Abstract
More than two-thirds of breast cancers express the estrogen receptor (ER) and depend on estrogen for growth and survival. Therapies targeting ER function, including aromatase inhibitors that block the production of estrogens and ER antagonists that alter ER transcriptional activity, play a central role in the treatment of ER+ breast cancers of all stages. In contrast to ER− breast cancers, which frequently harbor mutations in the p53 tumor suppressor, ER+ breast cancers are predominantly wild type for p53. Despite harboring wild-type p53, ER+ breast cancer cells are resistant to chemotherapy-induced apoptosis in the presence of estrogen. Using genome-wide approaches, we have addressed the mechanism by which ER antagonizes the proapoptotic function of p53. Interestingly, both ER agonists such as estradiol and the selective ER modulator (SERM) tamoxifen promote p53 antagonism. In contrast, the full ER antagonist fulvestrant blocks the ability of ER to inhibit p53-mediated cell death. This inhibition works through a mechanism involving the modulation of a subset of p53 and ER target genes that can predict the relapse-free survival of patients with ER+ breast cancer. These findings suggest an improved strategy for the treatment of ER+ breast cancer using antagonists that completely block ER action together with drugs that activate p53-mediated cell death.
Keywords: cistrome, DNA damage, nuclear receptor, nutlin, doxorubicin
Although breast cancer is often curable when it is treated at an early stage, metastatic disease is almost uniformly fatal due to the development of therapeutic resistance (1). Approximately two-thirds of all breast cancer cases are attributed to the dysregulation of estrogen receptor (ER) signaling (2, 3). ER is also the most significant therapeutic target for ER+ breast cancer (4). Among drugs that are approved for the treatment of ER+ breast cancers are tamoxifen and fulvestrant (4). Tamoxifen, a selective ER modulator (SERM), competes with estradiol (E2) for ER binding and induces a conformation that favors corepressor binding rather than coactivator binding by ER, thus blocking the expression of most estrogen-responsive genes (5) and resulting in growth inhibition (6). Although treatment of women with ER+ metastatic breast cancer with tamoxifen is useful in controlling their disease, resistance almost invariably develops, leading to recurrence (6). Unlike tamoxifen, which is a partial antagonist, fulvestrant is considered a full antagonist (7). ER binding by fulvestrant both blocks the agonist effects of E2 and leads to degradation of the ER through an ubiquitin-mediated mechanism (8–10). Although fulvestrant has activity in some women with tamoxifen-resistant disease, resistance to fulvestrant also limits its utility (11).
Adjuvant treatment of early stage ER+ breast cancers most commonly includes chemotherapy followed by tamoxifen or, in postmenopausal women, an aromatase inhibitor that blocks the production of estrogen from androgens. Chemotherapy for breast cancer often includes drugs such as doxorubicin that induce DNA damage and subsequently the p53-mediated apoptotic response. p53 is a tumor suppressor gene that plays a role in cell cycle control and apoptosis, and is found to be mutated in >50% of cancers in which its loss facilitates transformation (12, 13). Curiously, despite its high mutation rate in other forms of cancer, p53 has been shown to be mutated in just 20–30% of all breast malignancies, and for the most part these are ER− (14).
Prior studies have suggested that ER interacts with p53 and that this association could lead to the suppression of p53-mediated repression (1, 15). Here, we explored the functional interplay between p53 and ER in ER+ breast cancers on a genome-wide scale through an integrative analysis of the gene expression programs mediated by each and their cistromes. We find that ER represses the p53-mediated apoptotic response induced by DNA damage through a mechanism that involves the recruitment of ER to a subset of proapoptotic p53 target genes at sites that are distinct from those bound by p53.
Results
E2 Protects Cells Against p53-Mediated Cell Death.
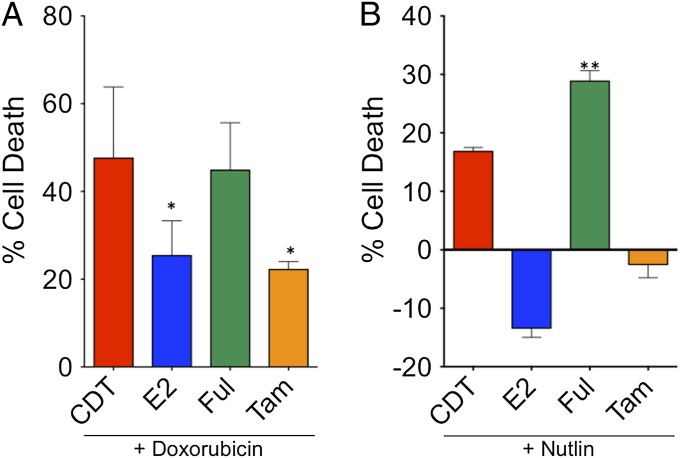
Because estrogen is required for the growth and survival of MCF7 ER+ breast cancer cells, we explored whether E2 had an effect on MCF7 cell survival in the presence of an apoptotic stimulus. To test this hypothesis, MCF7 cells were treated with the proapoptotic chemotherapeutic drug doxorubicin, and cell viability was determined in the presence or absence of estrogen after 3 d (Fig. 1A). In cells treated with doxorubicin under vehicle (i.e., charcoal-dextran-treated; CDT) conditions, only ∼50% survived, whereas the addition of E2 increased survival to ∼75%. Interestingly, tamoxifen was also able to block cell death induced by doxorubicin to the same extent as E2. In contrast, treatment with fulvestrant led to a similar loss in survival as with doxorubicin alone. We further examined the effects of E2, fulvestrant, and tamoxifen on cells treated with nutlin (16), a drug that stabilizes p53 by inhibiting the interaction between p53 and the E3 ubiquitin ligase mdm2 (Fig. 1B). Treatment with nutlin in the absence of E2 led to cell death, albeit to a lesser extent than doxorubicin. This cell death was significantly augmented in the presence of fulvestrant. In contrast, in the presence of E2 or tamoxifen, cell death was completely blocked, and there was increased cell number compared with baseline. These data demonstrate that E2 is protective against both doxorubicin- and nutlin-stimulated p53-mediated apoptosis in breast cancer cells.
Fig. 1.
E2 and tamoxifen protect MCF7 cells against doxorubicin- and nutlin-mediated cell death. MCF7 cells were seeded in hormone-depleted medium containing vehicle (EtOH), E2, fulvestrant, or tamoxifen for 2 d and then treated with and without doxorubicin (A) or nutlin (B) for 3 d. Afterward, the cell viability was measured. The data shown were performed in triplicate and are representative of three independent experiments. These data demonstrate that E2 is protective against doxorubicin- and nutlin-mediated apoptosis in breast cancer cells. *P < 0.05 compared with doxorubicin alone treatment; **P < 0.005 compared with nutlin alone treatment.
E2 and Doxorubicin Regulate a Set of Common Genes.
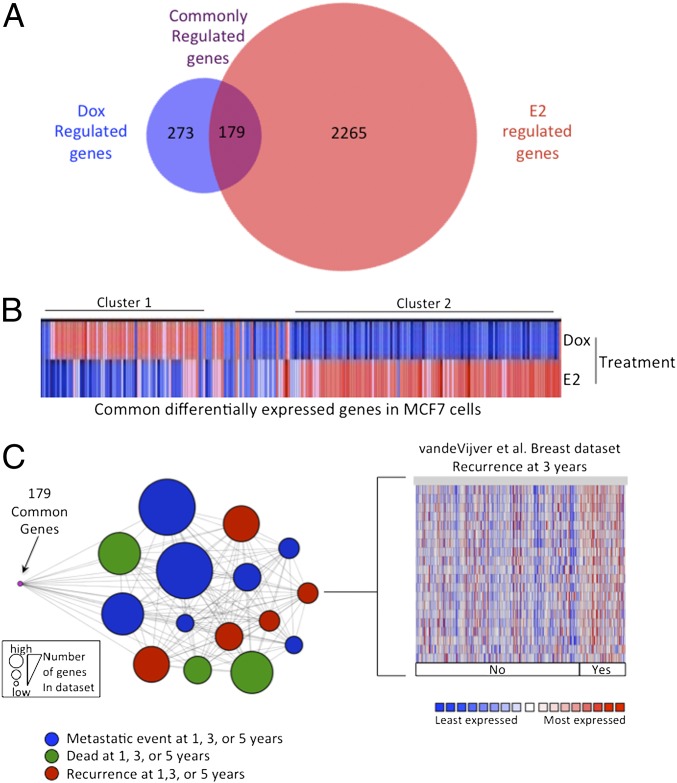
To begin to elucidate the mechanisms involved in the ER-mediated protection of doxorubicin-induced cell death, we first examined the genes differentially expressed in response to both stimuli. We compared the expression of genes altered in response to 12-h doxorubicin treatment using Affymetrix Human Genome U133A 2.0 arrays and compared them to an E2-regulated MCF7 dataset (17). We found that there were 452 genes significantly regulated (P < 0.01) by doxorubicin treatment and 2,444 genes regulated by E2 treatment (Fig. 2). Of these genes, 179 were differentially regulated by both stimuli. These commonly regulated genes were associated with Gene Ontology (GO) categories, including those associated with cell growth and the DNA damage response (Fig. S1). When we clustered the 179 genes commonly regulated by both stimuli, we observed two major clusters that displayed disparate regulation: genes that were up-regulated by doxorubicin and down-regulated by E2 (cluster 1, 49 genes) and genes that were down-regulated by doxorubicin and up-regulated by E2 (cluster 2, 97 genes; Fig. 2B). GO analysis of these subsets revealed that the genes in cluster 1 were primarily associated with the apoptotic categories, and genes in cluster 2 were mainly associated with cell growth process categories (Fig. S2). These data suggest that doxorubicin and estrogen have opposing effects on a common set of genes that regulate cell growth and apoptosis.
Fig. 2.
ER and p53 regulate a common subset of genes that is associated with poor outcome in patients with breast cancer. (A) We compared the 12-h doxorubicin-mediated gene expression profile in MCF7 cells, which was generated by using Affymetrix Human Genome U133A 2.0 arrays, with 12-h E2-mediated gene expression and found that 179 genes were shared by both stimuli. (B) We clustered these genes and found two main clusters: cluster 1 contained genes that were up-regulated by doxorubicin and down-regulated by E2, and cluster 2 comprised genes that were up-regulated by E2 and down-regulated by doxorubicin. (C) The 179 commonly regulated genes were interrogated by using Oncomine Concepts analysis (Compendia Biosciences) against publicly available primary breast tumor datasets, and significant associations were graphically represented in an interaction network by using Cytoscape (http://www.cytoscape.org). In this network, a node represents a dataset, and each edge represents a significant association with P < 0.01 and an odds ratio > 4. The 179 genes (shown in purple) demonstrated significant association with patient datasets exhibiting metastasis (blue circle), death (green circles), and recurrence (red circles) within 1, 3, or 5 y. The node size is proportional to the number of associated genes in each dataset. The van de Vijver dataset is shown on the right as an example of genes up-regulated in patients with recurrence at 3 y.
To determine whether the 179 genes were clinically relevant in patients with breast cancer, we used Oncomine Concepts Map (18) to determine whether these same genes were differentially regulated in samples from patients with breast cancer. We found that the genes regulated by both stimuli were significantly associated with poor outcome, including metastasis, death, and recurrence within 5 y, in several independent ER+ breast cancer datasets (Fig. 2C).
Determination of the p53 Cistrome.
To explore whether the commonly regulated genes are directly regulated by both p53 and ER, we determined the p53 cistrome in MCF7 cells by chromatin immunoprecipitation (ChIP) coupled with next-generation sequencing (ChIP-seq) following doxorubicin treatment and compared these data with an updated E2-induced ER cistrome. For the p53 cistrome, cells were stimulated with doxorubicin for 4 h, a time at which sufficient stable p53 protein levels are observed and where we observed sufficient DNA binding by directed ChIP. We found 1,210 doxorubicin-induced p53 binding sites in MCF7 cells (P < 1 × 10−5; Table S1). Computational analysis revealed that the p53 binding regions were enriched for the M00272 and M00761 TRANSFAC-derived p53 motifs (Table S2), particularly in the center of the binding sites (Fig. S3A). The p53 M00034 matrix was also significantly enriched in the center of the binding sites, although at a lower frequency (Fig. S3A). These data are in agreement with SeqPos motif analysis (19) of the binding sites, which demonstrated significant enrichment for these matrices (Table S2). De novo sequence analysis of the collective sites found a motif similar to these matrices (Fig. S3B) (20).
We found that a number of established p53 targets, including apaf1, p21, mdm2, and fas, contain p53-binding sites (Fig. S4A). Moreover, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed an enrichment for the p53 pathway (Fig. S4B), and GO analysis of genes near p53 binding sites (<30 kb) revealed a considerable enrichment for apoptosis biological process categories with significant P values (Fig. S5A). The distribution profile of the p53 binding sites was similar to other transcription factors where a majority of sites are found at distal intergenic regions (56%) and a minority at promoters (4%) (Fig. S5B). There was also a significant enrichment for p53 binding sites within introns (34%) and a small number of sites in the 5′ and 3′ UTRs (1% each) and immediately downstream of genes (2%) (Fig. S5B).
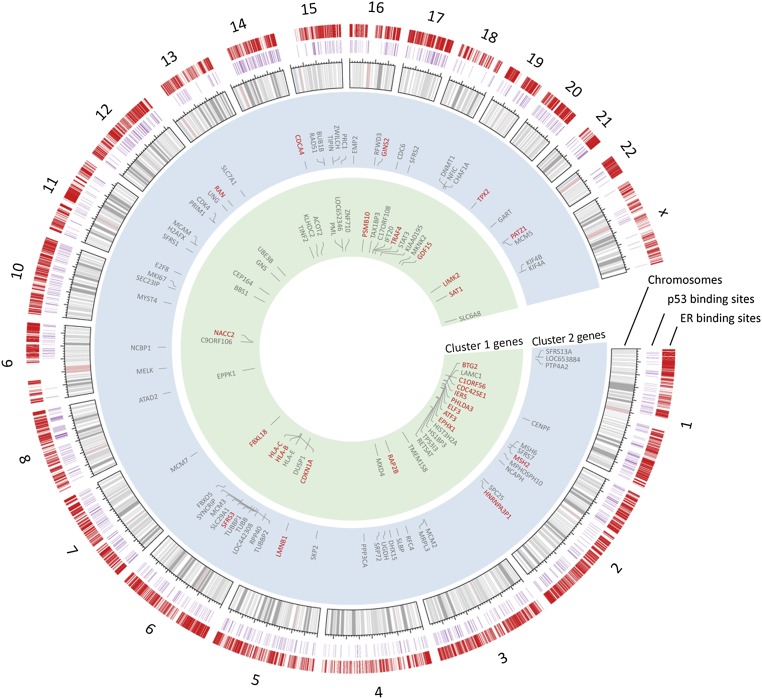
Using an integrative approach, we compared the p53 and ER cistromes (Fig. 3) with the 179 differentially expressed genes to determine which are directly affected by both transcription factors. We observed that there were 155 genes with an ER binding site and 38 genes with a p53 binding site within 100 kb of the transcription start site (Fig. S6). Of these genes, 36 had both a p53 and ER binding site that may directly influence their expression. These genes were enriched for cell cycle and apoptotic GO term categories (Fig. S7) and included targets previously reported to be associated with apoptosis and cell growth control, including cyclin-dependent kinase inhibitor 1 (p21), tumor necrosis factor (TNF) receptor-associated factor 4 (TRAF4), B-cell translocation gene 2 (BTG2), and activating transcription factor 3 (ATF3).
Fig. 3.
ER and p53 regulate a common set of gene targets. Circos plot demonstrating the ER and p53 cistromes, their relative genomic location, genes found within 100 kB of each transcription factor, and the clusters in which they reside. The outer hashes (shown in red) demonstrate the relative genomic location of the sites comprising the ER cistrome. The purple hashes display the location of the p53 cistrome. Genes possessing both an ER and p53 binding sites are also shown in red.
ER Blocks p53-Mediated Transcription.
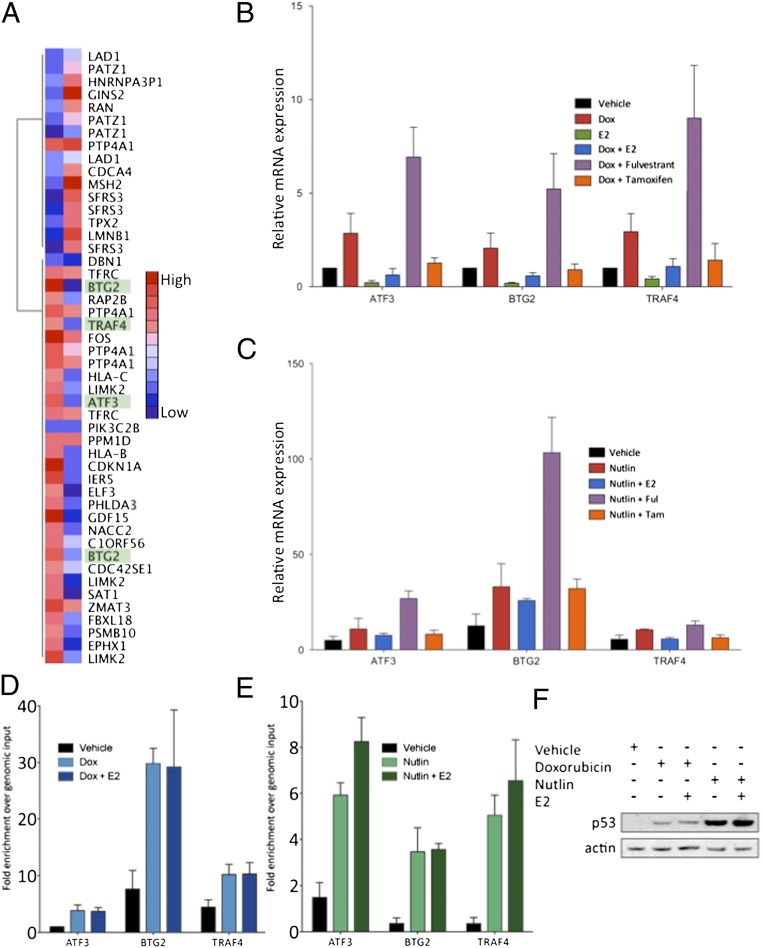
We compared the expression of the 36 genes that were differentially expressed and possessed an ER and p53 binding site using gene expression microarray data from MCF7 cells that were treated with E2 or doxorubicin (Fig. 4A). GO term analysis of this gene set demonstrated enrichment for genes correlated with the cell cycle and apoptosis.
Fig. 4.
p53 activation combined with ER depletion leads to greater target gene expression. (A) Heat map demonstrating the relative expression level of each of the 36 regulated genes from gene expression microarray analysis of MCF7 cells treated with doxorubicin and E2. (B) ER loss leads to more efficient putative apoptotic gene expression. MCF7 cells were treated with doxorubicin alone or in combination with E2, fulvestrant, and tamoxifen to evaluate the expression of putative apoptosis target genes. Cotreatment with fulvestrant led to greater expression of these apoptotic genes, whereas treatment with tamoxifen inhibited in a manner similar to E2. (C) Similar effects were observed when treating with nutlin. (D and E) MCF7 cells were hormone starved and stimulated with doxorubicin (D) or nutlin (E). After a 4-h treatment, E2 was added, and a targeted p53 ChIP assay was performed. No difference was observed in the p53 recruitment level in response to doxorubicin (Left) or nutlin (Right) in the presence of E2. (F) Protein lysate from MCF7 cells treated under ChIP assay conditions were assayed by immunoblot for the level of p53 protein expression, and β-actin was blotted as a control.
We examined the consequence of dual treatment with E2 and doxorubicin on the expression of genes that have been reported to participate in cell cycle arrest and apoptosis induction, i.e., genes with common sites from cluster 1. For this purpose, we examined the expression of the ATF3, BTG2, and TRAF4 genes. ATF3 is an ATF/CCAAT enhancer-binding protein (CREB) family member that can heterodimerize with Jun AP-1 family members to activate transcription (21–23). It has been reported to interact with and stabilize p53 by blocking its ubiquitination. BTG2 is a gene that was found to be suppressed in breast cancer (24, 25), and its up-regulation was shown to be correlated with better survival in patients with breast cancer (26). This gene was also found to be regulated in response to DNA-damaging stimuli in a p53-dependent manner (27, 28). TRAF4 is a TNF protein family member that was identified as a p53 target and shown to be proapoptotic when overexpressed in breast cancer cell lines (29, 30).
Although treatment with doxorubicin induced the expression of the ATF3, BTG2, and TRAF4 genes, these genes were repressed by E2 (Fig. 4B). Combined treatment with E2 and doxorubicin led to an inhibition of the expression of these genes, demonstrating that E2 plays an inhibitory role for these genes even in the presence of doxorubicin. To test the effects of ER antagonists, we treated the cells in the presence of doxorubicin with either fulvestrant or tamoxifen. Strikingly, whereas the full antagonist fulvestrant facilitated the expression of ATF3, BTG2, and TRAF4, the partial antagonist tamoxifen inhibited their expression in a manner that is similar to E2 treatment. We observed similar results when stimulating with E2 in combination with nutlin-3, demonstrating that elevated p53 levels are sufficient for the p53-mediated activation of these genes in MCF7 cells (Fig. 4C).
To address whether ER blocks p53-mediated expression through the prevention of p53 binding as has been suggested (15, 31), we examined the p53 binding sites near the ATF3, BTG2, and TRAF4 genes (Fig. 4D). Using a targeted ChIP approach, we found strong p53 binding at each of these sites upon doxorubicin treatment which was unaffected by the addition of E2. A similar effect was observed with nutlin treatment (Fig. 4E). In addition, we measured the effect of E2 on the p53 protein level by performing a Western blot using lysates from MCF7 cells treated under the ChIP conditions. We found that, whereas doxorubicin and nutlin led to an increase in p53 protein, E2 had no effect on p53 protein levels. These data suggest that although ER does not displace p53 binding or affect the p53 expression level, it independently targets and represses p53 target gene expression.
Differentially Regulated Genes Are Correlated with Patient Survival.
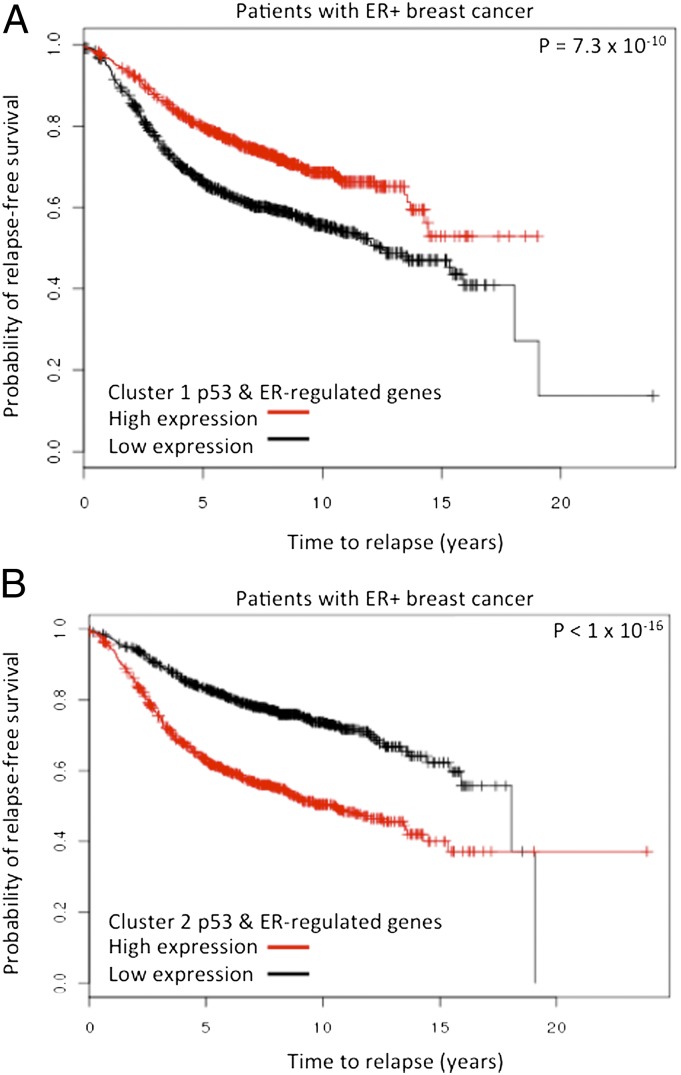
We next wanted to determine whether these genes, which are directly regulated by both p53 and ER, but in opposite directions, are associated with patient outcome. Using a Kaplan-Meier analysis (Fig. 5) of eight ER+ breast cancer patient datasets derived from Gyorffy et al. (32), we found that patients whose tumors had a high level of expression of genes that are up-regulated by doxorubicin and down-regulated by E2 had a better overall survival rate than those whose tumors expressed low levels of these genes. In contrast, patients whose tumors exhibited a high level of expression of the E2 up-regulated and doxorubicin down-regulated genes had a worse survival rate (Fig. 5). These data suggest that the genes oppositely regulated directly by both p53 and ER in MCF7 cells are relevant to the clinical behavior of ER+ breast cancers in general.
Fig. 5.
Kaplan-Meier analysis of patient datasets. Kaplan-Meier analysis reveals that the common regulated genes are predictors of relapse-free survival. To determine the role of these genes in predicting relapse-free survival in patients with ER+ breast cancer, we interrogated breast cancer datasets using the clusters 1 and 2 genes containing p53 and ER binding sites. (A) Kaplan-Meier analysis reveals that the cluster 1 genes that regulated by p53 and ER demonstrate better survival when they are highly expressed in patients with ER+ breast cancer. (B) The opposite result is observed when the p53 and ER-regulated cluster 2 genes are examined.
Discussion
In ER+ breast cancer, inhibiting ER action through the reduction in estrogen levels with aromatase inhibitors or the use of antagonists such as tamoxifen and fulvestrant leads to significant clinical benefit for both early stage patients and those with advanced disease. Despite frequently harboring wild-type p53, ER+ breast cancers are relatively insensitive to chemotherapy-induced apoptosis. We explored the mechanism by which ER protects ER+ breast cancers from DNA-damage– induced cell death by determining the genome-wide set of genes directly regulated by p53 and ER. We identified a small set of proapoptotic p53 target genes that are also directly targeted by ER. These genes are induced upon p53 binding, and this induction is blocked by ER binding. Interestingly, this is the case for full agonists such as E2 and the partial agonist tamoxifen. Clinical treatment of patients with breast cancer typically involves chemotherapies that lead to an increase in p53 expression followed by treatment with adjuvant therapy, which includes tamoxifen. Our data suggest that there are inherent deficiencies in this type of treatment. We demonstrate that treatment with E2 or tamoxifen suppresses the apoptotic response in MCF7 cells; thus, p53 activation by means of chemotherapeutic drugs before antiestrogen treatment in patients may be less effective. Moreover, our results demonstrate that tamoxifen may be insufficient for the treatment of ER-α– positive breast cancers that express wild-type p53. Although tamoxifen inhibits the ER transcriptional program, it is also known to act in a similar manner to the ER because it can activate a common subset of genes (6). Our data show that, in addition, tamoxifen treatment suppresses genes in a manner that is similar to E2 treatment, and these genes include those that are pro-cell death. Furthermore, this study suggests that effective chemotherapeutic treatment should include drugs that remove the influence of the ER completely, such as fulvestrant, which is a drug that leads to the down-regulation and degradation of the ER and results in increased p53 activity and ultimately apoptosis (33–35).
Among the p53 target genes that we found to be suppressed by ER, several—including ATF3, BTG2, and TRAF4—have been implicated in the p53-mediated cell death response. Interestingly, ATF3 is a transcription factor that was reported to negatively regulate cell-cycle progression and augment the transcription of p53 target genes (37), highlighting its importance in downstream p53 responses. Its direct repression by ER would thus contribute to further inhibition of the p53 response. BTG2 is also regulated in response to DNA-damaging stimuli in a p53-dependent manner (26, 27). It has previously been found to be down-regulated by ER and to inhibit breast cancer cell growth (37). Loss of expression of this gene is correlated with a poor prognosis in patients with breast cancer and was predictive of tumor grade and size, invasion, recurrence, and overall survival (38). These findings suggest that BTG2 might be a useful pharmacodynamic biomarker for the action of ER antagonists such as fulvestrant that potentiate p53-dependent cell death. Interestingly, the p53-dependent cell-death mediator TRAF4 was originally cloned from metastatic breast cancer samples and was found to reside near the HER2 locus on chromosome 17q11-12 (39, 40). Its role in modulating cell survival in HER2+ breast cancers that frequently harbor p53 mutations and whether ER status plays a role remain to be elucidated.
It has been reported that the ER binds to p53 and that this interaction occurs at the C terminus (31, 41). It has also been shown that ER binds and represses p53 on the p21, survivin, and MDR1 gene promoters along with their gene expression, and this inhibition is relieved with γ-irradiation– mediated p53 activation (15). However, we did not observe ER binding at the locations reported for these sites in this study or other published ER datasets (17, 42), including one dataset that measured ER binding under basal conditions (43). Moreover, although we observed ER and p53 binding at the same loci for a subset of the sites, the p53 chromatin interaction was not inhibited by the ER at these sites. This result suggests that the mechanism by which ER antagonizes p53 action does not involve blocking the access of p53 for the genome. Other studies have proposed recruitment of corepressors by ER to explain its ability to repress p53-mediated activation (11). This inhibition may involve AP-1 factors because AP-1 motifs are enriched in the ER binding sites found near the common set of ER and p53 regulated genes. AP-1 was previously shown to be capable of tethering the ER to sites in the genome, and these sites tend to be enriched for down-regulated genes (17).
In conclusion, we describe a transcriptional program that is directly regulated by both ER and p53 that explains the resistance of ER+/p53 wild-type breast cancers to apoptosis. Because the ER suppresses genes that are responsible for executing the p53-mediated apoptotic program, there is no selection for cells harboring mutant p53; rather, the selection that occurs in these cancers may actually be for increased ER signaling (44). Thus, effective treatment for these tumors may require the concomitant complete inhibition of ER signaling with drugs such as fulvestrant together with treatments that promote p53-mediated apoptosis (33–35).
Materials and Methods
Cell Culture and Reagents.
MCF7 cells were grown in high-glucose DMEM (Invitrogen) supplemented with 2 mmol/L l-glutamine, 10% (vol/vol) heat-inactivated FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin (Invitrogen) in a humidified incubator at 37 °C and 5% CO2. Doxorubicin was purchased from Sigma and dissolved in water.
Cell Viability and Apoptotic Assays.
Approximately 5 × 104 MCF7 cells were seeded in a 96-well plate in hormone-depleted medium and treated with vehicle alone, 10 nM E2, 100 nM fulvestrant, or 10 nM 4-OH tamoxifen. After a 2-d incubation, the medium was removed, and fresh ligand or vehicle containing medium was added with or without 10 μM doxorubicin or 5 μM nutlin. The cells were incubated for an additional 3 d, and a cell viability assay was performed by using the CellTiter 96 Non-Radioactive Cell Proliferation Assay (MTT) (Promega). This experiment was performed in triplicate and repeated three times.
ChIP and Sequencing.
ChIP was performed as described (45). Briefly, ∼5–10 × 106 MCF7 cells were seeded in hormone-depleted medium. After 3 d, the cells were treated with E2 or doxorubicin for 45 min or 4 h, respectively. The cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The cells were washed in PBS and then lysed in 1% SDS lysis buffer, pH 8.0, and sonicated to an average size of 250 bp. The fragmented chromatin was immunoprecipitated overnight by using antibodies directed against p53 (FL-393; Santa Cruz) or ERα (HC-20; Santa Cruz). For directed quantitative PCR, the DNA was purified by using the QIAquick PCR Purification kit (Qiagen), and the samples were subjected to RT-PCR by using the SYBR green method. For ChIP-Seq, after repairing the DNA ends, sequencing adaptors (Illumina) were ligated to the purified chromatin, and the DNA was amplified for sequencing as described. To assay simultaneous recruitment, hormone-depleted MCF7 cells were treated with doxorubicin for 4 h before a 45-min incubation in the presence of E2. The cells were then processed for ChIP as described above.
Immunoblotting.
MCF7 cells were stimulated for 4 h with either doxorubicin or nutlin before a 45-min incubation in the presence of E2. The cells were then lysed in Nonidet P-40 lysis buffer (1% Nonidet P-40, 150 mM NaCl, 50 mM Tris, pH 8.0), and the protein concentrations were determined by using the micro BCA kit (Pierce). A total of 50 μg of protein was then subjected to SDS/PAGE followed by transfer to a PVDF membrane. The membrane was immunoblotted with p53 (FL-393; Santa Cruz) and β-actin (4967; Cell Signaling Technologies) antibodies followed by secondary antibody incubation with a donkey anti-rabbit antibody (Pierce). The blots were developed by using Western Blotting Luminol Reagent (Santa Cruz).
Gene Expression Analysis.
Hormone-depleted MCF7 cells were treated with PBS or doxorubicin for 12 h. The total RNA was isolated by using a combination of TRIzol (Sigma) and the RNeasy Mini Kit (Qiagen) and hybridized to Affymetrix Human Genome U133A 2.0 arrays. The expression data were normalized by using the robust multichip average (RMA) method and processed by using linear models for microarray data (Limma) using R. Genes with P < 0.01 were considered statistically significant. The doxorubicin-treated cells were compared with a previously generated 12-h E2-treated dataset (17). The raw data from this series were also RMA-normalized and processed by using Limma with the same cutoff value. For k means clustering, the log2 fold change of corresponding genes from each dataset was clustered. For hierarchical clustering, Pearson correlation using pairwise complete linkage was used.
GO and KEGG pathway analyses were performed by using the Database for Annotation, Visualization, and Integrated Discovery bioinformatics resource (46, 47). SeqPos analysis was performed by using the Cistrome analysis pipeline (48), and the motif distribution analysis was performed as described (17).
RT-PCR.
For RT-PCR, total RNA was isolated from using a combination of TRIzol (Sigma) and the RNeasy Mini Kit (Qiagen). First-strand cDNA was created by using the Quantitect Reverse Transcription Kit (Qiagen) following the manufacturer’s protocol.
Oncomine Concepts Map.
The 179 genes that were found to be commonly regulated by E2 and doxorubicin were compared with genes expressed in tumors from patients with breast cancer by using the Oncomine Concepts Map (Compendia Bioscience). Significant associations between these genes and clinical outcome were established, and Cytoscape (49) was used to generate the node connections, which represent dataset associations with P < 0.01 and odds ratio > 4. The data represented in Fig. 2C are derived from the following independent datasets: van de Vijver et al. (50), Kao et al. (51), Loi et al. (52, 53), Wang et al. (54), Hatzis et al. (55), Sotiriou et al. (56), and Desmedt et al. (57).
Supplementary Material
Acknowledgments
This work was supported by a Terri Brodeur Breast Cancer Foundation fellowship (to S.T.B.) and National Institutes of Health Grants P01 CA080111 (to M.B.), R01 DK074967 (to M.B.), and R01 HG004069 (to X.S.L.).
Footnotes
Conflict of interest statement: M.B. serves as a consultant to Novartis and receives sponsored research support from Novartis and Pfizer.
This article is a PNAS Direct Submission. K.P. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE39870).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018858109/-/DCSupplemental.
References
- 1.Kanavos P. The rising burden of cancer in the developing world. Ann Oncol. 2006;17(Suppl 8):viii15–viii23. doi: 10.1093/annonc/mdl983. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100(14):8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne CK, Zhao H, Fuqua SA. Selective estrogen receptor modulators: Structure, function, and clinical use. J Clin Oncol. 2000;18(17):3172–3186. doi: 10.1200/JCO.2000.18.17.3172. [DOI] [PubMed] [Google Scholar]
- 5.Osborne CK, Schiff R. Estrogen-receptor biology: Continuing progress and therapeutic implications. J Clin Oncol. 2005;23(8):1616–1622. doi: 10.1200/JCO.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 6.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2(2):101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 7.Wakeling AE, Dukes M, Bowler J. A potent specific pure antiestrogen with clinical potential. Cancer Res. 1991;51(15):3867–3873. [PubMed] [Google Scholar]
- 8.Dauvois S, White R, Parker MG. The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J Cell Sci. 1993;106(Pt 4):1377–1388. doi: 10.1242/jcs.106.4.1377. [DOI] [PubMed] [Google Scholar]
- 9.Wijayaratne AL, McDonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276(38):35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 10.Marsaud V, Gougelet A, Maillard S, Renoir JM. Various phosphorylation pathways, depending on agonist and antagonist binding to endogenous estrogen receptor alpha (ERalpha), differentially affect ERalpha extractability, proteasome-mediated stability, and transcriptional activity in human breast cancer cells. Mol Endocrinol. 2003;17(10):2013–2027. doi: 10.1210/me.2002-0269. [DOI] [PubMed] [Google Scholar]
- 11.Konduri SD, et al. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci USA. 2010;107(34):15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vousden KH. p53: Death star. Cell. 2000;103(5):691–694. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 13.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187(1):112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Turpin E, et al. Increased incidence of ERBB2 overexpression and TP53 mutation in inflammatory breast cancer. Oncogene. 2002;21(49):7593–7597. doi: 10.1038/sj.onc.1205932. [DOI] [PubMed] [Google Scholar]
- 15.Sayeed A, et al. Estrogen receptor alpha inhibits p53-mediated transcriptional repression: Implications for the regulation of apoptosis. Cancer Res. 2007;67(16):7746–7755. doi: 10.1158/0008-5472.CAN-06-3724. [DOI] [PubMed] [Google Scholar]
- 16.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 17.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, et al. Molecular concepts analysis links tumors, pathways, mechanisms, and drugs. Neoplasia. 2007;9(5):443–454. doi: 10.1593/neo.07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sbisà E, et al. p53FamTaG: A database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinformatics. 2007;8(Suppl 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang G, Wolfgang CD, Chen BP, Chen TH, Hai T. ATF3 gene. Genomic organization, promoter, and regulation. J Biol Chem. 1996;271(3):1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson M, Ford J, Bohm S, Toftgard R. Characterization of a nuclear factor that binds juxtaposed with ATF3/Jun on a composite response element specifically mediating induced transcription in response to an epidermal growth factor/Ras/Raf signaling pathway. Cell Growth Differ. 1997;8(8):913–920. [PubMed] [Google Scholar]
- 23.Hsu JC, Bravo R, Taub R. Interactions among LRF-1, JunB, c-Jun, and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol Cell Biol. 1992;12(10):4654–4665. doi: 10.1128/mcb.12.10.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouault JP, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14(4):482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 25.Kawakubo H, et al. Loss of B-cell translocation gene-2 in estrogen receptor-positive breast carcinoma is associated with tumor grade and overexpression of cyclin d1 protein. Cancer Res. 2006;66(14):7075–7082. doi: 10.1158/0008-5472.CAN-06-0379. [DOI] [PubMed] [Google Scholar]
- 26.Möllerström E, et al. Up-regulation of cell cycle arrest protein BTG2 correlates with increased overall survival in breast cancer, as detected by immunohistochemistry using tissue microarray. BMC Cancer. 2010;10:296. doi: 10.1186/1471-2407-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes U, et al. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol Carcinog. 2000;27(2):57–64. [PubMed] [Google Scholar]
- 28.Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497(2-3):67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 29.Régnier CH, et al. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor-associated protein family, which is expressed in breast carcinoma. J Biol Chem. 1995;270(43):25715–25721. doi: 10.1074/jbc.270.43.25715. [DOI] [PubMed] [Google Scholar]
- 30.Sax JK, El-Deiry WS. Identification and characterization of the cytoplasmic protein TRAF4 as a p53-regulated proapoptotic gene. J Biol Chem. 2003;278(38):36435–36444. doi: 10.1074/jbc.M303191200. [DOI] [PubMed] [Google Scholar]
- 31.Liu G, Schwartz JA, Brooks SC. Estrogen receptor protects p53 from deactivation by human double minute-2. Cancer Res. 2000;60(7):1810–1814. [PubMed] [Google Scholar]
- 32.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 33.Kansra S, Yamagata S, Sneade L, Foster L, Ben-Jonathan N. Differential effects of estrogen receptor antagonists on pituitary lactotroph proliferation and prolactin release. Mol Cell Endocrinol. 2005;239(1-2):27–36. doi: 10.1016/j.mce.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Somaï S, et al. Antiestrogens are pro-apoptotic in normal human breast epithelial cells. Int J Cancer. 2003;105(5):607–612. doi: 10.1002/ijc.11147. [DOI] [PubMed] [Google Scholar]
- 35.Hur J, et al. Regulation of expression of BIK proapoptotic protein in human breast cancer cells: p53-dependent induction of BIK mRNA by fulvestrant and proteasomal degradation of BIK protein. Cancer Res. 2006;66(20):10153–10161. doi: 10.1158/0008-5472.CAN-05-3696. [DOI] [PubMed] [Google Scholar]
- 36.Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J. 2005;24(13):2425–2435. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karmakar S, Foster EA, Smith CL. Estradiol downregulation of the tumor suppressor gene BTG2 requires estrogen receptor-alpha and the REA corepressor. Int J Cancer. 2009;124(8):1841–1851. doi: 10.1002/ijc.24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi F, et al. Breast tumor progression induced by loss of BTG2 expression is inhibited by targeted therapy with the ErbB/HER inhibitor lapatinib. Oncogene. 2011;30(27):3084–3095. doi: 10.1038/onc.2011.24. [DOI] [PubMed] [Google Scholar]
- 39.Bièche I, et al. Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res. 1996;56(17):3886–3890. [PubMed] [Google Scholar]
- 40.Tomasetto C, et al. Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics. 1995;28(3):367–376. doi: 10.1006/geno.1995.1163. [DOI] [PubMed] [Google Scholar]
- 41.Yu CL, et al. The tumor suppressor p53 is a negative regulator of estrogen receptor signaling pathways. Biochem Biophys Res Commun. 1997;239(2):617–620. doi: 10.1006/bbrc.1997.7522. [DOI] [PubMed] [Google Scholar]
- 42.Welboren WJ, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross-Innes CS, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24(2):171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 46.Huang da W, et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 48.Liu T, et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 51.Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: Implications for treatment optimization. BMC Cancer. 2011;11:143. doi: 10.1186/1471-2407-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loi S, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 53.Loi S, et al. Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics. 2008;9:239. doi: 10.1186/1471-2164-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 55.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sotiriou C, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desmedt C, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.