Abstract
Recognition of methylated histone tail lysine residues by tudor domains plays important roles in epigenetic control of gene expression and DNA damage response. Previous studies revealed the binding of methyllysine in a cage of aromatic residues, but the molecular mechanism by which the sequence specificity for surrounding histone tail residues is achieved remains poorly understood. In the crystal structure of a trimethylated histone H3 lysine 4 (H3K4) peptide bound to the tudor-like domains of Spindlin1 presented here, an atypical mode of methyllysine recognition by an aromatic pocket of Spindlin1 is observed. Furthermore, the histone sequence is recognized in a distinct manner involving the amino terminus and a pair of arginine residues of histone H3, and disruption of the binding impaired stimulation of pre-RNA expression by Spindlin1. Our analysis demonstrates considerable diversities of methyllysine recognition and sequence-specific binding of histone tails by tudor domains, and the revelation furthers the understanding of tudor domain proteins in deciphering epigenetic marks on histone tails.
Keywords: methylation, rRNA expression
Histone lysine methylation imparts epigenetic information in chromatin biology, and the transduction of epigenetic signal is mediated by a diverse group of proteins containing methyllysine recognition domains (1). Some of the best known methyllysine recognition modules include chromo, tudor, MBT (Malignant Brain Tumor), and PHD (Plant Homeodomain) domains (2–4). A recent addition of this family of histone methyllysine “readers” is the BAH domain ORC1 (5). These methyllysine “readers” can recognize histone lysine methylation in a site-specific and methylation state-specific manner. A common feature of methyllysine recognitions involves a cage of aromatic residues, whereas the ways for discriminating the surrounding amino acid sequence of histones appear to be divergent among different methyllysine “readers.” In this study, we focus on the recognition of trimethylated lysine-4 of histone H3 (H3K4me3) by the tandem tudor-like domains of Spindlin1.
Mammalian Spindlin1 is a nucleolar protein that localizes to the active ribosomal DNA (rDNA) repeats locus, where it is normally enriched with histone H3K4 and H3K36 methylations, and facilitates rRNA expression (6, 7). Spindlin1 contains three tandem tudor-like domains (8), and we previously demonstrated that the tudor-like domains bind to H3K4me3 in vitro (7). Furthermore, Spindlin1 mutants with impaired H3K4me3 binding showed reduced stimulation of rRNA expression (7). Hence, these observations directly link the ability of H3K4me3 binding with the transcription function of Spindlin1. However, several important aspects of the molecular mechanism of H3K4me3 recognition by Spindlin1 remain outstanding; foremost ones include how Spindlin1 achieves its H3K4me3 specificity and what features distinguish the H3K4me3-binding tudor-like domain from others in Spindlin1. Previous structural studies of tudor domains have yielded some understandings of how they recognize methylated histone tails (9–13). Among the tudor domain structures with methylated histone peptides, the two most relevant ones to this study are the double tudor domains of JMJD2A and Sgf29 in complex with H3K4me3 peptides (9, 12).
Our cocrystal structure of Spindlin1 in complex with an H3K4me3 peptide encompassing residues 1–8 shows that the H3K4me3 is exclusively bound to the second tudor-like domain. Importantly, both the binding mode of the methyllysine in the aromatic cage and the manner by which the histone sequence specificity is achieved differ from all known structures. Our results have revealed intrinsic properties of Spindlin1 critical for H3K4me3 recognition and stimulation of rRNA expression and have uncovered an interesting binding mode of methylated histone tails by tudor-like domains.
Results
Second Tudor-Like Domain of Spindlin1 Binds the H3K4me3 Peptide.
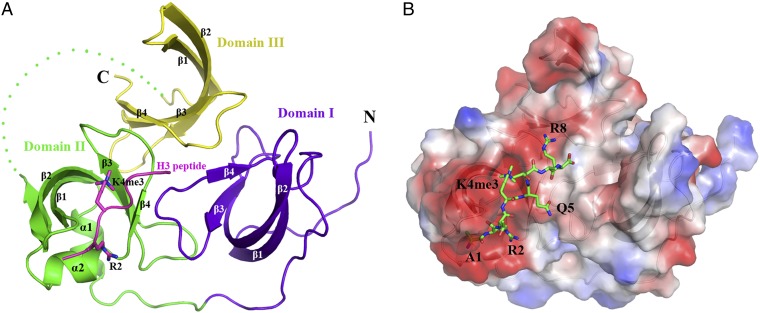
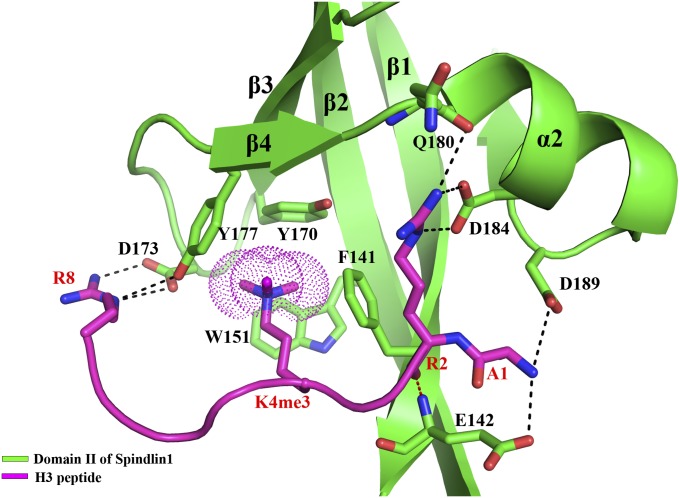
The 2.1 Å structure of human Spindlin1 in complex with a histone H3K4me3 peptide (aa 1–8) shows that it comprises three juxtaposed tudor-like domains (Fig. 1). Each tudor-like domain is composed of four antiparallel β-strands, β1 to β4, whereas the second tudor-like domain (domain II) contains an additional strand, β5, and two small alpha helices, α1 and α2 (Fig. 1A). The overall arrangement of the three tudor-like domains is similar to that of the apo-Spindlin1 structure (PDB ID: 2NS2) (8). The two structures can be superimposed with a root-mean-square deviation (rmsd) of 1.1 Å (Fig. S1). One H3K4me3 peptide is bound to domain II (Fig. 1 A and B and Fig. S2). Four aromatic residues—Phe141, Trp151, Tyr170, and Tyr177—located on β1, β2, β3, and β4 of domain II, respectively, form a cage that traps the trimethylated H3K4 residue (Fig. 2). No significant conformational changes of these residues are observed upon the binding of the H3K4me3 peptide, and the peptide binding does not seem to alter the organization of the three tudor-like domains in solution or in the crystal (Figs. S1 and S3). Typically, methyllysine binding aromatic cages, such as those found in chromo and other tudor domains, are formed by three aromatic residues (3, 14). An interesting case is the PHD finger of BPTF, which also uses an aromatic cage of four residues to bind H3K4me3 (15). Nevertheless, similar types of interactions, namely hydrophobic and charged (cation-π) interactions, are involved in the binding of H3K4me3 in the atypical aromatic cage of domain II of Spindlin1.
Fig. 1.
Overall structure of Spindlin1. (A) A ribbon diagram of the Spindlin1 tudor-like domains in complex with an H3K4me3 peptide. The three tudor-like domains of Spindlin1, following the order from the N-terminal to the C-terminal end, are colored blue, green, and yellow, respectively. The histone H3 peptide containing residues 1–8 with trimethylated K4 residue is shown in magenta. The trimethylated H3K4 sidechain is shown in a stick model. (B) Electrostatic potential on the surface of Spindlin1. The structure is viewed from the same direction as in A, and the peptide is shown in a stick representation (carbon, green; nitrogen, blue; oxygen, red).
Fig. 2.
A close view of histone H3K4me3 peptide binding by domain II of Spindlin1. The Spindlin1 domain is shown in a ribbon representation colored green, and the residues involved in interaction with the histone peptide are superimposed as a stick model (carbon, green; nitrogen, blue; oxygen, red). The histone peptide is shown as a magenta coil, and the residues interacting with Spindlin1 are superimposed as a stick model (carbon, magenta). The trimethyl group of H3K4me3 is highlighted with dots. Dashed lines indicate intermolecular hydrogen bonds (red dashed lines indicate those between mainchain atoms).
The histone peptide is bound in an extended negatively charged region encompassing β1, β2, and α2 of domain II (Fig. 1B). Besides the methylated H3K4, only two other histone residues, H3R2 and H3R8, are involved in sidechain interactions with Spindlin1. The guanidino moiety of H3R2 forms two hydrogen bonds with the sidechain hydroxyl groups of Asp184 located on α2 and one to the carbonyl group of Gln180. The guanidino moiety of H3R8 forms two hydrogen bonds with Asp173 and another with the hydroxyl group of Tyr177 of Spindlin1. In addition, the terminal amino group of H3A1 makes hydrogen bonds with the hydroxyl groups of Glu142 and Asp189 (Fig. 2). These interactions determine the binding specificity of Spindlin1 toward H3K4.
Structural Differences Between the Histone Binding and Nonbinding Tudor-Like Domains.
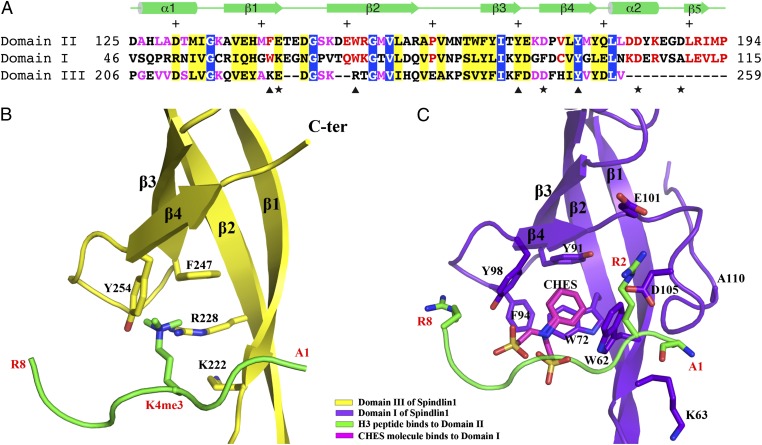
The three tudor-like domains of Spindlin1 enjoy a significant degree of sequence conservation (23%/59% and 26%/56% sequence identity/similarity between domain II and domains I and III, respectively) and share similar overall structures (rmsd deviations of 1.9 and 2.1 Å between domain II and domains I and III, respectively), yet only domain II binds the histone H3 peptide (Figs. 1 and 3). With the structure in hand we seek an explanation of the asymmetric histone-binding behavior of the tudor-like domains. Sequence comparison and structure alignment indicate that domain III cannot accommodate methyllysine binding, as it lacks critical residues that can form an intact aromatic cage. Domain III residues corresponding to the aromatic cage residues Phe141, Trp151, Tyr170, and Tyr177 of domain II are Lys222, Arg228, Phe247, and Tyr254, respectively (Fig. 3 A and B). In contrast, the corresponding residues in domain I are Trp62, Trp72, Tyr91, and Tyr98, respectively, and they form an aromatic cage (Fig. 3 A and C). However, instead of a methyllysine, a crystallization solvent molecule, CHES, occupies the aromatic cage of domain I (Fig. 3C). Domain I residues corresponding to histone H3–interacting Glu142, Asp173, Asp184, and Asp189 of domain II are Lys63, Phe94, Asp105, and Ala110, respectively. Obviously, domain I residues Lys63, Phe94, and Ala110 cannot make hydrogen bonds to the histone peptide as domain II residues E142, Asp173, and Asp189 do. Superficially, Asp105 of domain I can serve the function of Asp184 in histone binding. However, proper positioning of Asp184 depends on the α-helix (α2) on which it locates, and a corresponding helix is lacking in domain I (Figs. 2 and 3C). Thus, domain I contains an intact aromatic cage that can potentially bind a methylated peptide, but it cannot be the H3K4me3 peptide due to unsuitable or improperly positioned amino acids. To probe whether domain I is involved in binding other histone methyllysine marks, we tested interactions between Spindin1 and mononucleosomes mimicking H4K20, H3K79, or H3K36 methylation. The results of mononucleosome pull-down experiments were all negative (Fig. S4A). Previously, we demonstrated that Spindlin1 does not bind nucleosomes mimicking H3K9 or H3K27 methylation (7). We further tested the binding of an isolated domain I to histone tail peptides with H3K4me3, H3K9me3, H3K27me3, H3K36me2, or H4K20me3 modifications by isothermal titration calorimetry (ITC) measurements, and no binding was detected (Fig. S4B). Hence, domain I appears not to be a methyllysine binder.
Fig. 3.
The first and third tudor-like domains of Spindlin1 are incapable of binding H3K4me3 peptide. (A) Sequence alignment of the three tudor-like domains. The positions of aromatic residues forming the H3K4me3-binding aromatic cage of domain II are indicated with triangles at the bottom of the sequences. The positions of four dicarboxylic amino acids that interact with the terminal amino group, H3R2, and H3R8 are indicated with stars. Identical and similar residues among all three domains are shown in white letters over blue background and black letters over yellow background, respectively. Identical and similar residues between domains II and I are shown in red letters, and those between domains II and III are shown in magenta. Secondary structural elements of domain II are shown above the sequence, and every 10 even residues are indicated with +. (B) Domain III of Spindlin1 shown in the same orientation as that of domain II in Fig. 2. The H3K4me3 peptide bound to domain II is superimposed to indicate that such a peptide cannot bind domain III due to lack of an intact aromatic cage. (C) Domain I of Spindlin1 shown in the same orientation as that of domain II in Fig. 2. The H3K4me3 peptide bound to domains II is superimposed to show that domain I lacks appropriate dicarboxylic amino acids and α2 required for H3K4me3 binding.
Structural Determinants for Histone Binding Specificity Are Important for in Vitro and in Vivo Functions of Spindlin1.
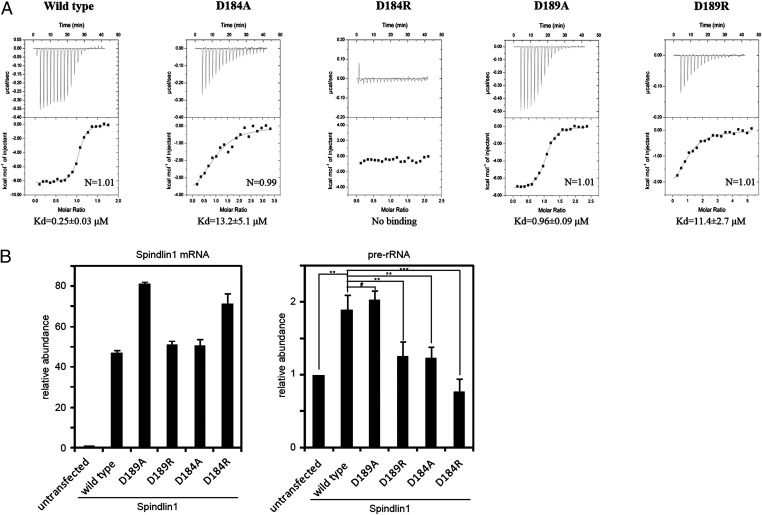
To correlate the structural findings with the biochemical and in vivo properties of Spindlin1, we made several mutants of Spindlin1, measured their bindings to a 10-residue H3K4me3 peptide (aa 1–10) by ITC, and tested their effect in rRNA expression. We have previously shown that an intact aromatic cage of domain II is critical for binding H3K4me3 and stimulation of rRNA expression (7). Here we test the functions of Asp184 and Asp189, which interact with Arg2 and the terminal amino group of histone H3, respectively (Fig. 2). ITC measurements show that Asp184 and Asp189 mutations affect the binding of the H3K4me3 peptide to different degrees (Fig. 4A). Although change of Asp189 to an alanine (D189A) resulted in ∼fourfold poorer binding to the histone peptide (Kd of 0.96 μM) in comparison with that of the wild-type Spindlin1 (Kd of 0.25 μM), an Asp184 to alanine (D184A) mutation reduced the binding by ∼50-fold (Kd of 13.2 μM). Replacing Asp184 or Asp189 with the oppositely charged arginine either abrogates or significantly impairs the ability to bind the H3K4me3 peptide: the D189R mutant binds with a dissociate constant of 11.4 μM, and the binding of the D184R mutant is below the detection level of ITC (Fig. 4A). The different contributions of Asp184 and Asp189 in H3K4me3 binding may be understood from the number of hydrogen bonds they engage in interaction with the histone peptide (Fig. 2).
Fig. 4.
Asp184 and Asp189 are important for Spindlin1’s H3K4me3-binding activity. (A) ITC measurements of binding affinities between the wild-type, D184A, D184R, D189A, and D189R mutants of Spindlin1 and a 10-residue H3K4me3 peptide (aa 1–10). Kd values with fitting errors are indicated. (B) RT-PCR detection of prerRNA expression in cells expressing the wild-type and Asp184 and Asp189 mutants of Spindlin1 (Right). The expression levels of the wild-type and mutant Flag-Spindlin1 mRNA were also determined by RT-PCR (Left). Error bars represent the SD calculated from three independent experiment repeats. ***P < 0.002; **P < 0.01; and #P > 0.1.
We have previously demonstrated that Spindlin1 stimulates the expression of rRNA, and the stimulation effect depends on the H3K4me3 binding ability of Spindlin1 (7). To assess the significance of structural and biochemical results, we tested the effect of the Asp184 and Asp189 mutants of Spindlin1 in pre-rRNA expression. Fig. 4B shows that, in HeLa cells expressing wild-type or mutant Spindlin1, the abilities to stimulate pre-RNA expression by these mutants correlate well with their abilities to bind H3K4me3 peptides in vitro. Therefore, we conclude that the aspartic residues identified by structural analyses are critical for histone binding specificity and important for in vitro and in vivo functions of Spindlin1.
Diverse Methyllysine Binding Modes and Distinct Sequence Recognition Mechanisms.
To date, several tudor domain structures in complex with methylated histone peptides have been determined. They include the double tudor domain of JMJD2A complexed with H3K4me3 and H4K20me3 peptides (9, 10), the double tudor domain of 53BP1/Crb2 in complex with a H4K20me2 peptide (11), the tandem tudor domain of UHRF1 in complex with a H3K9me3 peptide (13), and the double tudor domain of Sgf29 in complex with a H3K4me3 peptide (12). We choose the structures of JMJD2A and Sgf29 for close comparison with our Spindlin1 structure for two reasons: first, they all recognize methylated H3K4, and second, they are the only reported tudor domain structures with a significant number of ordered histone H3 residues, which are important for analysis of sequence-specific tudor domain–histone H3 interactions.
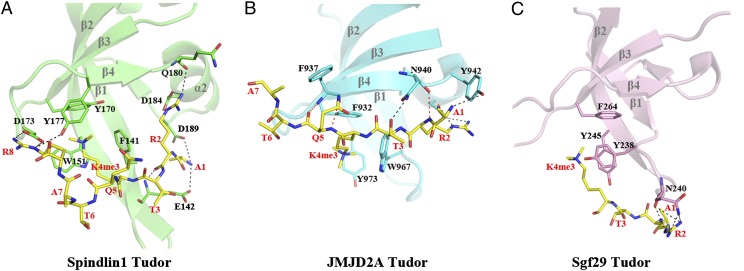
The methyllysine binding pockets of the tudor domains of Spindlin1, JMJD2A, and Sgf29 share three spatially defined aromatic residues, one each on β1, β2, and β3 or their close vicinities (Fig. 5). A fourth aromatic residue, Tyr177 on β4, is specific to Spindlin1. The three aromatic residues of JMJD2A are arranged in a configuration similar to that of Spindlin1. However, the two H3K4me3 residues are almost perpendicularly positioned in the two complexes (Fig. 5 A and B). In JMJD2A the trimethyl group is directed toward Tyr973 on β2 (which corresponds to Trp151 of Spindlin1), whereas in Spindlin1 it faces Tyr170 on β3 (equivalent to Phe932 of JMJD2A). The residue that corresponds to Tyr170 of Spindlin1 is Phe264 in Sgf29, and oddly, its aromatic sidechain turns away from the trimethyl group, making the binding site an aromatic wedge instead of a cage (Fig. 5C). Hence, Spindlin1 recognizes trimethylated H3K4 via a binding mode different from other H3K4me3-binding tudor domains.
Fig. 5.
Histone H3K4me3 binding modes of Spindlin1, JMJD2A, and Sgf29 tudor domains. Similarly oriented histone-binding tudor domains of Spindlin1, JMJD2A, and Sgf29, are shown in ribbon representations colored green, cyan, and pink in A, B, and C, respectively. Their residues interacting with their cognate H3K4me3 peptide are superimposed as a stick model, with the carbon bonds colored the same as that of the ribbon model, and covalent bonds involving nitrogen and oxygen atoms colored blue and red, respectively. The H3K4me3 peptides are shown in stick models (carbon, yellow; nitrogen, blue; oxygen, red).
It was described earlier that apart from the contacts by the trimethylated lysine, the terminal amino group and the sidechains of Arg2 and Arg8 interact via hydrogen bonds with sidechain atoms of domain II residues of Spindlin1 (Fig. 5A). Arg2 also makes a hydrogen bond to the carbonyl of Gln180 of Spindlin1, but there is only one mainchain–mainchain interaction, involving the carbonyl of H3R2 and the amide group of Glu142. In the JMJD2A complex, the H3K4me3 peptide is bound next to β4 of JMJD2A in a pseudo-antiparallel β-sheet pairing fashion with three intermolecular carbonyl-amide hydrogen bonds (Fig. 5B) (9). Both the amino terminus and H3R2 are also involved in interactions with JMJD2A sidechains, but H3R8 is disordered in the structure and Ala7 is not involved in intermolecular interactions. However, both Thr3 and Gln5 of histone H3 interact with JMJD2A, via hydrogen bonds with the sidechain amino group of Asn940 and the carbonyl of Phe937, respectively (Fig. 5B). In the Sgf29 structure, only residues 1–4 of histone H3 are ordered (12). Besides the methyllysine, the only sidechain involved in intermolecular interaction is H3R2, whose guanidino moiety makes four hydrogen bonds with the sidechain amino group and the mainchain carbonyl of Gln240 (Fig. 5C). The above comparisons clearly demonstrate that distinct mechanisms are used by these tudor domains to achieve sequence specificity for the same region of histone H3.
Discussion
The crystal structure of Spindlin1 in complex with the H3K4me3 peptide shows that only one of its tudor-like domains, domain II, binds the histone peptide. Our analysis reveals that an intact aromatic cage and several aspartic acid residues that interact with the terminal amino group, H3R2, and H3R8 are critical for domain II’s ability to bind the H3K4me3 peptide. Other tudor-like domains of Spindlin1 lack some of these features, rendering them unable to bind the H3K4me3 peptide. More and more examples have emerged that only one tudor domain in proteins with tandem tudor domains binds methylated histone tails, whereas others serve as packing scaffolds. Comparison of the histone binding and nonbinding tudor-like domains presented here should help the analysis of histone binding ability of tudor domains in general.
An extremely interesting finding is that even among tudor domains recognizing the same methyllysine mark, the methyllysine binding mode and the mechanism for sequence-specific recognition are very different. The varied binding modes of the aromatic cages can be mainly attributed to the context in which they are positioned. In particular, β4 and its preceding loop play important roles in determining the manner of histone peptide binding. In the JMJD2A tudor domain complex, the H3K4me3 peptide forms an extensive intermolecular hydrogen bond network through sidechain–sidechain, mainchain–sidechain, and mainchain–mainchain interactions with the tudor domain. In the Spindlin1 structure, β4 residues Tyr177 and Tyr179 point outward from the β-sheet edge, preventing the binding of H3K4me3 peptide in a manner resembling that with the JMJD2A tudor domain, highlighting the importance of sidechain interactions via H3R2 and H3R8. Sgf29 has an unusually long loop preceding β4, and Leu277 takes a conformation similar to Tyr177 of Spindlin1. Hence, the histone peptide cannot bind like that found in the JMJD2A complex. The peptide does not bind like that in the Spindlin1 complex either, because Pro239 significantly altered the positioning of Tyr238, an aromatic cage residue. These observations indicate diverse and context-dependent modes of methyllysine recognition by tudor domain.
Tudor domains have been shown to recognize histone lysine residues located at different positions and methylated to different degrees (3). In addition, some tudor domains recognize methylated arginine residues of histone and nonhistone proteins (16–20). These methylation-dependent interactions impact a wide spectrum of biological processes in eukaryotes. Therefore, it is imperative to understand the recognition modes of tudor domains in detail. The results obtained here unveil an intricate picture of sequence-specific recognition of methylated histone tails by tudor domains and, together with the dynamic and complex pattern of histone methylation, highlight the elaborate pattern of epigenetic control of gene expression.
Materials and Methods
Protein Expression, Purification, and Crystallization.
GST-fused Spindlin1 encompassing the three tudor-like domains (aa 27–262) was expressed in Escherichia coli using a pGEX-2T vector. Protein expression, GST tag cleavage, and purification were carried out following the published protocol (8, 21). Purified Spindlin1 was concentrated to ∼24 mg/mL in a buffer containing 20 mM Tris⋅HCl, pH 8.0, and 100 mM NaCl. Chemically synthesized H3K4me3 peptide (aa 1–8) with the sequence ARTK(me3)QTAR was purchased from SciLight Biotechnology, dissolved in water to a stock concentration of 100 mM, and added to the concentrated protein sample at a peptide-to-protein molar ratio of 10:1. The protein–peptide mixture was incubated on ice for an hour to allow efficient complex formation before crystallization trials. Crystals used for X-ray diffraction were obtained from a condition containing 0.1 M CHES (pH 9.5), 2.1 M ammonium sulfate, and 0.2 M lithium sulfate using the hanging drop diffusion method at 16 °C.
Crystal Structure Determination.
The X-ray diffraction data were collected at 100K using the crystallization buffer plus 15% (vol/vol) glycerol as cryoprotectant. A 2.1 Å dataset was collected at a wavelength of 0.9789 Å at beamline BL17U of Shanghai Synchrotron Radiation Facility using a Quantum 315r CCD detector (ADSC). The diffraction data were processed with the HKL2000 software (22). The crystal belongs to P21212 space group, and the structure was solved by molecular replacement using Molrep of the CCP4 program suite (23). The apo Spindlin1 structure (PDB ID: 2NS2) was used as the searching model. The peptide model of was built using Coot, and the complex structure was refined using PHENIX and Coot (24, 25). TLS refinements were applied to improve the electron density map. Detailed statistics of crystallographic analyses are shown in Table S1. Structure figures were prepared using PyMol (www.pymol.org), and the coordinates were deposited in PDB under ID code 4H75.
ITC Measurements and rRNA Rranscription Assay.
Spindlin1 mutants were generated by site-directed mutagenesis. The H3K4me3 peptide used for ITC analysis spans residues 1–10 of histone H3. Peptide binding ITC measurements and RT-PCR detection of rRNA expression were carried out following the procedures we previously described (7). For each ITC measurement, 1 × 0.5 μL followed by 19 × 2 μL of H3K4me3 peptide were titrated into a 250 μL protein sample. Each protein sample, except those that showed no bindings, was measured at least twice. The data were analyzed using the ORIGIN software (MicroCal Inc.), and the ones with the best fittings are represented here.
Supplementary Material
Acknowledgments
We thank the beamline scientists at Shanghai Synchrotron Radiation Facility for technical support during data collection and Menglong Hu and Ping Chen for assistance with ITC and analytic ultracentrifugation experiments. This work was supported by Ministry of Science and Technology Grants 2009CB825501 and 2010CB944903 (to R.M.X.), 2012CB910702 (to N.Y.), and 2011CB812700 and 2011CB965300 (to B.Z.); the Natural Science Foundation of China (90919029 and 3098801), and the Chinese Academy of Sciences (CAS). R.M.X. holds a CAS-Novo Nordisk Great Wall Professorship. B.Z. is an International Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4H75).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208517109/-/DCSupplemental.
References
- 1.Wang Y, et al. Beyond the double helix: Writing and reading the histone code. Novartis Found Symp. 2004;259:3–17. discussion 17–21, 163–169. [PubMed] [Google Scholar]
- 2.Maurer-Stroh S, et al. The tudor domain ‘royal family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28(2):69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 3.Adams-Cioaba MA, Min J. Structure and function of histone methylation binding proteins. Biochem Cell Biol. 2009;87(1):93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 4.Kutateladze TG. SnapShot: Histone readers. Cell. 2011;146(5):842–842 e841. doi: 10.1016/j.cell.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuo AJ, et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484(7392):115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12(11):1160–1166. doi: 10.1038/embor.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Q, et al. Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation. J Biol Chem. 2007;282(1):647–656. doi: 10.1074/jbc.M604029200. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312(5774):748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Thompson JR, Botuyan MV, Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat Struct Mol Biol. 2008;15(1):109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127(7):1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian C, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30(14):2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nady N, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem. 2011;286(27):24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blus BJ, Wiggins K, Khorasanizadeh S. Epigenetic virtues of chromodomains. Crit Rev Biochem Mol Biol. 2011;46(6):507–526. doi: 10.3109/10409238.2011.619164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442(7098):91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selenko P, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8(1):27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, et al. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24(17):1876–1881. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106(48):20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripsianes K, et al. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol. 2011;18(12):1414–1420. doi: 10.1038/nsmb.2185. [DOI] [PubMed] [Google Scholar]
- 20.Liu K, et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS ONE. 2012;7(2):e30375. doi: 10.1371/journal.pone.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang F, et al. Expression, purification, crystallization and preliminary X-ray analysis of human spindlin1, an ovarian cancer-related protein. Protein Pept Lett. 2006;13(2):203–205. doi: 10.2174/092986606775101661. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.