1 mM MgCl2. Recombinant Vam7(1-125), Vam7(1-125, Y42A), PX(SNX3), or FYVE(Hrs) (1.1 μM) were added when specified. The maximal, early rate of dequenching was calculated as the increased fluorescence at any time divided by the fluorescence at the first minute: [(Ft − F0) / F0]. An increase of 1 in this parameter is defined as one unit. Unless otherwise specified, mean values of the maximal rate of dequenching are presented and error bars represent SDs from three repeat experiments.
Acknowledgments
We thank Amy Orr, Holly Jakubowski, and Deborah Douville for excellent technical support and Dr. Mark Lemmon (University of Pennsylvania) for sending the plasmids encoding recombinant GST-PX(SNX3) and GST-FYVE(Hrs). This work was supported by National Institutes of Health Grant GM23377.
Footnotes
The authors declare no conflict of interest.
References
- 1.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15(7):658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizo J, Südhof TC. Snares and Munc18 in synaptic vesicle fusion. Nat Rev Neurosci. 2002;3(8):641–653. doi: 10.1038/nrn898. [DOI] [PubMed] [Google Scholar]
- 4.Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 5.Wickner W. Membrane fusion: 5 lipids, 4 SNAREs, 3 chaperones, 2 nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 6.Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267(26):18665–18670. [PubMed] [Google Scholar]
- 7.Haas A, Conradt B, Wickner W. G-protein ligands inhibit in vitro reactions of vacuole inheritance. J Cell Biol. 1994;126(1):87–97. doi: 10.1083/jcb.126.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136(2):307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97(17):9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroupe C, Hickey CM, Mima J, Burfeind AS, Wickner W. Minimal membrane docking requirements revealed by reconstitution of Rab GTPase-dependent membrane fusion from purified components. Proc Natl Acad Sci USA. 2009;106(42):17626–17633. doi: 10.1073/pnas.0903801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21(13):2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Seeley ES, Wickner W, Merz AJ. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108(3):357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Merz AJ, Collins KM, Wickner W. Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J Cell Biol. 2003;160(3):365–374. doi: 10.1083/jcb.200209095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167(6):1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95(26):15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409(6822):839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 17.Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85(1):83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 18.Nichols BJ, Ungermann C, Pelham HRB, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387(6629):199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- 19.Ungermann C, Nichols BJ, Pelham HRB, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140(1):61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungermann C, Sato K, Wickner W. Defining the functions of trans-SNARE pairs. Nature. 1998;396(6711):543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29(12):1948–1960. doi: 10.1038/emboj.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheever ML, et al. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3(7):613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 23.Stroupe C, Collins KM, Fratti RA, Wickner WT. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25(8):1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda R, et al. Functional architecture of an intracellular membrane t-SNARE. Nature. 2000;407:198–202. doi: 10.1038/35025084. [DOI] [PubMed] [Google Scholar]
- 25.Mima J, Hickey CM, Xu H, Jun Y, Wickner W. Reconstituted membrane fusion requires regulatory lipids, SNAREs and synergistic SNARE chaperones. EMBO J. 2008;27(15):2031–2042. doi: 10.1038/emboj.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu H, Wickner W. Phosphoinositides function asymmetrically for membrane fusion, promoting tethering and 3Q-SNARE subcomplex assembly. J Biol Chem. 2010;285(50):39359–39365. doi: 10.1074/jbc.M110.183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mima J, Wickner W. Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc Natl Acad Sci USA. 2009;106(38):16191–16196. doi: 10.1073/pnas.0908694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu JW, Lemmon MA. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276(47):44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 29.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107(52):22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen W, et al. Identification of the yeast R-SNARE Nyv1p as a novel longin domain-containing protein. Mol Biol Cell. 2006;17(10):4282–4299. doi: 10.1091/mbc.E06-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonin W, et al. The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J Biol Chem. 2002;277:36449–36456. doi: 10.1074/jbc.M204369200. [DOI] [PubMed] [Google Scholar]
- 32.Dulubova I, et al. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21(14):3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi T, et al. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2(3):295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- 34.Dulubova I, Yamaguchi T, Wang Y, Südhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8(3):258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- 35.Laage R, Ungermann C. The N-terminal domain of the t-SNARE Vam3p coordinates priming and docking in yeast vacuole fusion. Mol Biol Cell. 2001;12(11):3375–3385. doi: 10.1091/mbc.12.11.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz ML, Merz AJ. Capture and release of partially zipped trans-SNARE complexes on intact organelles. J Cell Biol. 2009;185(3):535–549. doi: 10.1083/jcb.200811082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15(1):34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 38.Boeddinghaus C, Merz AJ, Laage R, Ungermann C. A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J Cell Biol. 2002;157(1):79–89. doi: 10.1083/jcb.200112098. [DOI] [PMC free article] [PubMed] [Google Scholar]