Abstract
In the presence of extensive DNA damage, eukaryotes activate endonucleases to fragment their chromosomes and induce apoptotic cell death. Apoptotic-like responses have recently been described in bacteria, but primarily in specialized mutant backgrounds, and the factors responsible for DNA damage-induced chromosome fragmentation and death have not been identified. Here we find that wild-type Caulobacter cells induce apoptotic-like cell death in response to extensive DNA damage. The bacterial apoptosis endonuclease (BapE) protein is induced by damage but not involved in DNA repair itself, and mediates this cell fate decision. BapE fragments chromosomes by cleaving supercoiled DNA in a sequence-nonspecific manner, thereby perturbing chromosome integrity both in vivo and in vitro. This damage-induced chromosome fragmentation pathway resembles that of eukaryotic apoptosis. We propose that damage-induced programmed cell death can be a primary stress response for some bacterial species, providing isogenic bacterial communities with advantages similar to those that apoptosis provides to multicellular organisms.
Cell survival depends on the reliable transmission of intact genomes to daughter cells. When DNA is damaged, broken DNA must be accurately repaired to protect progeny from the aberrant transmission of incorrect genetic information. What do cells do if DNA damage accumulates beyond repair? In animal cells, DNA damage activates a p53-dependent cell cycle checkpoint that stimulates repair in the presence of low levels of damage and induces apoptotic cell death in the presence of high levels of damage (1, 2). By contrast, unicellular organisms such as bacteria were generally thought to simply arrest division upon DNA damage (3). Recent studies have suggested that Escherichia coli can undergo an apoptotic-like response to bactericidal antibiotics, including those that cause DNA damage (4, 5). However, similar effects were also observed with non–DNA-damaging drugs, such that the relatedness of the apoptotic response to DNA damage itself remained unclear (4, 5). Furthermore, the factors that mediate the chromosome fragmentation observed during bacterial apoptosis remained unidentified.
Here we focus on the DNA damage response of Caulobacter crescentus because it lacks known lesion bypass pathways for copying extremely damaged DNA (6) and it has a eukaryotic-like cell cycle in which it replicates its DNA once and only once per cell division (7). Although the DNA damage response is less well characterized in Caulobacter than in other bacteria such as E. coli (8), a recent study identified SidA as an early SOS-induced division inhibitor in C. crescentus (9). However, SidA was found to only partially suppress damage-induced division (9), suggesting the existence of additional regulators of the C. crescentus DNA damage response. Here we demonstrate that wild-type Caulobacter cells undergo an apoptotic-like response to extensive DNA damage. We also identify BapE as a unique DNA endonuclease that is induced by the SOS DNA damage response pathway and leads to DNA fragmentation both in vivo and in vitro.
Results
DNA Damage Stimulates Apoptotic-Like Death in Caulobacter.
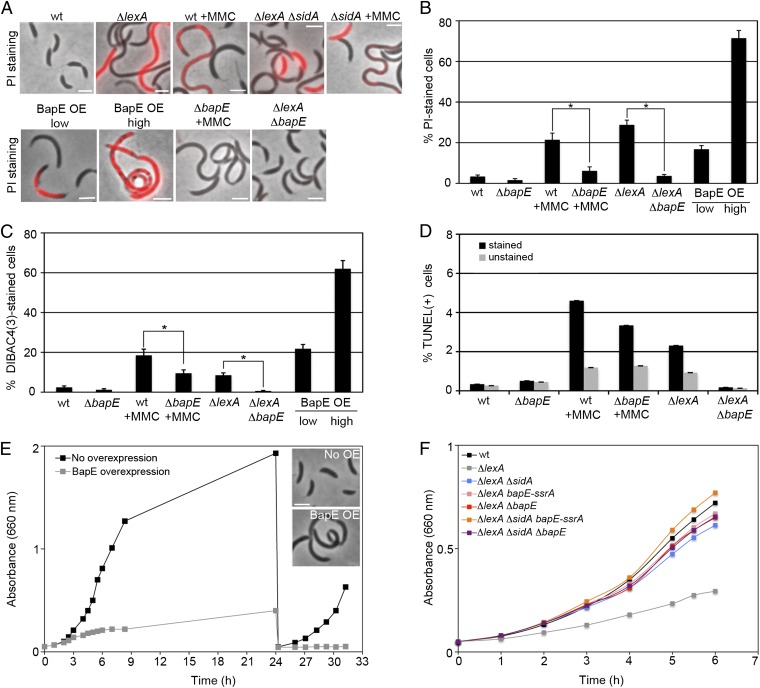
To quantitatively analyze the phenotypic consequences of activating the C. crescentus SOS response, we used two different strategies: deleting the lexA SOS repressor and treating cells with the DNA-crosslinking drug mitomycin C (MMC). C. crescentus ΔlexA mutants are viable and generate a heterogeneous population of cell sizes (8). In addition to displaying a significant number of filamentous cells, both the ΔlexA and MMC-treated cells displayed a significant subpopulation of cells that died (28 ± 2% and 21 ± 3%, respectively), as evidenced by their inability to exclude the membrane-impermeable DNA-binding dye propidium iodide (PI) (Fig. 1 A and B). Cell death is surprising given that SOS-induced bacterial lysis has generally been associated with prophage induction (10, 11), but Caulobacter laboratory strains do not host any temperate phage (12). Therefore, we tested the possibility that Caulobacter induced an apoptotic-like cell death pathway by staining these cells with two markers of apoptotic death, DIBAC4(3), a voltage-sensitive fluorescent dye that is stimulated by the membrane depolarization associated with eukaryotic apoptosis (13, 14), and TUNEL, which labels the ends of fragmented chromosomes and is also characteristic of eukaryotic apoptosis (15). We found that both MMC treatment and loss of lexA stimulated DIBAC4(3) and TUNEL staining (Fig. 1 A–D and Fig. S1), indicating that DNA damage stimulates an apoptotic-like response in these wild-type bacteria. A ΔsidA mutant did not significantly suppress the death phenotype, suggesting that this apoptosis-like response is induced by a novel factor (Fig. 1A and Fig. S2).
Fig. 1.
DNA-damage–induced BapE protein mediates apoptotic-like death. (A and B) Visualization and quantification of cell shape and PI-stained cells in mutant and MMC-treated cells. Asterisks denote significant differences of the percentage of PI-stained cells between MMC-treated wild-type and bapE mutant strains (P = 0.005) and ΔlexA and ΔlexA ΔbapE strains (P = 0.004). (C) Quantification of DIBAC4(3)-stained cells in mutant and MMC-treated cells. Asterisks denote significant differences of the percentage of DIBAC4(3)-stained cells between MMC-treated wild-type and bapE mutant strains (P = 0.01) and between ΔlexA and ΔlexA ΔbapE strains (P = 0.01). Microscopy images of DIBAC4(3)-stained cells are shown in Fig. S1A. (D) Quantification of TUNEL assays performed by fluorescence flow cytometry in mutant and MMC-treated cells. Due to a high-fluorescence background of nonapoptotic cells, control samples with no d-Transferase enzyme (unstained) were performed in parallel to samples treated with d-Transferase enzyme (stained). Flow cytometry plots are shown in Fig. S1B. (E) Growth curves of a BapE high-overexpression strain with and without xylose induction. ZG484 was grown in PYE supplemented with 0.3% xylose to induce BapE. After 24 h of growth, cultures were rediluted to an OD660 of 0.05 in fresh PYE medium with or without xylose. (Inset) Phase-contrast images from both cultures at time (t) = 24 h. (F) Growth curves of wild-type and ΔlexA mutant strains. (Scale bar, 2 μm in all images.) In all panels, error bars indicate SE of proportion.
BapE Is Sufficient and Necessary to Induce Apoptotic-Like Death.
To identify the factor that mediates DNA damage-induced apoptotic-like death, we screened for an SOS-induced protein (8) whose overexpression would induce death. We identified one such previously uncharacterized protein, CC0627, which we have renamed BapE for Bacterial Apoptosis Endonuclease. We confirmed that BapE is induced upon MMC treatment and in the absence of lexA (Fig. S3). BapE overexpression mimics the ΔlexA phenotype, causing reduced growth rate, cell filamentation, and cell death in a subpopulation of the cells (Fig. 1 A, B, E, and F and Fig. S1). BapE-induced cell death also resembled apoptosis in that it stimulated DIBAC4(3) staining (Fig. 1 C and D and Fig. S1).
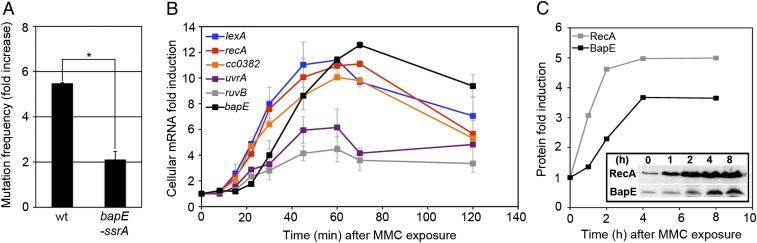
Because BapE overexpression is sufficient to induce apoptotic-like death, we next determined whether the presence of BapE is necessary for damage-induced apoptotic-like death. To lower BapE levels, we both dramatically reduced BapE levels by tagging the BapE C terminus with an SsrA tag that targets peptides for rapid proteolysis (bapE-ssrA) (16) and deleted the bapE gene (ΔbapE). BapE does not appear to function in the absence of DNA damage, because reducing BapE abundance with bapE-ssrA had no phenotype on its own and we could delete bapE under normal growth conditions (Fig. S4). BapE does, however, play a significant role in the C. crescentus DNA damage response because both bapE-ssrA and ΔbapE significantly suppressed the ΔlexA growth and cell death defects (Fig. 1 A–D and F and Fig. S1). ΔbapE also suppressed the PI, DIBAC4(3), TUNEL-positive, and CFU-measured cell death induced by either MMC treatment or ΔlexA, indicating that BapE is critical for the Caulobacter apoptotic-like response to DNA damage (Fig. 1 A–D and Fig. S1). The similarity among the results from these four different cell death assays supports the conclusion that BapE-related cell death is not an artifact of any particular method of measuring cell viability. Importantly, bapE-ssrA did not exhibit an elevated mutation frequency (Fig. 2A), indicating that BapE does not function as a DNA repair factor and thus appears to be a specific cell death-inducing factor.
Fig. 2.
BapE is induced after prolonged DNA damage but does not affect DNA repair. (A) Mutation frequency (fold increase) of wild-type and bapE-ssrA strains after MMC treatment. Error bars indicate SD. Asterisk denotes significant difference between MMC-treated wild-type and bapE-ssrA mutant strains (P = 0.04) (B) Dynamics of SOS-induced gene induction as measured by qRT-PCR. Fold induction of each gene as a function of time after the addition of MMC. Error bars represent SD. (C) The endogenous levels of BapE and RecA protein were determined by Western blotting after 0, 1, 2, 4, and 8 h of MMC treatment (Inset shows blot and plot shows fold induction).
As SidA is the only other Caulobacter SOS-induced regulator to be described, we examined the genetic interactions between sidA and bapE. The ΔsidA bapE-ssrA double mutant fully suppressed the ΔlexA filamentation and growth defect (Fig. 1F), whereas ΔsidA had no effect on the cell death induced by BapE overexpression (Fig. S2). Thus, BapE and SidA act redundantly to inhibit division and BapE stimulates cell death independently of SidA. Interestingly, the ΔsidA ΔbapE mutant only partially suppressed the ΔlexA growth defect (Fig. 1F), suggesting that yet more SOS-induced growth regulators remain to be identified.
BapE Is a Cell Fate Determinant That Induces Cell Death in a Concentration-Dependent Manner.
To determine when C. crescentus cells make the decision to promote repair or to induce cell death after DNA damage, we investigated the dynamics of the SOS response in C. crescentus (Fig. 2B). We used quantitative RT-PCR (qRT-PCR) to monitor the expression levels of multiple LexA-regulated genes involved in distinct repair pathways over time after exposure to MMC. Different subsets of SOS genes turn on with distinct kinetics and distinct induction levels, indicating that C. crescentus cells activate a graded response to DNA damage (Fig. 2B). By 15 min after MMC exposure, lexA, recA, and CC0382 (base excision repair pathway) genes are expressed, followed by uvrA (nucleotide excision repair pathway) and ruvB (homologous recombination after double-strand break pathway). Interestingly, bapE expression is induced even later and cannot be robustly detected until 30 min after MMC treatment. Although the expression levels of the other SOS genes plateau or decline by 45 min after MMC exposure, bapE induction continues to rise until 70 min after MMC treatment and remains high throughout the course of the experiment (Fig. 2B). The induction kinetics of the RecA and BapE protein levels upon MMC treatment mirror their gene expression kinetics (Fig. 2C). The late induction of BapE is consistent with a function in inducing death rather than in promoting repair.
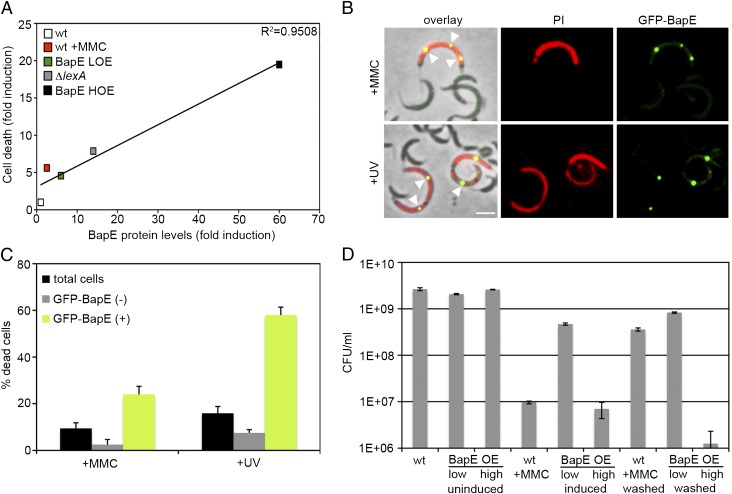
To test whether BapE induction has a differential effect on cell fate depending on the levels to which it accumulates, we determined the phenotypic consequences of weakly or strongly overexpressing BapE. Low-level BapE overexpression from a low-copy-number plasmid led to division arrest and significant cell death (∼4.5-fold increase compared with a wild-type strain). This extent of cell death is similar to that induced by ΔlexA (∼7.5-fold increase) or MMC treatment (∼5.5-fold increase). High-level BapE overexpression from a high-copy-number plasmid led to more severe division arrest and dramatic cell death (∼19-fold increase) (Fig. 3A and Fig. S5). Quantification of these findings revealed a strong positive correlation between the level of BapE protein induction and the frequency of cell death (Fig. 3A). Furthermore, using a strain expressing a GFP-BapE fusion from the endogenous bapE promoter, we found that individual cells that expressed high levels of BapE after prolonged MMC treatment or UV irradiation were significantly more likely to die (24 ± 3% and 58 ± 3% of dead cells after MMC and UV treatment, respectively) than individuals expressing low levels of BapE (2.5 ± 2% and 7.5 ± 1.5% of dead cells after MMC and UV treatment, respectively) (Fig. 3 B and C). Cell viability assays demonstrated that BapE consistently induced reversible division arrest at low induction levels, whereas high BapE induction led to irreversible cell death (Fig. 3D). Thus, BapE appears to induce cell death in a concentration-dependent manner.
Fig. 3.
BapE induces cell death in a concentration-dependent manner. (A) Plot of cell death extent (based on PI staining) as a function of BapE protein levels in various strains (raw data for each are shown in Figs. S3 and S5). (B) Images of cells expressing GFP-BapE from its endogenous promoter (ZG683), grown either in the presence of MMC or after UV irradiation (12 h) and stained with PI to assess death. Arrowheads point to GFP-BapE foci in dead cells. (C) Quantification of the number of dead cells described in B after MMC treatment (n = 119) or after UV irradiation (n = 542) among all cells (black bars), among cells without localized GFP-BapE (gray bars), and among cells with GFP-BapE localized to discrete foci (green bars). Error bars represent SE. (Scale bar, 2 μm.) (D) Cell viability counts for the wild-type, low-BapE overexpressing and high-BapE overexpressing strains after 4 h of growth in the presence of either MMC or xylose to induce BapE expression (induced) and after the strains are shifted in fresh medium with no inducer (washed) for 2 h. Control cultures (uninduced) were grown in parallel. All error bars represent SD.
BapE Is an Endonuclease That Fragments DNA in Vitro and in Vivo.
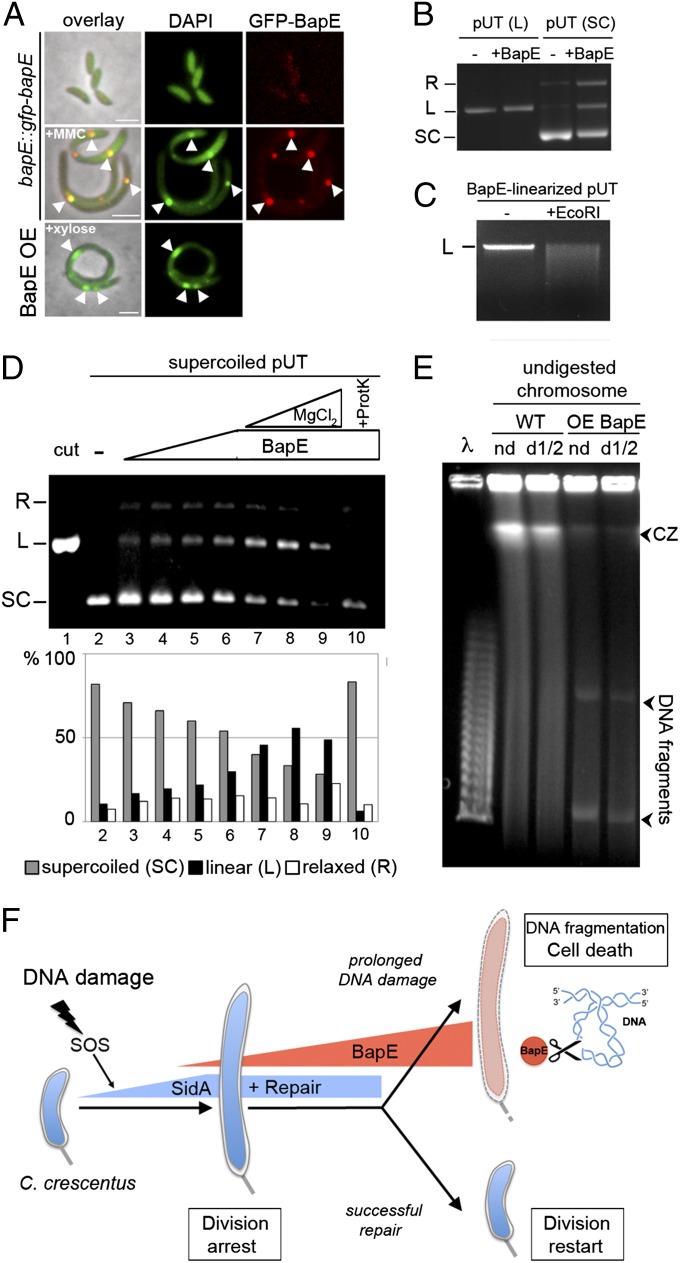
The damage-induced stimulation of TUNEL staining indicates that Caulobacter apoptotic-like death involves chromosome fragmentation (Fig. 1), but no apoptotic-like DNA endonucleases have been identified in bacteria. We thus tested the possibility that the BapE protein might represent this missing endonuclease. We first examined the effect of BapE on chromosome organization in vivo. Consistent with an effect on chromosome integrity, BapE overexpression dramatically altered chromosome morphology (Fig. 4A). Whereas Caulobacter cells normally homogeneously distribute their DNA throughout the cell, BapE overexpression caused cells to form heterogeneously distributed large foci of accumulated DNA (Fig. 4A). The ΔlexA mutant displayed a similar perturbation in chromosomal organization, and the bapE-ssrA mutant largely suppressed this defect (Fig. S6 A and B). MMC treatment also induced the accumulation of DNA foci, and these foci colocalized with GFP-BapE (Fig. 4A). To further support the conclusion that BapE induces chromosome rearrangement, we examined the localization of PopZ-mCherry, a marker for DNA-free regions of the cell. In wild-type cells, PopZ is found only at the cell poles (Fig. S6C). However, PopZ-mCherry accumulated at midcell regions in cells overexpressing BapE, indicating that these cells developed DNA-free subcellular domains (Fig. S6C).
Fig. 4.
BapE affects DNA integrity in vivo and in vitro. (A) Examination of chromosome morphology (DAPI staining) in cells expressing GFP-BapE from its endogenous promoter (bapE::gfp-bapE) grown either with or without MMC and in cells overexpressing BapE. White arrowheads indicate colocalization of GFP-BapE and bright foci of DNA. (B) Cut pUT plasmid (linearized with EcoRI) and uncut plasmid (supercoiled) DNA samples were incubated either with or without BapE protein. The topological states of pUT plasmid are indicated: SC, supercoiled; L, linear; and R, relaxed. (C) Purifed BapE-linearized pUT plasmid was incubated either with or without EcoRI restriction enzyme. The linear (L) state of pUT plasmid is indicated. (D) Supercoiled plasmid DNA (pUT) was incubated alone (lane 2), with increasing concentrations of BapE protein (0.16, 0.32, 0.54, and 0.82 μM) (lanes 3–6), with BapE protein (0.82 μM), and with increasing concentrations of MgCl2 (0.1, 0.5, and 1 mM) (lanes 7–9), and with BapE (0.82 μM) and proteinase K (lane 10). Cut (linear) plasmid is shown as a control (lane 1). Quantifications of the resulting plasmid topological states (SC, supercoiled; L, linear; and R, relaxed) are shown below the gel. (E) Analysis of in vivo chromosome fragmentation by pulsed-field gel electrophoresis of undigested Caulobacter chromosomes [nondiluted (nd) or 1/2 diluted (d1/2)] from cells with and without BapE overexpression. Arrowheads at the bottom denote linearized DNA fragments that are specifically obtained when BapE is overexpressed; arrowhead at the top indicates the compression zone (CZ) of intact high-molecular-weight DNA likely due to overloading of DNA. Lambda concatemers are used as molecular size markers. (F) Model of BapE-dependent DNA damage cell fate determination. In response to DNA damage, C. crescentus induces a graded expression of LexA-dependent genes. Early in the SOS response, cells induce the expression of the division inhibitor SidA (9) and DNA repair pathways to arrest cell division and give cells time to attempt to fix the damage. If DNA damage persists, cells induce the expression of the unique endonuclease, BapE, which functions as a cell fate determinant in a concentration-dependent manner: low levels of BapE reinforce the SidA-mediated reversible division arrest until cells succeed in repairing the damage and re-enter the cell cycle, whereas high levels of BapE promote apoptotic-like cell death by stimulating chromosome fragmentation in conditions of prolonged DNA damage.
To determine whether BapE can directly affect the chromosome, we purified a recombinantly expressed BapE-His6 fusion (Fig. S7) and examined its effects on DNA in vitro. Remarkably, although the BapE amino acid sequence does not resemble that of known nucleases, BapE nonetheless cleaved supercoiled plasmid DNA (Fig. 4B). Cleavage was specific to supercoiled DNA as BapE had no effect on linear DNA (Fig. 4B). BapE does not appear to cut at specific DNA sequences because digesting the BapE-cleaved plasmid with a site-specific endonuclease resulted in an even smear of differently sized fragments (Fig. 4C). BapE has all of the hallmarks of a nuclease in that it is dependent on both BapE and Mg2+ concentration and completely abolished by Proteinase K treatment (Fig. 4D). As a negative control, we also performed the same purification protocol on a strain expressing an empty vector, rather than on BapE, and demonstrated that these fractions exhibit no endonuclease activity (Fig. S8). Thus, we conclude that BapE is a unique supercoiled DNA-specific, sequence-nonspecific nuclease.
The ability of BapE to cleave supercoiled DNA in vitro suggested that BapE might function by disrupting genome integrity in vivo. Indeed, pulsed-field gel electrophoresis of undigested Caulobacter chromosomes revealed that BapE overexpression caused significant chromosome fragmentation in vivo (Fig. 4E). The pattern observed is similar to that seen upon overexpression of other proteins that cleave DNA (17), supporting the conclusion that BapE fragments chromosomes in vivo. Similar levels of chromosome fragmentation induced by topoisomerase inhibition have previously been reported to lead to bacterial cell death (18, 19), such that this activity is sufficient to explain why Caulobacter cells die upon extensive BapE induction.
Discussion
Our results indicate that, in response to DNA damage, Caulobacter cells first induce division arrest and damage repair pathways in an effort to fix the damage, but that when damage persists, hey eventually induce the BapE endonuclease, which clusters, fragments chromosomes, and promotes apoptotic-like death (see model in Fig. 4F). Eukaryotic apoptosis factors also cluster (20), suggesting that protein clustering may be a mechanism to tightly control the potentially catastrophic consequences of unregulated cell death proteins. BapE mutants do not increase mutation frequency, suggesting that BapE does not contribute to the repair process itself. In fact, mutation frequencies decrease upon loss of BapE, which may reflect less DNA damage caused by BapE-mediated double-strand breaks. Thus, BapE is a bacterial DNA endonuclease whose function appears dedicated to promoting cell death.
Apoptosis is generally thought to benefit multicellular organisms by eliminating damaged cells and thereby preventing the accumulation of oncogenic mutations that could threaten the viability of the entire organism (1). In unicellular organisms, however, the selective advantage of promoting cell death is less obvious. One possibility is that BapE-induced death protects Caulobacter populations from the activation of lysogenic phages or an extracellular death factor. In E. coli, for example, the SOS response induces both the excision of dormant phages (10, 11) and the secretion of an extracellular death factor (21–23). Caulobacter may prevent the release of such population-endangering factors by killing severely damaged cells, thereby sacrificing a small number of damaged bacteria rather than releasing a large number of agents that could threaten the entire population. The potential role for BapE in eliminating phage induction may thus explain why no prophages have been found in wild-type Caulobacter strains (12).
A second exciting possibility is that isogenic bacterial communities effectively act as multicellular organisms. In this scenario, there would be a selective advantage for extensively damaged cells to die. Such death would prevent damaged cells from consuming resources and would release their contents for consumption by the remaining members of the community. Caulobacter thrives in nutrient-poor environments such as freshwater lakes where few other competing species exist. By contrast, cell lysis could be counterproductive in multispecies environments as it would provide the other species with a selective advantage and supply them with additional resources. Such ecological differences may explain why Caulobacter cells undergo apoptotic-like death whereas other species have evolved the lesion bypass pathway in which they excise and replace irreparably damaged DNA with an error-prone polymerase (6, 24), trading off cell survival for the cost of passing on dramatically altered genomes. Further support for the analogy between the damage responses of Caulobacter and multicellular eukaryotes comes from the fact that both systems promote cell death by inducing chromosome fragmentation. The presence of such similar apoptotic cell death responses in both bacteria and eukaryotes suggests that programmed cell death may be an ancient process that confers a wide range of cells with a selective advantage. Indeed, C. crescentus is an α-proteobacterium that is related to the bacterial ancestor of mitochondria (25), which are themselves essential for eukaryotic apoptosis.
Materials and Methods
All Caulobacter strains were grown at 30 °C in peptone yeast extract (PYE) medium. When needed, MMC was added at 1 μg/mL. For BapE overexpression, 0.3% xylose was used (see SI Materials and Methods for details of strain growth and strain construction). Strains, plasmids, and primers used in this study are listed in Tables S1–S3, respectively.
Apoptotic-cell death was visualized by fluorescence microscopy on PYE-containing agarose pad using PI (1 μg/mL) staining, DIBAC4(3) (1 μg/mL) staining, and by FACS using the standard TUNEL assay (see SI Materials and Methods for details of experimental protocol). In vivo DNA fragmentation in the BapE-overexpressing strain was visualized using pulsed-field gel electrophoresis (see SI Materials and Methods for details of experimental method).
Kinetics of SOS gene expression was quantified by real-time quantitative PCR analysis. To induce SOS response, MMC was applied at midlog phase, and samples were withdrawn at 8, 15, 30, 45, 60, and 120 min after MMC exposure (see SI Materials and Methods for details of experimental protocol and the list of primers used for this study).
Recombinant Caulobacter BapE protein was expressed in E. coli using the pET system and purified by a His-tag affinity. In vitro nuclease assays were performed by incubating purified BapE protein with the pUT plasmid as a DNA template for 10 min at 37 °C before analysis on a 1% agarose gel (see SI Materials and Methods for details of the method).
Supplementary Material
Acknowledgments
We thank Mike Laub for the generous gift of Caulobacter strains (MT1758 and MT1760) and Tina de Coste and the Princeton FACS facility for assistance with flow cytometry. We also thank the members of the Z.G. laboratory, Tom Silhavy, Thomas Gregor, Coleen Murphy, Rich Losick, and Alison Gammie, for helpful discussions. This work was supported in part by National Institutes of Health New Investigator Award 1DP2OD004389-01 (to Z.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213332109/-/DCSupplemental.
References
- 1.Vousden KH. Outcomes of p53 activation—spoilt for choice. J Cell Sci. 2006;119(Pt 24):5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 2.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18(53):7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 3.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 4.Erental A, Sharon I, Engelberg-Kulka H. Two programmed cell death systems in Escherichia coli: An apoptotic-like death is inhibited by the mazEF-mediated death pathway. PLoS Biol. 2012;10(3):e1001281. doi: 10.1371/journal.pbio.1001281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dwyer DJ, Camacho DM, Kohanski MA, Callura JM, Collins JJ. Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol Cell. 2012;46(5):561–572. doi: 10.1016/j.molcel.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pham P, Rangarajan S, Woodgate R, Goodman MF. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc Natl Acad Sci USA. 2001;98(15):8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marczynski GT. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol. 1999;181(7):1984–1993. doi: 10.1128/jb.181.7.1984-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Rocha RP, Paquola AC, Marques MdoV, Menck CF, Galhardo RS. Characterization of the SOS regulon of Caulobacter crescentus. J Bacteriol. 2008;190(4):1209–1218. doi: 10.1128/JB.01419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25(12):1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozanov DV, D’Ari R, Sineoky SP. RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J Bacteriol. 1998;180(23):6306–6315. doi: 10.1128/jb.180.23.6306-6315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro L. Differentiation in the Caulobacter cell cycle. Annu Rev Microbiol. 1976;30:377–407. doi: 10.1146/annurev.mi.30.100176.002113. [DOI] [PubMed] [Google Scholar]
- 13.Adams DS, Levin M. 2012. Measuring resting membrane potential using the fluorescent voltage reporters DiBAC4(3) and CC2-DMPE. Cold Spring Harb Protoc 2012(4):459–464.
- 14.Düssmann H, Rehm M, Kögel D, Prehn JH. Outer mitochondrial membrane permeabilization during apoptosis triggers caspase-independent mitochondrial and caspase-dependent plasma membrane potential depolarization: A single-cell analysis. J Cell Sci. 2003;116(Pt 3):525–536. doi: 10.1242/jcs.00236. [DOI] [PubMed] [Google Scholar]
- 15.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119(3):493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271(5251):990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 17.Liu QY, et al. Regulation of DNaseY activity by actinin-alpha4 during apoptosis. Cell Death Differ. 2004;11(6):645–654. doi: 10.1038/sj.cdd.4401401. [DOI] [PubMed] [Google Scholar]
- 18.Couturier M, Bahassi el-M, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6(7):269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 19.Malik M, Zhao X, Drlica K. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol Microbiol. 2006;61(3):810–825. doi: 10.1111/j.1365-2958.2006.05275.x. [DOI] [PubMed] [Google Scholar]
- 20.Mace PD, Riedl SJ. Molecular cell death platforms and assemblies. Curr Opin Cell Biol. 2010;22(6):828–836. doi: 10.1016/j.ceb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belitsky M, et al. The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol Cell. 2011;41(6):625–635. doi: 10.1016/j.molcel.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Kolodkin-Gal I, Engelberg-Kulka H. The extracellular death factor: Physiological and genetic factors influencing its production and response in Escherichia coli. J Bacteriol. 2008;190(9):3169–3175. doi: 10.1128/JB.01918-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolodkin-Gal I, Hazan R, Gaathon A, Carmeli S, Engelberg-Kulka H. A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science. 2007;318(5850):652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- 24.Rattray AJ, Strathern JN. Error-prone DNA polymerases: When making a mistake is the only way to get ahead. Annu Rev Genet. 2003;37:31–66. doi: 10.1146/annurev.genet.37.042203.132748. [DOI] [PubMed] [Google Scholar]
- 25.Esser C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21(9):1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.