Abstract
Cytosolic bacterial pathogens require extensive metabolic adaptations within the host to replicate intracellularly and cause disease. In phagocytic cells such as macrophages, these pathogens must respond rapidly to nutrient limitation within the harsh environment of the phagosome. Many cytosolic pathogens escape the phagosome quickly (15–60 min) and thereby subvert this host defense, reaching the cytosol where they can replicate. Although a great deal of research has focused on strategies used by bacteria to resist antimicrobial phagosomal defenses and transiently pass through this compartment, the metabolic requirements of bacteria in the phagosome are largely uncharacterized. We previously identified a Francisella protein, FTN_0818, as being essential for intracellular replication and involved in virulence in vivo. We now show that FTN_0818 is involved in biotin biosynthesis and required for rapid escape from the Francisella-containing phagosome (FCP). Addition of biotin complemented the phagosomal escape defect of the FTN_0818 mutant, demonstrating that biotin is critical for promoting rapid escape during the short time that the bacteria are in the phagosome. Biotin also rescued the attenuation of the FTN_0818 mutant during infection in vitro and in vivo, highlighting the importance of this process. The key role of biotin in phagosomal escape implies biotin may be a limiting factor during infection. We demonstrate that a bacterial metabolite is required for phagosomal escape of an intracellular pathogen, providing insight into the link between bacterial metabolism and virulence, likely serving as a paradigm for other cytosolic pathogens.
Subversion of the hostile phagosomal environment is required for the survival of intracellular bacteria. Although bacterial strategies to resist antimicrobial phagosomal defenses have been studied in great detail (1, 2), the ways in which bacteria counter phagosomal nutrient limitation are largely unknown. This is especially true for cytosolic pathogens that are often in the phagosome for a very limited time (15–60 min), before escaping this compartment to reach their replicative niche in the cytoplasm. During this brief and dynamic time, it is unclear if cytosolic pathogens require sequestration of nutrients or synthesis of de novo metabolites to promote their virulence strategies and escape the toxic phagosome.
Francisella tularensis is a cytosolic intracellular Gram-negative bacterial pathogen that uses a multitude of mechanisms to evade phagosomal host defenses (3). This pathogen is highly virulent and causes the potentially fatal disease tularemia. Francisella novicida U112 and Francisella holarctica LVS (live vaccine strain) are less virulent yet highly related strains that are often used as models to study F. tularensis. Like other cytosolic bacterial pathogens, after initial contact with the host macrophage, Francisella spp. are taken up into a phagosome and rapidly escape (30–60 min) this compartment to reach and replicate within the cytosol (3–5). The mechanism by which Francisella escapes the Francisella-containing phagosome (FCP) is unknown; however, this process requires expression of the Francisella pathogenicity island (FPI), a cluster of 17 genes encoding a putative type VI secretion system (T6SS) (6–8).
We previously identified FTN_0818, a hypothetical protein with no known function, as one of the most critical genes for F. novicida replication in mouse macrophages (9). We also identified FTN_0818 as being required for infection of mice using an unbiased genome-wide, in vivo negative selection screen (10), a finding later supported by another group as well (11). FTN_0818 was also identified in an intracellular replication screen in arthropod-derived cells (12). Here, we characterize FTN_0818 and highlight an adaptation of Francisella to the FCP by linking intraphagosomal metabolic requirements with rapid escape from this compartment.
Our studies demonstrate that FTN_0818 is required for growth in nutrient-limiting environments, and by use of a phenotypic microarray, we identified the enzymatic cofactor biotin as being able to fully complement the growth defect of the FTN_0818 mutant. The addition of exogenous biotin alleviated the requirement of FTN_0818 for rapid FCP escape, intracellular replication, and pathogenesis in mice. Our data suggest that biotin may be a limiting factor that, when absent, restricts cytosolic pathogens to the phagosome, blocking their escape and preventing them from reaching their replicative niche in the cytoplasm. We show that bacterial metabolism within the phagosome is vital for rapid phagosomal escape and likely serves as a paradigm for other cytosolic bacterial pathogens.
Results
FTN_0818 Is Required for Rapid Escape from the FCP and Intracellular Replication.
The screens that identified FTN_0818 as being required for Francisella virulence used transposon insertion mutants that can have defects in genes other than the one targeted. We therefore wanted to validate the identification of FTN_0818 and constructed a clean deletion mutant in F. novicida (ΔFTN_0818). We infected macrophages and found that at 7.5 h postinfection (pi), wild-type (WT) bacteria replicated almost 10-fold, whereas ΔFTN_0818 was unable to replicate (Fig. S1). To ensure that this phenotype was attributable solely to deletion of FTN_0818 and not an unknown second-site mutation, we complemented the deletion strain with a WT copy of FTN_0818. The complemented strain replicated to levels similar to the WT (Fig. S1). These data confirm that FTN_0818 is indeed required for F. novicida replication in macrophages.
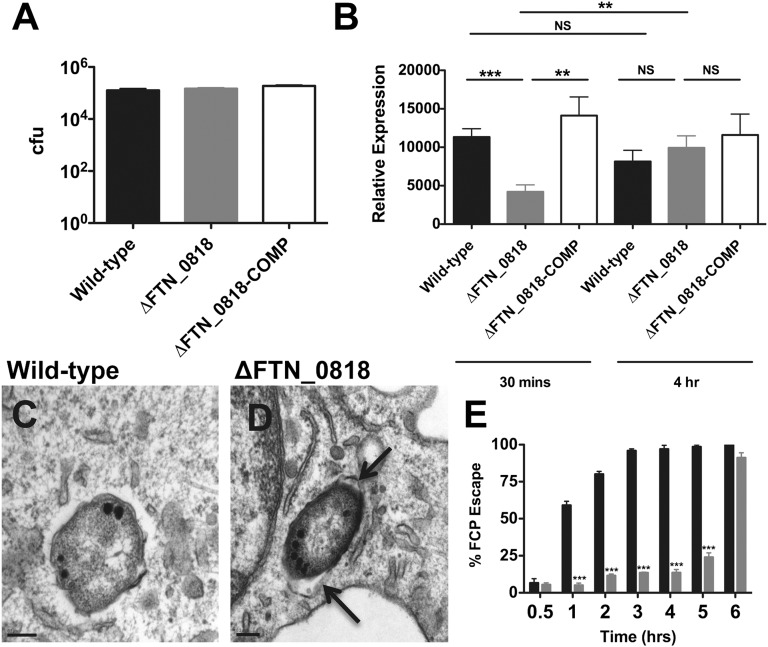
Several steps are required for Francisella replication in macrophages including passage through the highly nutrient-limiting FCP (13), and we set out to determine at which step ∆FTN_0818 was defective. To test whether ∆FTN_0818 had a deficiency in entry, we infected macrophages and determined the levels of intracellular colony-forming units at 30 min pi, before any bacterial replication occurs. WT, ∆FTN_0818, and the complemented strain were present at similar levels (Fig. 1A), demonstrating that FTN_0818 is not required for initial uptake of F. novicida by macrophages.
Fig. 1.
FTN_0818 is required for rapid phagosomal escape. (A and B) Macrophages were infected with the indicated strains, and colony-forming units were quantified at 30 min pi (A) or qRT-PCR was used to measure the expression of iglA and normalized to the expression of uvrD at 30 min and 4 h pi (B). (C and D) Transmission electron microscopy of infected macrophages at 3 h pi (arrows, intact FCP). (E) Phagosomal escape of WT (black) and ΔFTN_0818 (gray) was quantified 30 min to 6 h pi. One hundred bacteria per condition were viewed and the percentage of phagosomal escape was determined for three independent experiments. *P < 0.05; **P < 0.001; ***P < 0.0001.
Escape from the FCP is essential for Francisella to evade this nonpermissive environment to successfully replicate in the cytosol (4), and this process requires the expression of Francisella pathogenicity island (FPI) genes. We, therefore, measured the expression of the FPI gene iglA during macrophage infection with either the WT or ∆FTN_0818 strain. At 30 min pi, iglA expression in the ∆FTN_0818 mutant was significantly lower than that in the WT strain, although its expression increased by 4 h pi (Fig. 1B). The kinetics of FCP escape correlated with this iglA expression defect. At 30 min pi, both WT and ΔFTN_0818 were almost exclusively (>95%) within phagosomes (Fig. 1E and Fig. S2). At 3 h pi, WT had largely escaped as >95% of the bacteria were cytosolic (Fig. 1 C and E), whereas ΔFTN_0818 was still almost completely retained within the FCP (Fig. 1 D and E). However, ΔFTN_0818 escaped the FCP at 6 h pi after iglA expression increased in this strain (Fig. 1E). These results indicate that FTN_0818 is required for WT expression of an FPI gene early in infection and subsequent rapid escape from the FCP, correlating with the severe growth defect of the ∆FTN_0818 mutant during macrophage infection.
FTN_0818 Plays a Role in Biotin Metabolism.
Because FTN_0818 is required for regulation of iglA in the nutrient-limiting FCP, a process critical for escape from this compartment (Fig. 1B), and recent literature has emphasized the importance of the metabolic state of Francisella for virulence (14), we hypothesized that FTN_0818 may play a role in the acquisition of nutrients or production of metabolites. To determine whether FTN_0818 might be involved in these processes, we compared the growth of ΔFTN_0818 in rich [tryptic soy broth (TSB)] and defined minimal medium [Chamberlain’s medium (CHB; Table S1)] (15). We found that ΔFTN_0818 replicated to WT levels in TSB (Fig. S3A); however, it exhibited a severe growth defect in CHB in comparison with the WT and complemented strains (Fig. S3B). These data demonstrate that FTN_0818 is specifically required for growth in a nutrient-limiting environment (13), suggesting that it may contribute to the acquisition and/or biosynthesis of nutrients that are required for growth in these conditions.
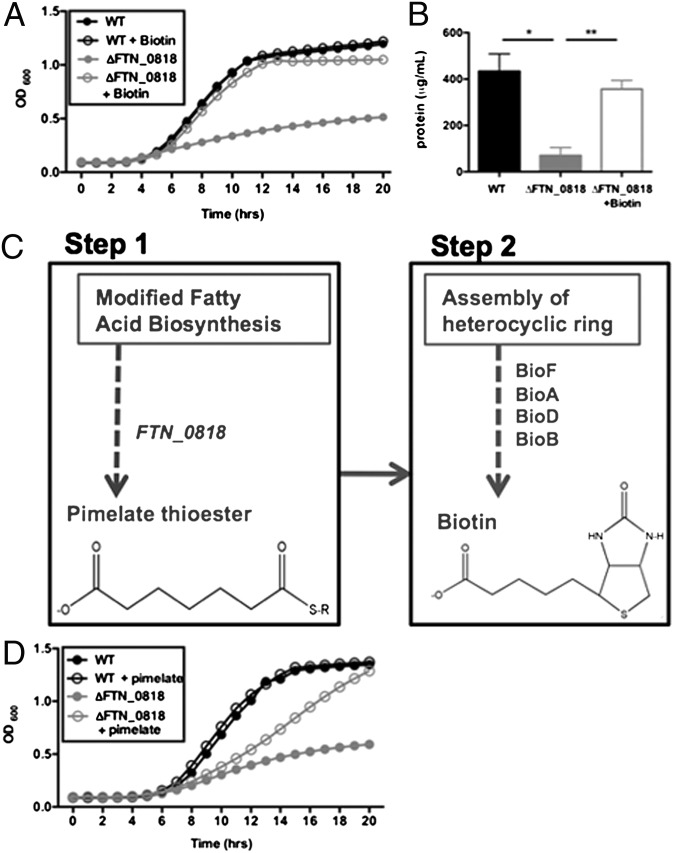
To determine whether a specific metabolite could complement the growth defect of ∆FTN_0818 in minimal media, we used a Biolog Phenotypic Microarray. As expected, the WT strain grew well in minimal medium (modified CHB), whereas the FTN_0818 mutant did not (Fig. S4). Only biotin was able to complement growth of the FTN_0818 mutant (Fig. S4). We further validated these results, showing that biotin complemented ΔFTN_0818 growth in CHB (Fig. 2A). These data suggest that the FTN_0818 mutant has insufficient levels of biotin and that FTN_0818 is involved in the acquisition or synthesis of biotin in F. novicida.
Fig. 2.
Biotin complements the ∆FTN_0818 growth defect in minimal media. (A) WT and ΔFTN_0818 were grown in CHB with or without biotin and the OD600 was measured every hour. (B) The concentration of biotinylated proteins in whole cell lysates of all strains was quantified after immunoprecipitation with anti-biotin antibodies. *P < 0.05; **P < 0.001. (C) Schematic of the biotin biosynthesis pathway with the proposed placement of FTN_0818 (steps before the generation of pimelate have not been defined in Francisella). (D) WT and ΔFTN_0818 were grown in CHB with or without pimelate and the OD600 was measured every hour.
Biotin is required for numerous metabolic pathways and is covalently attached (biotinylation) to proteins to facilitate their activity. Therefore, one method for quantifying biotin levels in bacteria is to measure the level of biotinylated proteins. Using immunoprecipitation with streptavidin, we quantified the total concentration of biotinylated proteins and detected much lower levels in the FTN_0818 mutant compared with WT (Fig. 2B). Furthermore, exogenous addition of biotin to CHB restored the levels of biotinylated proteins in ΔFTN_0818 to those of the WT. Therefore, these data further suggest that the FTN_0818 mutant has a biotin deficiency.
Biotin biosynthesis in E. coli consists of two major steps: the well-characterized latter step involves the synthesis of two fused heterocyclic rings on a valeryl side chain, and the first step is dedicated to the acquisition of a pimelate moiety, which is required to generate the aforementioned valeryl side chain (Fig. 2C) (16). To gain an indication of where FTN_0818 is required in the pathway, we tested whether pimelate could complement the growth defect of ∆FTN_0818 in CHB. Interestingly, when pimelate was added to CHB, it rescued the ∆FTN_0818 growth defect with a minor delay (Fig. 2D). These data suggest that FTN_0818 is required for the production of pimelate and subsequent biotin biosynthesis.
Biotin Alleviates the Requirement of FTN_0818 for Phagosomal Escape and Replication in Macrophages.
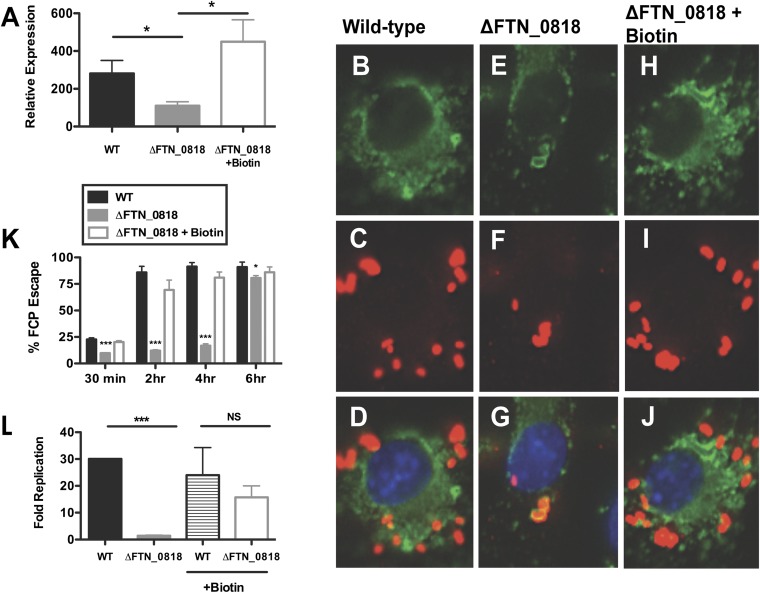
We next tested whether exogenous biotin could also rescue the intracellular defects of the ΔFTN_0818 mutant. At 30 min pi, exogenous biotin complemented iglA expression in the ∆FTN_0818 mutant (Fig. 3A). We used immunofluorescence microscopy to determine whether FCP escape kinetics correlated with the rescue of iglA expression in the presence of biotin. We observed that ∆FTN_0818 had a phagosomal escape defect (Fig. 3 B–G and K), similar to our previous results using electron microscopy (Fig. 1 C–E). At 30 min pi, biotin-supplemented ΔFTN_0818 localized to the FCP (Fig. 3K). However, at 2 h pi, this strain was within the cytosol (Fig. 3 H–K), similar to WT. These data clearly demonstrate that biotin is required for the rapid escape of Francisella from the FCP.
Fig. 3.
Biotin rescues rapid phagosomal escape and the ΔFTN_0818 replication defect in macrophages. (A–J) Macrophages were infected, and qRT-PCR was used to measure the expression of iglA and normalized to the expression of uvrD at 30 min pi (*P < 0.05) (A) and immunofluorescence microscopy was used to determine escape kinetics of WT (B–D), ΔFTN_0818 (E–G), and ΔFTN_0818 supplemented with biotin (H–J) 2 h pi (FITC-stained LAMP-1, green; anti-Francisella, red; DAPI, blue). (K) Two hundred bacteria were counted per sample, and colocalization with lysosomal-associated membrane protein 1 (LAMP-1) was used as a marker for phagosomal localization. *P < 0.05; ***P < 0.0001. (L) Macrophages were infected with WT or ΔFTN_0818 strains in media with or without biotin. Colony-forming units were quantified 30 min and 6 h pi, and fold replication was calculated.
Because biotin rescued iglA gene expression and subsequent escape of the FTN_0818 mutant, and escape is required for intracellular replication, we tested whether biotin could also rescue replication. During macrophage infection, the WT strain replicated nearly 30-fold, whereas ΔFTN_0818 exhibited a severe replication defect (Fig. 3L), in agreement with our previous data (Fig. S1). However, when biotin was added to the macrophages at the time of infection, the ΔFTN_0818 replication defect was significantly complemented (Fig. 3L). We further tested whether pretreatment with biotin before infection would rescue the intracellular growth defect of the FTN_0818 mutant, or whether biotin had to be present during the infection. ΔFTN_0818 grown in CHB supplemented with biotin overnight, but without exogenous biotin during infection, was unable to replicate in macrophages (Fig. S5). This demonstrates that biotin must be present at the time of infection to facilitate replication. These data show that biotin is required to promote escape when the bacteria are present within the FCP.
FTN_0818 Is Required for FCP Escape in Multiple Francisella Species.
To determine whether the role of FTN_0818 was conserved in other Francisella species, we first generated a deletion mutant lacking the FTN_0818 ortholog, FTT_0941 (99% amino acid identity), in the human pathogenic Francisella tularensis strain SchuS4. Similar to our findings with F. novicida, the FTT_0941 mutant in F. tularensis had a defect in escape from the FCP (Fig. S6). However, when biotin was added to the media, the FTT_0941 mutant escaped with WT kinetics (Fig. S6). We also generated and tested a mutant in the live vaccine strain (LVS), a derivative of highly pathogenic F. holarctica. We found that the FTN_0818 ortholog, FTL_1266 (99% amino acid identity), was also required for LVS escape from the phagosome, as well as growth in minimal media, and that these phenotypes were complemented by biotin (Fig. S7 A–D). Furthermore, FTL_1266 was also required for replication in macrophages (Fig. S7E), in agreement with the role of FTN_0818 in F. novicida. Together, these data highlight the conserved role of FTN_0818 in multiple Francisella species.
FTN_0818 Is Necessary for Pathogenesis in Mice, and This Requirement Is Alleviated by Biotin.
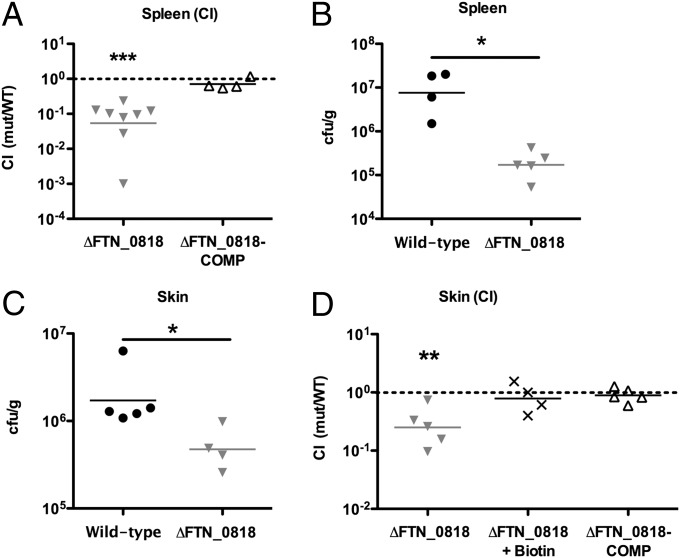
We and others identified FTN_0818 as being required for Francisella virulence in mice using in vivo screens (10, 11). To validate these findings, we performed competition experiments in which a 1:1 mixture of the WT and ΔFTN_0818 or the complemented strain was used to infect mice. Forty-eight hours pi, ΔFTN_0818 levels were 1–2 logs lower in spleens compared with WT (Fig. 4A). In contrast, the complemented strain colonized the spleen of mice similarly to WT bacteria (Fig. 4A). We also infected mice with the WT or ΔFTN_0818 strain separately and determined that ΔFTN_0818 was attenuated 100-fold in the spleen (Fig. 4B) and almost 10-fold in the skin (Fig. 4C), compared with WT. In agreement, the FTN_0818 ortholog, FTL_1266, was required to reach WT LVS levels in spleens 48 h pi (Fig. S7F). Together, these results demonstrate the requirement of FTN_0818 for Francisella virulence in vivo.
Fig. 4.
Biotin rescues the ∆FTN_0818 virulence defect in vivo. (A) Mice were infected s.c. with a 1:1 mixture of WT with the ΔFTN_0818 or the ΔFTN_0818 complemented strain (ΔFTN_0818-COMP). At 48 h pi, spleens were harvested to quantify bacterial levels, and the CI was calculated. (B and C) Mice were s.c. infected with 106 cfu of WT or ΔFTN_0818. At 48 h pi, the spleen (B) and skin at the site of infection (C) were harvested and bacterial levels quantified. (D) A competition assay was performed with WT and ΔFTN_0818 in the absence of biotin or WT and the ΔFTN_0818 complemented strain in the presence of biotin. At 24 h pi, the skin at the site of infection was harvested to quantify bacterial levels, and the CI was calculated. *P < 0.05; **P < 0.001; ***P < 0.0001.
To determine whether exogenous biotin could rescue the attenuation of the FTN_0818 mutant during in vivo infection, as we observed during macrophage infection, we added biotin to the inoculum. ΔFTN_0818 without biotin was attenuated nearly 10-fold compared with WT in the skin at the site of infection, whereas when biotin was added, ΔFTN_0818 was present at WT levels (Fig. 4D). Furthermore, addition of biotin resulted in rescue to levels similar as genetic complementation, as observed with the complemented strain (Fig. 4D). These results confirm that FTN_0818 is required for virulence in mice and that biotin can alleviate this requirement. Taken together, we have characterized a metabolic protein that links the requirement for biotin in the phagosome with rapid phagosomal escape and virulence in vivo.
Discussion
Evasion of the harsh phagosomal environment is imperative for the survival of intracellular bacterial pathogens. We have characterized a metabolic protein, FTN_0818, revealing a unique link between metabolism and rapid escape from the FCP during F. novicida infection of macrophages. Exogenous biotin overrode the requirement of FTN_0818 for rapid phagosomal escape, replication in macrophages, and in vivo pathogenesis. Pretreatment with biotin before infection of macrophages was unable to complement the mutant strain. However, when the mutant was microinjected with biotin into the host cytosol (bypassing the phagosome), or when biotin was added at 6 h (after the mutant escaped the phagosome), the mutant's replication defect was rescued (Fig. S8 A and B). This suggests that Francisella requires biotin in the FCP to promote rapid escape and in the cytosol for intracellular replication. These data contribute to current literature highlighting the link between Francisella metabolism and virulence (3). Specifically, it has been shown that utilization of glutathione as a cysteine source is required for intracellular replication (14). Similarly, utilization of uracil has been shown to be required for inhibition of the neutrophil respiratory burst (17). It would be interesting to delineate the full metabolic requirements of Francisella within host cells and, specifically, to determine how these control phagosomal escape and other virulence traits.
In support of our current data, biotin biosynthetic genes have been identified as being important for Francisella replication in vitro and in vivo (9, 10, 18). In addition, Wehrly et al. previously published a transcriptional profile of F. tularensis within the macrophage and identified bioB, a gene required for step 2 (Fig. 2C) of biotin biosynthesis, as being up-regulated (19). Additionally, Asare and Abu Kwaik published a screen for mutants with defects in phagosomal escape and identified birA, a biotin associated gene (18). Taken together, these data provide additional evidence that biotin, and biotin-associated genes, play important roles during intracellular infection by Francisella.
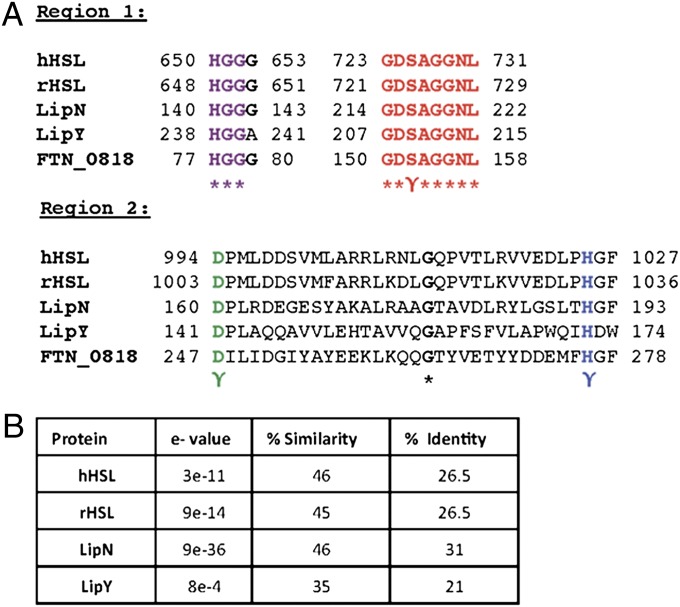
Bioinformatic analysis revealed that FTN_0818 shares high sequence similarity with the hormone-sensitive lipase (HSL) superfamily of proteins. Mammalian HSL family proteins hydrolyze triacylglycerols for release into the circulation to provide energy for other tissues (20). They are also the rate-limiting enzyme in the mobilization of free fatty acids and are, therefore, critical for lipid metabolism and energy homeostasis (21, 22). Interestingly, most HSL family proteins characterized in Mycobacterium tuberculosis are also required for utilization of stored triacylglycerols under starvation conditions (23). Alignment of the amino acid sequence of FTN_0818 with human HSL, rat HSL, and the M. tuberculosis HSL family proteins LipN and LipY revealed two regions that contained conserved active site residues (Fig. 5A) (22, 23). Additionally, these HSL family proteins have between 21–31% identity and 35–46% similarity with FTN_0818 (Fig. 5B). The first region (FTN_0818, amino acids 77–158) contains the characteristic HGGG motif present in most HSL family proteins and the GDSAGGNL motif that includes the catalytic serine residue (Fig. 5A) (24). The second region (FTN_0818, amino acids 247–278) includes the conserved aspartate and histidine catalytic residues (Fig. 5A) (24). These data show that critical catalytic residues conserved in HSL family proteins are present in FTN_0818 and suggest that this protein may act as an HSL.
Fig. 5.
FTN_0818 is a putative HSL family protein. (A) CLUSTAL multisequence alignment including mammalian HSL proteins (human, GenBank accession no. NP_055348.2; rat, GenBank accession no. NP_036991.1) and the Mycobacterium tuberculosis HSL family proteins LipY (GenBank accession no. YP_177924.1) and LipN (GenBank accession no. CAB05441.1), in two regions, including the conserved residues (γ) of the catalytic triad (serine, red; aspartic acid, green; histidine, blue) and HSL family protein amino acid motif HGGG (purple). (B) Percentage identity, percentage similarity, and e value of HSL family proteins to FTN_0818.
Interestingly, we showed that when the putative catalytic serine (S151) in FTN_0818 was mutated to an alanine residue, Francisella could no longer grow in minimal media (this phenotype was rescued by exogenous biotin) (Fig. S9A). This was not attributable to a decrease in the level of expression of the point mutant compared with WT FTN_0818 (Fig. S9B). Furthermore, disruption of this catalytic residue led to retention of Francisella within the FCP (which could be rescued by the addition of biotin) (Fig. S9C), and inhibition of replication in macrophages and during in vivo infection (Fig. S9 D and E). These data provide further support for the hypothesis that FTN_0818 is an HSL family protein.
The role of FTN_0818 in biotin metabolism and its homology to HSL family lipases raises the question of how fatty acid metabolism might contribute to biotin biosynthesis. The link between fatty acid metabolism and biotin biosynthesis has long been unclear but recent insights have been made. Recently, Lin et al. demonstrated that pimelate is the product of a modified fatty acid synthesis pathway in E. coli (Fig. 2C) (16, 25). The fact that (i) fatty acid metabolism has been shown to play an important role in generating the pimelate intermediate required for biotin biosynthesis, (ii) FTN_0818 has homology to the HSL family of lipases that cleave triacylglycerols, (iii) the FTN_0818 catalytic serine point mutant abolishes function of the protein, and (iv) pimelate and biotin rescue the growth defect of the FTN_0818 mutant, together, strongly suggest that FTN_0818 is an HSL family member that acts early in the biotin metabolic pathway to liberate free fatty acids for biotin biosynthesis.
Taken together, the work presented here strongly suggests that biotin availability may be a limiting factor for Francisella spp., and likely other bacterial pathogens, during infection. Biotin has been reported as being required for Mycobacterium tuberculosis virulence in mice and Vibrio cholera colonization of the mouse intestine, both through unknown mechanisms (26–28). In addition, several antimicrobials target the biotin pathway by causing the degradation of biotin or biotin precursors including amiclenomycin, actithiazic acid, and the biotin analog α-dehydrobiotin (29–31), further demonstrating the importance of biotin during infection, as well as the therapeutic utility of limiting biotin availability to pathogens.
Our data show that biotin is required in the phagosome to promote rapid escape, suggesting that biotin is limiting in this compartment. Iron is also limiting in the phagosome and numerous host factors such as transferrin play a critical role in the control of infection by depleting phagosomal iron. Similarly, the host innate immune system has been shown to target biotin. Chicken embryo fibroblasts and yolk-sac macrophages induce the production of avidin, which binds and sequesters biotin in response to Escherichia coli infection, treatment with lipopolysaccharide (LPS), or interleukin-6 (32, 33). These data suggest that sequestration of biotin may be a form of nutritional immunity by the host innate immune system and support the idea that biotin might be a critical and limited commodity during infection. Sequestration of biotin could restrict cytosolic pathogens to the phagosome, blocking their escape and preventing them from reaching their replicative niche in the cytoplasm. Understanding more about how specific bacterial metabolites are generated and how the host attempts to sequester these compounds could provide insight into host-pathogen interactions and may reveal targets for the development of antimicrobials to inhibit bacteria at an early step in pathogenesis and combat infection.
Materials and Methods
WT F. novicida strain U112 and F. holarctica LVS growth conditions were described previously (9), and F. tularensis (SchuS4) growth conditions are described in SI Materials and Methods. Details of the construction of mutant/complemented strains and growth curve protocols are in SI Materials and Methods. Macrophage preparation and infections described in SI Materials and Methods. RNA was collected during macrophage infections as described previously (9). Quantitative (q)RT-PCR (real-time PCR) was performed with the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems) and primers (Table S2) using the StepOnePlus Real-time PCR System (Applied Biosystems). Immunoprecipitation and microscopy complete descriptions found in SI Materials and Methods. For mouse infections, female C57BL/6 mice (6–8 wk) (Jackson Laboratory) were housed under specific pathogen-free housing at Emory University. Experimental studies were performed in accordance with the Institutional Animal Care and Use Committee guidelines. Competitive index (CI) [(mutant output/WT output)/(mutant input/WT input)] and infections with single strains were carried out as described previously (9). Statistical analysis for CI experiments was as described previously (10). Macrophage experiments were analyzed by using the Student’s unpaired t test (in escape experiments, average percentage escape per strain for three independent experiments were compared).
Supplementary Material
Acknowledgments
We thank Colin Manoil and Beth Ramage for help with the Biolog array; Patrik Rydén (Umeä University) for statistical analysis of the microinjection data; and Hong Yi for help with electron microscopy (Emory Robert P. Apkarian Integrated EM Core). This work was supported by National Institutes of Health Grant U54 AI057157 [from the Southeast Regional Center of Excellence for Emerging Infections and Biodefense (SERCEB)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206411109/-/DCSupplemental.
References
- 1.Skeiky YA, Sadoff JC. Advances in tuberculosis vaccine strategies. Nat Rev Microbiol. 2006;4(6):469–476. doi: 10.1038/nrmicro1419. [DOI] [PubMed] [Google Scholar]
- 2.Flannagan RS, Cosío G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol. 2009;7(5):355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 3.Meibom KL, Charbit A. Francisella tularensis metabolism and its relation to virulence. Front Microbiol. 2010;1:140. doi: 10.3389/fmicb.2010.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71(10):5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong A, Celli J. The francisella intracellular life cycle: Toward molecular mechanisms of intracellular survival and proliferation. Front Microbiol. 2010;1:138. doi: 10.3389/fmicb.2010.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bruin OM, et al. The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology. 2011;157(Pt 12):3483–3491. doi: 10.1099/mic.0.052308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker JR, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74(6):1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meibom KL, Charbit A. The unraveling panoply of Francisella tularensis virulence attributes. Curr Opin Microbiol. 2010;13(1):11–17. doi: 10.1016/j.mib.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Llewellyn AC, Jones CL, Napier BA, Bina JE, Weiss DS. Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE. 2011;6(9):e24201. doi: 10.1371/journal.pone.0024201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiss DS, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci USA. 2007;104(14):6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su J, et al. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun. 2007;75(6):3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asare R, Akimana C, Jones S, Abu Kwaik Y. Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ Microbiol. 2010;12(9):2587–2612. doi: 10.1111/j.1462-2920.2010.02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Headley VL, Payne SM. Differential protein expression by Shigella flexneri in intracellular and extracellular environments. Proc Natl Acad Sci USA. 1990;87(11):4179–4183. doi: 10.1073/pnas.87.11.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alkhuder K, Meibom KL, Dubail I, Dupuis M, Charbit A. Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 2009;5(1):e1000284. doi: 10.1371/journal.ppat.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamberlain RE. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin S, Cronan JE. Closing in on complete pathways of biotin biosynthesis. Mol Biosyst. 2011;7(6):1811–1821. doi: 10.1039/c1mb05022b. [DOI] [PubMed] [Google Scholar]
- 17.Schulert GS, et al. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect Immun. 2009;77(4):1324–1336. doi: 10.1128/IAI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asare R, Abu Kwaik Y. Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ Microbiol. 2010;12(9):2559–2586. doi: 10.1111/j.1462-2920.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wehrly TD, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;11(7):1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeaman SJ. Hormone-sensitive lipase—new roles for an old enzyme. Biochem J. 2004;379(Pt 1):11–22. doi: 10.1042/BJ20031811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osterlund T. Structure-function relationships of hormone-sensitive lipase. Eur J Biochem. 2001;268(7):1899–1907. doi: 10.1046/j.1432-1327.2001.02097.x. [DOI] [PubMed] [Google Scholar]
- 22.Shen WJ, Sridhar K, Bernlohr DA, Kraemer FB. Interaction of rat hormone-sensitive lipase with adipocyte lipid-binding protein. Proc Natl Acad Sci USA. 1999;96(10):5528–5532. doi: 10.1073/pnas.96.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deb C, et al. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J Biol Chem. 2006;281(7):3866–3875. doi: 10.1074/jbc.M505556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaya S, Koyanagi T, Kanaya E. An esterase from Escherichia coli with a sequence similarity to hormone-sensitive lipase. Biochem J. 1998;332(Pt 1):75–80. doi: 10.1042/bj3320075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin S, Hanson RE, Cronan JE. Biotin synthesis begins by hijacking the fatty acid synthetic pathway. Nat Chem Biol. 2010;6(9):682–688. doi: 10.1038/nchembio.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100(22):12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salaemae W, Azhar A, Booker GW, Polyak SW. Biotin biosynthesis in Mycobacterium tuberculosis: Physiology, biochemistry and molecular intervention. Protein Cell. 2011;2(9):691–695. doi: 10.1007/s13238-011-1100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang SL, Mekalanos JJ. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27(4):797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 29.Kitahara T, Hotta K, Yoshida M, Okami Y. Biological studies of amiclenomycin. J Antibiot (Tokyo) 1975;28(3):215–221. doi: 10.7164/antibiotics.28.215. [DOI] [PubMed] [Google Scholar]
- 30.Eisenberg MA, Hsiung SC. Mode of action of the biotin antimetabolites actithiazic acid and alpha-methyldethiobiotin. Antimicrob Agents Chemother. 1982;21(1):5–10. doi: 10.1128/aac.21.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piffeteau A, Dufour MN, Zamboni M, Gaudry M, Marquet A. Mechanism of the antibiotic action of alpha-dehydrobiotin. Biochemistry. 1980;19(13):3069–3073. doi: 10.1021/bi00554a036. [DOI] [PubMed] [Google Scholar]
- 32.Elo HA, Korpela J. The occurrence and production of avidin: A new conception of the high-affinity biotin-binding protein. Comp Biochem Physiol B. 1984;78(1):15–20. doi: 10.1016/0305-0491(84)90137-8. [DOI] [PubMed] [Google Scholar]
- 33.Zerega B, et al. Avidin expression during chick chondrocyte and myoblast development in vitro and in vivo: Regulation of cell proliferation. J Cell Sci. 2001;114(Pt 8):1473–1482. doi: 10.1242/jcs.114.8.1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.