Abstract
Neural stem cells (NSCs) are considered to be the cell of origin of glioblastoma multiforme (GBM). However, the genetic alterations that transform NSCs into glioma-initiating cells remain elusive. Using a unique transposon mutagenesis strategy that mutagenizes NSCs in culture, followed by additional rounds of mutagenesis to generate tumors in vivo, we have identified genes and signaling pathways that can transform NSCs into glioma-initiating cells. Mobilization of Sleeping Beauty transposons in NSCs induced the immortalization of astroglial-like cells, which were then able to generate tumors with characteristics of the mesenchymal subtype of GBM on transplantation, consistent with a potential astroglial origin for mesenchymal GBM. Sequence analysis of transposon insertion sites from tumors and immortalized cells identified more than 200 frequently mutated genes, including human GBM-associated genes, such as Met and Nf1, and made it possible to discriminate between genes that function during astroglial immortalization vs. later stages of tumor development. We also functionally validated five GBM candidate genes using a previously undescribed high-throughput method. Finally, we show that even clonally related tumors derived from the same immortalized line have acquired distinct combinations of genetic alterations during tumor development, suggesting that tumor formation in this model system involves competition among genetically variant cells, which is similar to the Darwinian evolutionary processes now thought to generate many human cancers. This mutagenesis strategy is faster and simpler than conventional transposon screens and can potentially be applied to any tissue stem/progenitor cells that can be grown and differentiated in vitro.
Glioblastoma multiforme (GBM) is the most common form of malignant brain cancer in adults. Patients with GBM have a uniformly poor prognosis, with a mean survival of 1 y (1). Thus, advances on all fronts, both basic and applied, are needed to combat this deadly disease better. Recent studies have provided evidence for self-renewing, stem-like cells within human gliomas (2). These glioma-initiating cells constitute a small minority of neoplastic cells within a tumor and are defined operationally by their ability to seed new tumors (3). To target these rare glioma-initiating cells, a better understanding of the molecular mechanisms that regulate their formation is essential.
Considerable progress has been made in understanding the mutations responsible for GBM. The Cancer Genome Atlas network has cataloged the recurrent genomic abnormalities in GBM by genome-wide DNA copy number events and sequence-based mutation detection for 601 genes (4). Gene expression-based molecular classification has also defined four subtypes of GBM termed proneural, neural, classical, and mesenchymal (5). Proneural GBM is enriched for the oligodendrocyte gene signature, whereas the classical group is associated with the astrocytic signature. The neural class is enriched for genes differentially expressed by neurons, whereas the mesenchymal class is associated with the cultured astroglial signature (5). Several recurrent mutations, such as PDGFRA, IDH1, EGFR, and NF1, also correlate with these GBM subtypes, providing additional support for their existence. Numerous other, often rare, mutations have also been identified in GBM. Although these datasets are valuable for understanding the molecular pathogenesis of GBM, it is still difficult to distinguish between mutations that contributed to tumor initiation and those acquired later during tumor progression.
The cell of origin (COO) of GBM is still controversial. Neural stem cells (NSCs) are good candidates because the adult brain has very few proliferating cells capable of accumulating the numerous mutations required for gliomagenesis. NSCs are also more susceptible to malignant transformation than differentiated cells in the adult brain (6, 7). However, the genetic pathways that can transform NSCs into glioma-initiating cells still remain elusive. Transposon-based mutagenesis provides an unbiased, high-throughput method for identifying genes important for GBM (8). Here, we describe a unique two-step insertional mutagenesis strategy that makes it possible to identify genes and signaling pathways that are able to transform a NSC into a cancer-initiating cell for the mesenchymal subtype of GBM. In this two-step approach, NSCs are first mutagenized in vitro and the mutagenized cells are then transplanted into immunocompromised mice for subsequent tumor development following additional rounds of transposon-based mutagenesis. This makes it possible to discriminate between the genetic changes that occur early in tumor initiation and those required for tumor progression. In addition to identifying several previously undescribed GBM candidate cancer genes, our studies suggest that transposon-induced tumors mimic the evolutionary processes now thought to generate many human cancers, in which tumors have a branched cellular and genetic architecture reminiscent of Darwin’s iconic evolutionary tree.
Results
Transposon Mutagenesis of NSCs Promotes the Immortalization of Astroglial-Like Cells.
Mice homozygous for a conditional floxed-stop Sleeping Beauty (SB) transposase allele knocked in to the ubiquitously expressed Rosa26 locus (Rosa26-LSL-SB11) (9) and up to 350 copies of a mutagenic SB transposon (T2/Onc2 or T2/Onc3) all linked together at a single site in the genome (termed the donor transposon concatamer) (9, 10) were crossed to Nestin-Cre (Nes-cre) transgenic mice. Nes-cre is expressed in NSCs, and will thus activate SB transposase, and hence SB transposition, specifically in the NSC compartment (11). Brains were collected from embryos of various different genotypes between embryonic day 17 (E17) and postnatal day 1 (P1) of development (Fig. 1A). At this stage, the subventricular zone (SVZ) is a rich source of NSCs (12). By culturing SVZ cells with EGF, multipotent self-renewing NSCs could be propagated as clonal aggregates, denoted as neurospheres (13) (Fig. 1B). As expected, excision PCR (14) showed that transposons were being mobilized in these neurospheres (Fig. 1C). The NSCs were then induced to differentiate by removal of EGF and the addition of FBS, and were then cultured for 2 wk (Fig. 1A), which led to the appearance of cells that stained positive for neuronal, astrocytic, and oligodendrocytic markers (Fig. 1 D and E). The cells were then reseeded every week, counted, and plotted against time (Fig. 1F). During early passages, total cell numbers continuously decreased; however, after several passages, exponentially growing populations of cells were obtained. The cells were considered immortalized once the cumulative total cell number exceeded 108. A conditional dominant-negative mutant form of p53 (LSL-p53R172H) (15) was also included in many experiments as a sensitizing mutation because p53 is one of the most frequently mutated genes in GBM (4).
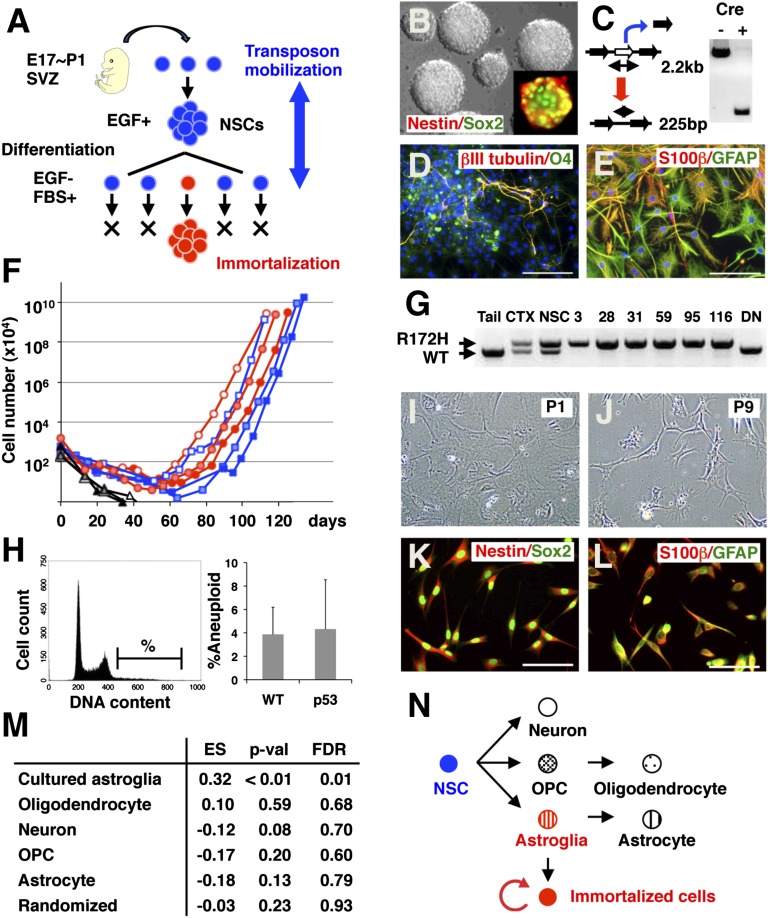
Fig. 1.
Transposon mutagenesis in NSCs promotes the immortalization of astroglial-like cells. (A) Mutagenesis strategy. NSCs isolated from the SVZ of mice with active SB transposition were expanded in vitro and induced to differentiate to select for immortalized cells. (B) Nestin+ and Sox2+ NSCs were expanded as neurospheres. (Magnification: 400×.) (C) Transposon excision from the transposon concatamer could be detected in neurospheres containing a nestin-cre allele by the presence of a 225-bp fragment. NSCs that were induced to differentiate stained positive for neuronal (βIII tubulin) and oligodendrocytic (O4) markers (D), as well as for astrocytic (GFAP and S-100β) markers (E). The nucleus is counterstained with DAPI. (Scale bars: 100 μm.) (F) Growth during serial passage of three Nes-cre/+; T2/Onc2,3/+; SBase/+ (red) and three p53R172H/+; Nes-cre/+; T2/Onc2,3/+; SBase/+ (blue) lines. Three additional lines in which the cells entered senescence are also shown (black). (G) PCR-based detection of WT and mutant p53 alleles. In nestin-cre transgenic animals, expression of the p53R172H mutant allele is induced in the cortex (CTX) and neurospheres (NSC) but not in the tail. The p53 WT allele is not expressed in six immortalized lines harboring a p53R172H mutant allele but is expressed in a cell line overexpressing p53DN (DN). (H) Fractions of cells with increased DNA content (%) were compared between p53 mutant (n = 5) and WT (n = 3) immortalized lines (Fig. S1A). Differentiating cells exhibit flat and polygonal morphology at passage 1 (I) but become spindle-shaped when immortalized at passage 9 (J). Immortalized lines expressed NSC markers (K; nestin and Sox2) and astrocyte markers (L; GFAP and S-100β). (Scale bars: 100 μm.) (M) GSEA enrichment score (ES) was calculated for 10 immortalized lines against control samples using five gene sets specific for neuron, oligodendrocyte, OPC, astrocyte, and cultured astroglia (19). Randomly selected gene sets were used as a control. The P value (p-val) and false discovery rate (FDR) are also shown for each gene set. (N) Schematic diagram of differentiation of NSCs into neuronal, oligodendrocytic, and astrocytic lineages.
The frequency of immortalization was higher in WT cells undergoing active SB transposition than in WT cells alone (Table 1; 19% vs. 2%; P < 0.05, Fisher’s exact test), indicating that transposon mobilization promotes immortalization. No difference in the immortalization frequency was observed between cells carrying T2/Onc2 or T2/Onc3; therefore, the data for these two transposons were grouped together. The frequency of immortalization was higher in p53R172H/+ mutant NSCs, and, once again, the immortalization frequency was higher in p53R172H/+ mutant NSCs undergoing active SB transposition (Table 1; 27% vs. 56%; P < 0.05, Fisher’s exact test). The R172H mutation affects the global structure of the p53 DNA binding domain. By oligomerizing with WT p53, p53R172H acts as a dominant-negative protein (16). Consistent with this, overexpression of a dominant-negative mutant p53 allele lacking the DNA binding domain (p53DN) (17) introduced into NSCs by retroviral transduction also promoted immortalization in cooperation with SB transposition (Table 1; 0% vs. 100%). Tumors that develop in humans with hereditary p53 mutations frequently lose the remaining WT allele. PCR analysis of DNA from p53R172H/+ immortalized lines showed that they have also lost the WT p53 allele (Fig. 1G), indicating that allele loss confers a growth advantage to these cells. Immortalized cells were also subjected to DNA analysis by flow cytometry to monitor for changes in DNA stability. Fractions of cells with increased DNA content were comparable between WT and p53 immortalized lines (Fig. 1H and Fig. S1A), indicating that the p53R172H mutation does not induce pronounced aneuploidy in immortalized cells, consistent with the findings of a previous study (18).
Table 1.
Immortalized cell lines and tumors used for insertion site analysis
| Genotype | Immortalization* | Tumor formation† | Cell samples‡ | Tumor samples§ |
| T2/Onc2,3/+; SBase/+ | 1 of 46 (2%) | 0 of 1 (0%) | ||
| Nes-cre/+; T2/Onc2,3/+; SBase/+ | 9 of 48 (19%) | 6 of 9 (67%) | 6 | 20 |
| p53R172H/+; Nes-cre/+ | 6 of 22 (27%) | 1 of 6 (17%) | ||
| p53R172H/+; Nes-cre/+; T2/Onc2,3/+; SBase/+ | 10 of 18 (56%) | 7 of 10 (70%) | 18 | 43 |
| p53DN¶ and T2/Onc2/+; SBase/+ | 0 of 4 (0%) | |||
| p53DN¶ and Nes-cre/+; T2/Onc2/+; SBase/+ | 2 of 2 (100%) | 2 of 2 (100%) | 1 | 4 |
| Total | 25 | 67 |
SBase, Rosa26-LSL-SB11; T2/Onc2,3, mutagenic SB transposons (Dataset S1).
*Number of immortalized cell lines/cultures.
†Number of tumorigenic cell lines/immortalized cell lines. Tumors were induced by s.c. injection.
‡Number of cell samples used for insertion analysis.
§Number of tumor samples used for insertion analysis.
¶p53DN was retrovirally transduced.
During early passages after the induction of differentiation, the cells showed a flat and polygonal morphology (Fig. 1I). However, after several passages, the growing cells showed an elongated morphology (Fig. 1J). Immortalized cells expressed NSC markers as well as astrocyte markers (Fig. 1 K and L) but were negative for neuronal (βIII tubulin) and oligodendrocytic (O4) markers (the same pattern was observed for 3 WT and p53 mutant immortalized lines). These data suggest that the immortalized cells are committed to the astrocyte lineage. To confirm these results, we used DNA microarrays to quantitate the levels of gene expression in 10 immortalized lines (4 WT and 6 p53 mutant lines). Hierarchical clustering showed a close similarity in gene expression across the 10 cell lines (Fig. S1B). We then compared their gene expression profiles with those present in the brain transcriptome database (19). Using five gene sets that are associated with neurons, oligodendrocytes, oligodendrocyte precursor cells (OPCs), astrocytes, and cultured astroglial cells, Gene Set Enrichment Analysis (GSEA) enrichment scores were calculated for the 10 lines against two control samples (NSCs) (Fig. S1 C–H). We found a strong enrichment for genes that are differentially expressed in cultured astroglial cells (Fig. 1M). Cultured astroglial cells can be generated from the neonatal cortex but not from the adult cortex, and they are considered to represent an immature stage of the astrocyte lineage (20). These data indicate that transposon mutagenesis in NSCs leads to the immortalization of a cultured astroglial-like cell (Fig. 1N).
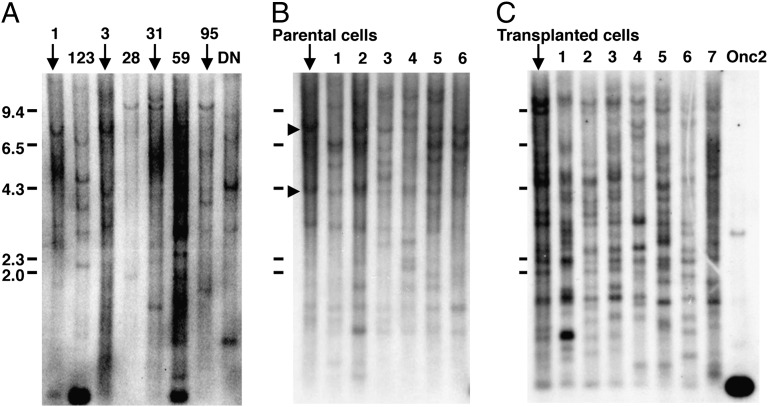
Southern blot analysis of genomic DNA isolated from eight immortalized lines showed that each line carries multiple clonal insertions (Fig. 2A). Southern blot analysis of six single-cell subcloned lines derived from one immortalized line also revealed the presence of clonal insertions that were conserved among all subcloned lines (arrowheads in Fig. 2B), in addition to insertions that were unique to each subcloned line. The presence of these unique insertions confirms that transposons are being continuously mobilized in immortalized cells, as expected, generating a large repertoire of immortalized cells with unique combinations of insertions over time (see Fig. 5K).
Fig. 2.
Southern blot analysis of transposon insertion sites in immortalized lines and tumors. (A) Genomic DNA isolated from eight immortalized lines was digested with BamHI and hybridized with a probe specific for the transposon. Each cell line shows a distinct pattern of transposon insertions. DN, a cell line overexpressing p53DN. (B) Southern blot analysis of the transposon insertion sites in six subcloned lines derived from line 3. (C) Seven tumors derived from cell line 31 all show unique patterns of insertions that are distinct from the transplanted cells. A strong signal is observed for the transposon concatamer (Onc2) in DNA isolated from mice that harbor the transposon concatamer but lack active SB transposase.
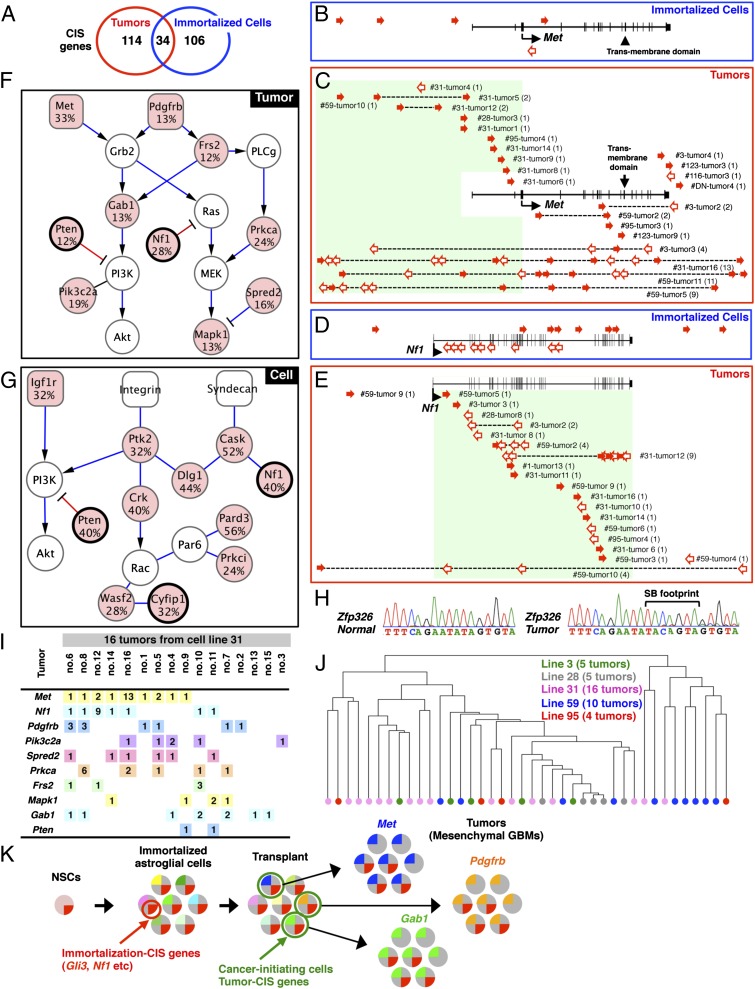
Fig. 5.
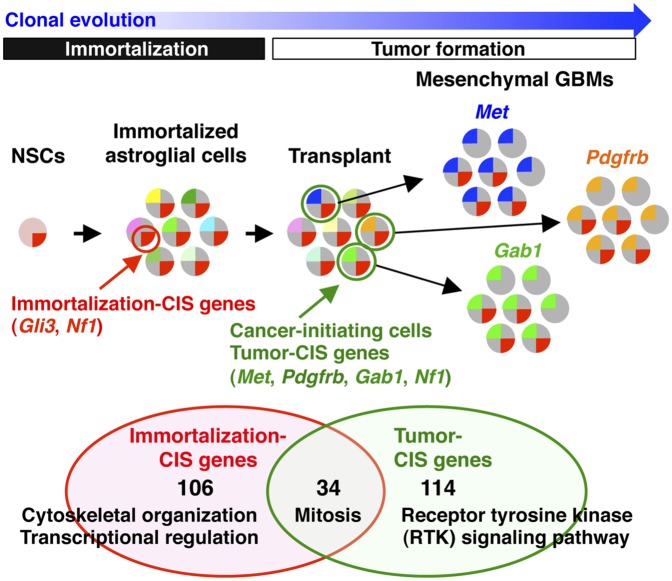
Genes and signaling pathways mutated in tumors and immortalized lines. (A) Overlap between genes mutated in tumors and immortalized lines. Five Met insertions were identified in 5 immortalized lines (B), whereas 60 Met insertions were identified in 22 tumors (C). Transposon insertions are located in the sense (solid red arrows) or antisense (open red arrows) orientation relative to Met transcription. (C) Patterns of Met insertions are shown for individual tumor samples. The number of insertions is indicated in parentheses for each tumor. Eighteen Nf1 insertions were identified in 10 immortalized lines (D), whereas 35 Nf1 insertions were identified in 19 tumors (E). Gene networks generated by IPA analysis are combined and illustrated for 10 tumor CIS genes (F) and 11 CIS genes from immortalized cells (G). Mutation frequencies are shown for each CIS gene (red). CIS genes found in both tumors and cell lines are thick-framed. (H) SB footprint was detected at the Zfp326 locus in a tumor derived from an immortalized line with the Zfp326 insertion (line 95) but not in a normal allele. (I) Combinations of RTK pathway genes were examined for tumors derived from line 31. The number of insertions in each tumor is shown in boxes. (J) Hierarchical clustering based on the Hamming distance was performed for 40 tumors derived from 5 cell lines with the same genotype (p53R172H/+; Nes-cre/+; T2/Onc2/+; SBase/+). The similarity of patterns of CIS genes among different samples is represented by vertical distance. (K) Model for transposon-induced tumor development. Immortalization CIS genes (e.g., Gli3, Nf1) promote immortalization of NSCs. Immortalized astroglial-like cells have a large repertoire of insertions represented by different colors. Transposons are continuously jumping after transplantation, and transplanted cells become cancer-initiating cells (green) by randomly acquiring new insertions in tumor CIS genes (e.g., Met, Pdgfrb, Gab1) and clonally expand to form tumors. Several immortalization CIS genes are lost during tumor formation by continuous mobilization of transposons.
Immortalized Cells Induce Tumors Resembling Mesenchymal GBM.
Immortalized astroglial-like cells with continuously mobilizing transposons provide a unique opportunity to identify genes that play essential roles in GBM. To determine whether immortalized cells are tumorigenic in transplanted hosts, the cells were injected s.c. into the flanks of SCID mice. We primarily used s.c. injection rather than intracranial injection because a previous study showed that the tumorigenic potential and tumor phenotypes of mouse gliomas were the same when induced by intracranial or s.c. injections (21) and because the extraction of genomic DNA was made easier due to clear tumor boundaries. Immortalized cells that lacked active SB transposition were mostly nontumorigenic in transplanted hosts, although more than half of the immortalized lines with active SB transposition induced tumors (Table 1). The average age of tumor onset was ∼2 mo (average of 66 d for 67 tumors; Table 1). In contrast, dissociated primary tumor cells induced tumors in secondary recipients within 3 wk (average of 21 d for 4 tumors), providing evidence for the selection of additional cancer-causing mutations during primary tumor formation.
Southern blot analysis showed that each tumor generated from a single immortalized line harbored a unique collection of insertions that differed from all other tumors generated from the same line as well as the parental line (Fig. 2C). This occurs because transposons present in immortalized cells that are not required for tumor formation can be lost by remobilization because there is no selective pressure to maintain them. Many new insertions will also be acquired during tumor formation; some will represent passenger insertions, whereas others will mark the location of genes important for tumor development.
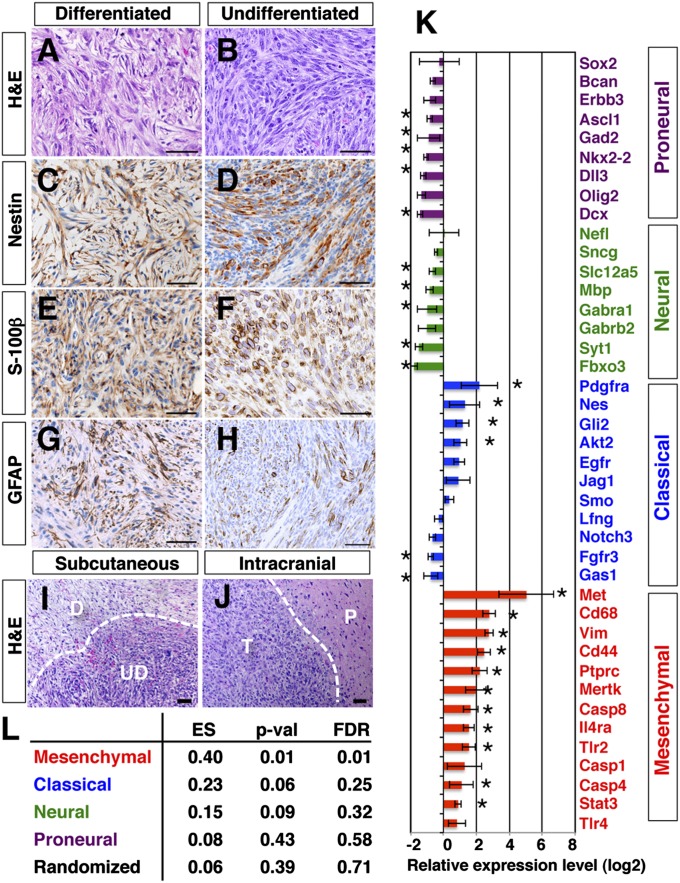
H&E staining of tumors revealed two morphologically distinct lesions: well-differentiated and poorly differentiated (Fig. 3 A and B). Differentiated lesions showed features of benign tumors, such as differentiated cellular morphology with more abundant tumor cytoplasm, whereas undifferentiated lesions showed histological features of high-grade tumors with hypercellularity, poor differentiation, and high mitotic activity. WT immortalized lines induced both differentiated and undifferentiated lesions in the same tumor (Fig. 3I), whereas p53 mutant lines only induced undifferentiated lesions. This tumor phenotype is likely to be intrinsic to the immortalized cells, because intracranial injection of these same cells induced tumors with similar morphologies (Fig. 3J, undifferentiated lesion). Tumors expressed classic glioma markers, such as nestin (Fig. 3 C and D; 13 of 13 cell lines), S-100β (Fig. 3 E and F; 11 of 13 cell lines), and GFAP (Fig. 3 G and H; 4 of 13 cell lines). These tumors were negative for markers typical for primitive neuroectodermal tumors (PNETs), such as βIII tubulin and synaptophysin (Fig. S2 A–D), ruling out the possibility of PNETs.
Fig. 3.
Tumors have characteristics of the mesenchymal subtype of GBM. Differentiated and undifferentiated lesions are stained with H&E (A and B) or immunostained with nestin (C and D), S-100β (E and F), or GFAP (G and H). (Scale bars: 50 μm.) (I) Differentiated lesion (D) locates adjacent to an undifferentiated lesion (UD). (J) Intracranial injection of tumorigenic cells generated a tumor (T) in brain parenchyma (P). (Scale bars: 50 μm.) (K) Expression levels of genes specific to the four subtypes of GBM (5) were examined for 12 tumors and compared with normal s.c. tissues. The relative gene expression level (log2 value of fold change) is shown for each gene. The asterisks indicate a difference from control (*P < 0.05, Student t test). (L) GSEA enrichment score (ES) was calculated for 12 tumors against control samples using the four gene sets specific to mesenchymal, classical, neuronal, and proneural subtypes of GBM (5). Randomly selected gene sets were used as a control. The P value (p-val) and false discovery rate (FDR) are also shown for each gene set.
To classify these tumors further, we used DNA microarrays to quantitate the expression levels of markers specific for each GBM subtype (5) in 6 WT and 6 p53 mutant tumors (Fig. 3K). Markers for proneural and neural GBM subtypes were expressed at low levels in these tumors. Some markers for the classical subtype were expressed, but most of the markers for the mesenchymal subtype were strongly expressed, indicating that the tumors have gene expression profiles most similar to mesenchymal GBM. We also performed GSEA analysis on the tumors. Hierarchical clustering of 12 tumors showed a close similarity among the tumors compared with control s.c. tissues (Fig. S2E). Using gene sets specific to the four defined GBM subtypes (5), a GSEA enrichment score was calculated for the 12 tumor samples against 3 control samples (Fig. S2 F–J). This analysis showed that the tumors were significantly enriched for genes specific for mesenchymal GBM (Fig. 3L), consistent with previous studies showing that this subclass of GBM is strongly associated with the cultured astroglial signature (5).
Analysis of Transposon Insertion Sites in Tumors and Immortalized Lines.
To identify genes responsible for immortalization and tumor development in transplanted mice, we PCR-amplified and -sequenced the transposon insertion sites from 25 immortalized lines and 67 tumors (Table 1 and Dataset S1). In total, 30,315 and 36,770 unique, nonduplicated insertions were sequenced from immortalized cells and tumors, respectively. Insertions were then analyzed using the Gaussian kernel convolution method (22) to identify common insertion sites (CISs). CISs are regions in the genome that harbor a higher number of transposon insertions than predicted by random chance, and therefore are most likely to harbor a driver gene for cancer. We also used Monte Carlo simulations to identify CISs as described previously (22). These CISs were then annotated to the nearest gene, and the lists were combined.
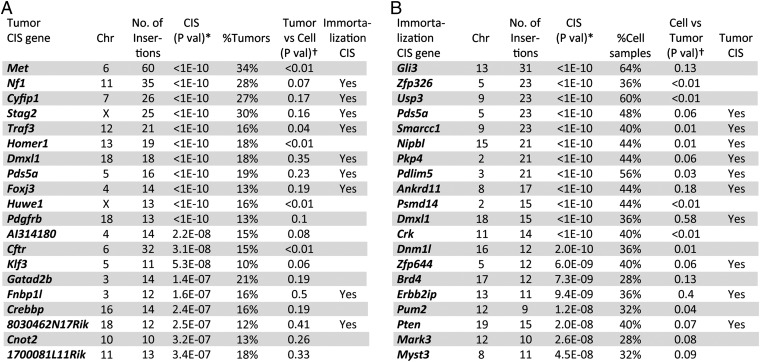
As a control, 5,699 insertions were sequenced from 12 histologically normal brain cortices obtained from 12 mice at 1 mo of age (Nes-cre/+; T2/Onc2,3/+; SBase/+). From this dataset, we identified 11 CIS genes (Dataset S2). Only 1 CIS gene identified in control brains, Sfi1, was identified as a CIS gene in immortalized cells and tumors, and it was removed from subsequent analysis. In the end, 140 CIS genes were identified in immortalized cells and 148 were identified from tumors (Datasets S3 and S4). The 20 most significantly mutated genes in tumors and immortalized cells are shown in Fig. 4 A and B, respectively. Thirty-four genes were CIS genes in both immortalized cells and tumors, whereas the rest were specific for either immortalized cells or tumors (Fig. 5A).
Fig. 4.
Top 20 CIS genes for tumors and immortalized lines. (A) Top 20 CIS genes for tumors (Dataset S3). (B) Top 20 CIS genes for immortalized lines (Dataset S4). Chr, chromosome. *P value (P-val) was determined as previously described (21). †Numbers of insertions at each CIS were compared between tumors and immortalized cells using Fisher’s exact test.
Identification of Driver Genes for Tumor Development.
The most frequently mutated gene in tumors was Met (Fig. 4A). Few Met insertions were identified in immortalized lines (Fig. 5B), however, indicating that Met insertions are only positively selected during tumor development (P < 0.01). Nearly half of the insertions are located upstream of Met or within intron 1, in the sense orientation (Fig. 5C), consistent with its known function as an oncogene. Flow cytometric analysis showed the presence of a Met-expressing subpopulation in a tumor with a Met insertion but not in a tumor without it (Fig. S3G). Sequence analysis of RNA from three such tumors showed that the murine stem cell virus (MSCV) promoter in the transposon was spliced to Met exon 2 in all cases, resulting in the overexpression of Met transcripts that retain the initiating ATG codon located in exon 3 (Fig. S3H). Several other genes were also identified that were highly mutated only in tumors, such as Homer1, Huwe1, and Cftr (P < 0.01; Fig. 4A), indicating that these genes are also specific for tumor development in transplanted mice. We also identified genes that are specific to immortalized lines, such as Zfp326, Usp3, Psmd14, and Crk (P < 0.01; Fig. 4B), raising the possibility that insertions in these genes might have been lost during tumor formation. To test this, we looked for an SB footprint at the Zfp326 locus in tumors that lacked insertions at Zfp326 but were derived from a cell line (line 95) containing an insertion at the locus. This analysis provided evidence for SB remobilization at the same nucleotide in the four tumors. Three tumors had a “TACAGTA” SB footprint (Fig. 5H), whereas the other had a “TACTGTA” footprint, further implicating clonal relationships between these four tumors.
The second most frequently mutated gene in tumors was Nf1 (Fig. 4A). Unlike Met, Nf1 was also frequently mutated in immortalized cells (Fig. 5D), suggesting that it functions during both immortalization and tumor development. Insertions in Nf1 were distributed throughout the gene, and there was little orientation bias (Fig. 5E), consistent with its known tumor suppressor functions. Most tumors (73%) have a single Nf1 insertion, suggesting a haploinsufficient mechanism for tumor formation. Consistently, the expression level of Nf1 was slightly decreased in tumors with Nf1 insertions (Fig. S3I). However, we cannot exclude the possibility that there are tumor subpopulations with inactivation of the second allele by additional SB insertions, loss of heterozygosity, or other epigenetic mechanisms. Mesenchymal GBM is characterized by frequent NF1 mutations and increased MET expression (5). Identification of Met overexpression and Nf1 loss as the two most frequent genetic changes in our mouse tumors once again shows how similar these tumors are to mesenchymal GBM.
Genetic Networks Involved in Immortalization and Tumor Formation.
Ingenuity pathway analysis (IPA; Ingenuity Systems) of CIS genes from immortalized lines identified three significant canonical signaling pathways; neuregulin signaling (P = 2.9E-03), IGF1 signaling (P = 3.13E-03), and Rac signaling (P = 4.77E-03). The CIS genes in these pathways regulate adherence junctions (Cask, Dlg1, and Nf1), cell polarity (Pard3 and Prkci), Rac signaling (Ptk2, Crk, Wasf2, and Cyfip1), and PI3K signaling (Igf1r and Pten) (Fig. 5G). These pathways regulate cytoskeletal organization, which is known to be essential for NSCs, where cell polarity plays a key role in regulating self-renewal vs. differentiation (23). These data suggest that transposons promote immortalization, in part, by inducing cytoskeletal alterations, consistent with the morphological changes we observed during the immortalization process (Fig. 1J). To provide additional evidence for this, we chose two candidate immortalization genes (Gli3 and Nf1) and asked whether shRNA-mediated knockdown (Fig. S3 A and B) had any effect on their morphology or survival, because the pattern of transposon insertions in both genes was suggestive of loss-of-function mutations (Fig. 5D and Fig. S3J). Gli3 can function as a repressor of hedgehog signaling (24), and thus could act as a tumor suppressor gene in certain contexts. Nf1 is a well-known tumor suppressor gene (25). NSCs expressing dominant-negative mutant p53 were retrovirally transduced with shRNAs against Gli3 or Nf1, induced to differentiate, and reseeded. shRNA-expressing cells showed elongated morphology (Fig. S3 N–R) and enhanced cell survival (Fig. S3S) relative to control cells. These results provide additional evidence that these genes function during immortalization, in part, through their effects on cell shape and the cytoskeleton.
IPA analysis of CIS genes identified in tumors identified two canonical signaling pathways important for tumor development: FGF signaling (P = 2.76E-05) and hepatocyte growth factor (HGF) signaling (P = 6.35E-05), in addition to molecular mechanisms of cancer (P = 1.04E-04). Tumor CIS genes function at multiple levels in these pathways (Fig. 5F), including at the receptor level (Met and Pdgfrb) and at downstream adaptor proteins (Gab1 and Frs2), in addition to being regulators of RAS (Nf1, Spred2, Mapk1, and Prkca) and PI3K signaling (Pten and Pik3c2a). With the exception of Nf1 and Pten, genes identified in tumors in these signaling pathways are not highly mutated in immortalized cells, suggesting these pathways play an essential role in later stages of tumor development.
The receptor tyrosine kinase (RTK) pathway is the most frequently mutated signaling pathway identified in human GBM (4). A comparison of our CIS gene list with the mutant genes identified in recent large-scale GBM exon-resequencing projects (4, 26) identified 12 tumor and 8 immortalization CIS genes (P = 3.5E-03 and 9.2E-02, respectively, Fisher’s exact test) (Dataset S5), indicating a significant overlap between tumor CIS genes and candidate driver genes for human GBM. Another comparison of our CIS genes with the Catalog of Somatic Mutations in Cancer database (27) identified 26 tumor and 23 immortalization CIS genes that have at least one nonsilent mutation in human glioma (P = 2.1E-04 and 1.4E-03, respectively, Fisher’s exact test) (Dataset S5).
We also used the Database for Annotation, Visualization, and Integrated Discovery functional annotation tool to delineate further the pathways regulated by CIS genes unique to tumors or immortalized cells, or which were identified in both tumors and immortalized cells (Dataset S6). Consistent with the IPA analysis, genes that function in RTK/MAPK signaling were enriched in the genes unique to tumors, whereas processes associated with transcription were enriched in the genes unique to immortalized cells. Transcriptional regulation is essential for cellular differentiation, supporting the notion that these genes promote immortalization by regulating astroglial differentiation. Finally, the genes common to both tumors and immortalized cells were enriched in processes associated with mitosis. Mitosis is a process essential for cell cycle progression, which may explain why these genes were retained in both immortalized cells and tumors.
Tumors Develop in Transplanted Hosts Largely Through New Insertions That Promote Tumor Development.
To confirm that tumors develop largely through new insertional mutations in genes that promote tumor development in transplanted hosts, we looked at the frequency of insertional mutations in CIS genes that function in the RTK pathway in 16 tumors derived from a single immortalized line. Notably, each tumor showed a distinct combination of insertional mutations in RTK pathway genes (Fig. 5I), even though all tumors were derived from a single line. We then looked to see if these insertions were present in immortalized cells or were newly acquired during tumor formation. Among 92 insertional mutations detected in RTK pathway genes in the 16 tumors, only 3 were detected in the parental cells, indicating that most of the insertions were acquired after transplantation. These data indicate that transplanted cells become tumorigenic by acquiring new insertions in genes that promote tumor development in transplanted hosts, followed by clonal expansion and the generation of tumors with distinct combinations of mutations in tumor CIS genes.
Because most of the insertions in tumor CIS genes were acquired after transplantation, we sought to determine whether they resulted from “local hopping” of preexisting insertions present in immortalized cells or by random insertions from transposons located elsewhere in the genome. Local hopping refers to the fact that remobilized transposons often reinsert near the donor transposon (8, 28). If local hopping plays a major role in the selection of CIS genes in tumors, clonally related tumors derived from the same cell line would be expected to have more similar patterns of insertions in CIS genes than nonclonally related tumors. We applied a statistical method of hierarchical clustering to clonally related tumors to classify tumors as either related or independent by measuring similarity based on Hamming distance between patterns of tumor CIS genes. We analyzed 40 tumors derived from five different cell lines, all with the same genotype (Fig. 5J). Of 12 closely related clusters of tumors, 9 clusters contained tumors that were largely derived from different cell lines. These data demonstrate that clonally related tumors do not form distinct pairs and clusters, indicating that random integrations contribute more than local hopping of preexisting insertions to the acquisition of new insertions in tumor CIS genes.
The distinct combinations of RTK pathway genes in clonally related tumors (Fig. 5I) raise a possibility that these genes may have functionally equivalent roles in tumor development. To test this, we made use of five p53-deficient (p53R172H/+; Nes-cre/+) immortalized lines that are nontumorigenic in transplanted hosts because they lack active SB transposition (Table 1). By retrovirally transducing cDNAs or shRNAs into these cell lines, we can mimic the effect of these mutations in tumors and ask if these cells become tumorigenic in transplanted hosts. A similar approach has been used to identify novel tumor suppressors and oncogenes in liver cancer (29, 30). Transposon insertions are primarily located upstream of Met (Fig. 5C) and Pdgfrb (Fig. S3K) or in intron 1 of Gab1 (Fig. S3L), where they drive the expression of full-length proteins that initiate in exon 3 (Met) or exon 2 (Pdgfrb) or an N-terminal truncated protein that initiates in exon 2 (Gab1). In contrast, the patterns of transposon insertions in Nf1 and Pten are consistent with their tumor suppressor functions (Fig. 5E and Fig. S3M). The five nontumorigenic cell lines were transduced with retroviral expression vectors expressing full-length Met, Pdgfrb, or a truncated Gab1 transcript that begins in exon 2 or shRNAs against Nf1 and Pten (Fig. S3 B–F). Transduced cells were then transplanted into SCID hosts, and the mice were monitored for tumor development. Met overexpression or Nf1 silencing rendered all five cell lines tumorigenic; Pdgfrb or Gab1 overexpression, or Pten silencing, rendered four of the lines tumorigenic (Fig. S3T). Together with the IPA analysis, these findings indicate that these genes have functionally overlapping roles in tumor development, suggesting that transplanted cells became cancer-initiating cells by generating a branched and diverse genetic architecture (Fig. 5K).
Discussion
Here, we describe a unique type of GBM model that uses transposons to mutagenize NSCs in culture. The mutagenized stem cells were then induced to differentiate, and cultures composed of immortalized, self-renewing, astroglial-like cells were selected and transplanted into mice, where, following additional rounds of transposon-based insertional mutagenesis, tumors resembling mesenchymal GBM were produced, consistent with the astroglial-like expression signature of this subclass of GBM (5). Analysis of the patterns of insertional mutations in tumors derived from a single immortalized line showed heterogeneity among the individual tumors. The constitutive transposase activity is, in part, responsible for this heterogeneity, because it allows transplanted subclones to compete for advantageous insertional mutations under the same selective pressure (the growth in the immunocompromised host). This competition among genetically variant subclones is similar to the Darwinian evolutionary processes now thought to generate many human cancers, which involve the selection of genetically variant cells (31). Until recently, it was generally thought that cancer progression occurs in a linear fashion, through the sequential accumulation of mutations, with genetic selection occurring after each new mutation, producing a more aggressive clone each time, until a clone is produced that has accumulated enough mutations to form a tumor. Recent data (32, 33), however, challenge this notion and indicate instead that some cancers have a branched cellular and genetic architecture reminiscent of Darwin’s iconic evolutionary tree diagram depicting speciation (31). The end result of this evolutionary process is that each tumor is composed of many subclones of self-renewing cells, each with a different combination of mutations. This intratumor heterogeneity and branched evolution could represent a significant roadblock to effective cancer therapy. This evolutionary process can be modeled in this type of cancer model, where one self-renewing immortalized cell line is used to generate many different tumors, with each tumor showing significant intra- and intertumor heterogeneity and combinations of transposon-induced mutations. This mutagenesis strategy is also much faster and simpler than conventional transposon screens, and it can generate hundreds or even thousands of tumors in a very short period for relatively little cost.
Precedence for these studies comes from retroviral insertional mutagenesis experiments showing that retroviral mutagenesis of primary bone marrow progenitor/stem cells can promote the immortalization of immature myeloid cell lines with neutrophil and macrophage differentiation potential (34). More than half of these immortalized lines had activating proviral insertional mutations in two well-known human myeloid leukemia oncogenes important for hematopoietic stem cell self-renewal, Evi1 and Prdm16 (35, 36). These immortalized immature myeloid cells were not tumorigenic in transplanted hosts, however, probably because the retrovirus used in these experiments was replication-defective and could not induce the additional rounds of insertional mutations required for tumor development in transplanted mice.
Recent studies have suggested that the cell acquiring the first cancer-promoting mutation might not be the COO for the tumor, which is operationally defined as the cell that initiates tumors in transplanted mice (37). Recent data have suggested that the COO for the proneural subtype of GBM is an OPC, even though the originating mutations occurred in the NSC (38). Likewise, our results suggest that the COO for the mesenchymal subtype of GBM is an astroglial-like cell, even when the originating mutations occurred in the NSC. These results also indicate how a cancer-initiating mutation in a relatively quiescent stem cell can be retained for many years and then end up in a tumor decades later. Nondifferentiating divisions of the mutant stem cell could generate additional long-lived stem cells that carry the mutation but do not respond to it. Subsequent differentiating divisions would then generate differentiated progeny that respond to the mutation, producing clones of differentiated cells with unlimited self-renewal, which then go on to accumulate the additional rounds of mutations required to become a cancer-initiating cell.
Cultured astroglial cells express many of the genes expressed by astrocytes but generally do not express genes that are enriched in neurons and oligodendrocytes; however, cultured astroglial cells and astrocytes are as different from each other as OPCs are from oligodendrocytes, and this difference cannot be explained by the presence of serum in the medium because it persists even in serum-free medium (19). Cultured astroglial cells can be generated from either the neonatal cortex or the adult subependymal zone (SEZ) but not from the adult cortex, and these cells partially maintain NSC-like immature properties, such as multipotent sphere formation (39). Cultured astroglial cells are therefore considered to be an immature stage of the astrocyte lineage (20). Although an in vivo counterpart of cultured astroglial cells in the adult brain remains elusive, there is a general consensus that adult mammalian NSCs have many astrocytic characteristics (12) and cultured astroglial-like cells can be isolated from the adult SEZ, raising the possibility that immature astrocytic cells in the SEZ may serve as the cancer COO for mesenchymal GBM.
The analysis of CIS genes in immortalized lines identified genes that regulate cytoskeletal organization and transcriptional regulation. These pathways also regulate the differentiation of NSCs (23). However, the reason why these pathways are selected for during immortalization is not obvious, because lines of evidence suggest that cellular immortalization is regulated by the G1-S control pathway (40). Cultured astroglial cells can undergo cell doubling several times but gradually enter senescence and finally differentiate into mature astrocytes (41). Previous studies showed that cultured astroglial cells can be immortalized by the deletion of either Ink4a/Arf or p53 (42). However, the overexpression of p53DN was not sufficient to immortalize NSCs on differentiation. This is probably because the astroglial cells induced from NSCs and the primary culture of astroglial cells are different. The removal of growth factor and the addition of serum put a strong selective pressure on NSCs to undergo terminal differentiation into mature astrocytes, which are mitotically inactive. Only cells harboring mutations that prevent terminal differentiation can continue proliferation under these culture conditions. Cell polarity and transcription also play essential roles in cellular differentiation. This may explain why genes involved in these pathways were positively selected during immortalization. Consistent with this, silencing of Nf1 or Gli3 induced continuous growth of NSCs on differentiation; however, the growth rate was slow and none of them reached our criteria of immortalization (cumulative total cell number exceeds 108), indicating that accumulation of additional mutations is likely required to phenocopy the immortalization.
We identified the RTK pathway as significantly enriched in the CIS genes identified in tumors. An interesting observation is that Pten insertions are observed less frequently in the tumor outgrowths compared with the in vitro immortalized cell lines (12% vs. 40% of all samples, respectively), raising the possibility that Pten loss may not be a strong driver mutation in tumor formation. PTEN is mutated in all subtypes of human GBM (the mutation frequency is between 20% and 30%) and is not specific to any GBM subtype (5). Pten inactivation can promote proliferation of NSCs (43). However, the genetic deletion of Pten and p53 alone is not sufficient to drive GBM. The genomic analysis of GBMs in Pten and p53 double-mutant mice showed the presence of additional events in RTK pathway genes, such as focal amplifications of Met and Egfr (44). These data are consistent with the notion that Pten deficiency can be an initiating event, but additional mutations in RTK pathway genes play essential roles in tumor progression in GBM.
Our mutagenesis screen identified a large number of CIS genes. To identify genes specific to GBM further, we compared our CIS genes with those identified in large-scale mutagenesis screens, such as intestinal tumors (1,010 genes) (22), pancreatic tumors (136 genes) (45), and medulloblastomas (64 genes) (46). This comparison showed that 70 tumor CIS genes (47%) and 66 immortalization CIS genes (47%) are unique to our screen (Datasets S3 and S4). These genes include the RTK pathway genes (Met, Nf1, and Pdgfrb) as well as previously undescribed genes, such as Stag2, Traf3, Dmxl1, Foxj3, and Faf1, which are the top five glioma-specific candidate cancer genes listed in Dataset S3. Stag2, a gene encoding a subunit of cohesin, plays essential roles in genomic instability (47). Traf3 is an adaptor protein that directly binds to a number of TNF receptors. Traf3 mutant mice show a lymphoproliferative phenotype (48), suggesting a tumor suppressor function for Traf3. Dmxl1 is a homolog of Drosophila DmX, which codes for a WD40 repeat protein. Nonsynonymous mutations of DMXL1 have been identified in patients with hepatitis C virus-associated hepatocellular carcinoma (49). Foxj3, a forkhead transcriptional factor expressed in neuroectoderm (50), has not been previously associated with cancer. Faf1 is a member of the Fas-mediated death-inducing signaling complex, and there is compelling evidence implicating FAF1 as a tumor suppressor (51).
The strategy described here can accelerate the utility and potential of insertional mutagenesis approaches. The in vitro propagation stage, which allows mutagenesis to select for distinct properties, such as immortalization, has much potential, although this strategy also has its limitations, because not all cell types can be propagated in vitro. Liu et al. (38) showed that OPCs are the COO for the proneural subtype of GBM. OPCs can be induced from NSCs by culturing the cells with defined media (52), raising the possibility that the proneural subtype of GBMs can be modeled by inducing OPCs from NSCs and then transplanting these cells into immunocompromised mice to generate tumors. Another limitation of this strategy is that tumor formation occurs in the absence of an adaptive immune system; therefore, the genes identified are cell-intrinsic regulators of tumor formation.
In the present study, we also used a standard protocol to amplify transposon insertion sites, which is based on ligation-mediated PCR amplification of junction fragments from restriction endonuclease-digested genomic DNA. Because uneven genomic distribution of restriction enzyme sites can cause amplification biases, read numbers are not presented in this study. Recently, a shear-splink method, which allows for semiquantitative analysis of the clonality of individual insertions, was developed (53). Application of this technology to heterogeneous tumors will further delineate the clonality of the SB insertions in the heterogeneous tumors.
In summary, this study demonstrates that transposon-based insertional mutagenesis of primary cultures of tissue stem cells can be an effective method for identifying genes responsible for the transformation of normal stem cells into cancer-initiating cells. This mutagenesis strategy can also provide important information regarding the origin of the cancer-initiating cell for a tumor and is not limited to NSCs but, in principle, can be applied to any stem/progenitor cells that can be cultured and differentiated in vitro.
Materials and Methods
Mouse Strains.
All manipulations were performed with Institutional Animal Care and Use Committee approval (Institute of Molecular and Cell Biology). Mouse lines used in this study are described in SI Materials and Methods.
Cell Culture.
Primary NSCs were isolated from the SVZ of embryos between E17 and P1. Dissociated single cells were cultured in DMEM/F12 medium supplemented with 1% N2 (Gibco), 20 ng/mL EGF (Peprotech), 0.6% glucose, 4 μg/mL heparin, and penicillin/streptomycin. Primary neurospheres were passaged by dissociation of the spheres into single cells by incubation with trypsin. Single cells were reseeded and incubated in the culture medium. Secondary neurospheres were collected and induced to differentiate by incubating cells in culture media depleted of EGF and supplemented with 10% (vol/vol) FBS for 2 wk (21). Then, cells were split, counted, and reseeded every 7–10 d.
Injection.
For s.c. tumor formation, 1 × 106 cells were injected s.c. into both flanks of 6- to 8-wk-old SCID mice in 100 μL of PBS using a 26-gauge needle. Animals were followed for development of a palpable mass. If mice did not develop tumors by 4 mo after injection, transplanted cells were considered nontumorigenic. Mice were euthanized when the diameter of the tumor exceeded 1 cm. For validation experiments, immortalized cell lines were infected with retroviruses expressing cDNA or shRNA, and were s.c. transplanted.
Cloning of Transposon Insertion Sites and Identification of CISs.
Linker-mediated PCR was performed as described (10). For secondary PCR, nested primers carrying the required fusion sequences for the titanium platform (Roche), as well as a unique 10-bp barcode recognition sequence for each sample, were used. CISs were identified and annotated according to the nearest mouse gene as described previously (22).
Retroviral Constructs.
cDNAs for Met, Pdgfrb, Gab1, and p53DN were cloned into retroviral expression vectors. Previously reported shRNAs were used for Nf1 (54) and Pten (55). Sequences for Gli3 shRNAs and nontargeting shRNA (luciferase) are provided in SI Materials and Methods.
Microarray and GSEA Analysis.
Array data are available in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE36897. Detailed procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Keith Rogers and Susan M. Rogers for rodent necropsy and histopathological analysis of tumor specimens. We also thank Pearlyn Cheok, Nicole Lim, and Dorothy Chen for their help with tumor monitoring. This work was supported by the Biomedical Research Council; Agency for Science, Technology, and Research, Singapore; and the Cancer Prevention Research Institute of Texas. D.J.A. and A.G.R. are supported by Cancer Research-United Kingdom and the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
Data deposition: The array data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession no. GSE3689).
See Author Summary on page 17746 (volume 109, number 44).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215899109/-/DCSupplemental.
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109(1):93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcantara Llaguno S, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marumoto T, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15(1):110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kool J, Berns A. High-throughput insertional mutagenesis screens in mice to identify oncogenic networks. Nat Rev Cancer. 2009;9(6):389–399. doi: 10.1038/nrc2647. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69(20):8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436(7048):221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 11.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 12.Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr Opin Cell Biol. 2006;18(6):704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175(1):1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 14.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436(7048):272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 15.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 16.Milner J, Medcalf EA. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991;65(5):765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- 17.Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: Negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12(12):5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36(1):63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- 19.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stiles CD, Rowitch DH. Glioma stem cells: A midterm exam. Neuron. 2008;58(6):832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 21.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1(3):269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 22.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43(12):1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doe CQ. Neural stem cells: Balancing self-renewal with differentiation. Development. 2008;135(9):1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 24.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100(4):423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264(1):19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- 26.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forbes S, et al. COSMIC 2005. Br J Cancer. 2006;94(2):318–322. doi: 10.1038/sj.bjc.6602928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruf S, et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43(4):379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- 29.Sawey ET, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell. 2011;19(3):347–358. doi: 10.1016/j.ccr.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6(12):924–935. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 32.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469(7330):356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 34.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106(12):3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shing DC, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117(12):3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goyama S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3(2):207–220. doi: 10.1016/j.stem.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laywell ED, Rakic P, Kukekov VG, Holland EC, Steindler DA. Identification of a multipotent astrocytic stem cell in the immature and adult mouse brain. Proc Natl Acad Sci USA. 2000;97(25):13883–13888. doi: 10.1073/pnas.250471697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14(23):3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holland EC, Hively WP, Gallo V, Varmus HE. Modeling mutations in the G1 arrest pathway in human gliomas: Overexpression of CDK4 but not loss of INK4a-ARF induces hyperploidy in cultured mouse astrocytes. Genes Dev. 1998;12(23):3644–3649. doi: 10.1101/gad.12.23.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23(2):379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 43.Gregorian C, et al. Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J Neurosci. 2009;29(6):1874–1886. doi: 10.1523/JNEUROSCI.3095-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann KM, et al. Australian Pancreatic Cancer Genome Initiative Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2012;109(16):5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482(7386):529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon DA, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333(6045):1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27(2):253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li M, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43(9):828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landgren H, Carlsson P. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev Dyn. 2004;231(2):396–401. doi: 10.1002/dvdy.20131. [DOI] [PubMed] [Google Scholar]
- 51.Menges CW, Altomare DA, Testa JR. FAS-associated factor 1 (FAF1): Diverse functions and implications for oncogenesis. Cell Cycle. 2009;8(16):2528–2534. doi: 10.4161/cc.8.16.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, et al. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2(5):1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- 53.Koudijs MJ, et al. High-throughput semiquantitative analysis of insertional mutations in heterogeneous tumors. Genome Res. 2011;21(12):2181–2189. doi: 10.1101/gr.112763.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Munchhof AM, et al. Neurofibroma-associated growth factors activate a distinct signaling network to alter the function of neurofibromin-deficient endothelial cells. Hum Mol Genet. 2006;15(11):1858–1869. doi: 10.1093/hmg/ddl108. [DOI] [PubMed] [Google Scholar]
- 55.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436(7051):725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]