Abstract
Aberrant activation of signaling by the Wnt pathway is strongly implicated in the onset and progression of numerous types of cancer. Owing to the persistent dependence of these tumors on Wnt signaling for growth and survival, inhibition of this pathway is considered an attractive mechanism-based therapeutic approach. Oncogenic activation of Wnt signaling can ensue from a variety of distinct aberrations in the signaling pathway, but most share the common feature of causing increased cellular levels of β-catenin by interfering with its constitutive degradation. β-Catenin serves as a central hub in Wnt signaling by engaging in crucial protein–protein interactions with both negative and positive effectors of the pathway. Direct interference with these protein–protein interactions is a biologically compelling approach toward suppression of β-catenin hyperactivity, but such interactions have proven intransigent with respect to small-molecule targeting. Hence β-catenin remains an elusive target for translational cancer therapy. Here we report the discovery of a hydrocarbon-stapled peptide that directly targets β-catenin and interferes with its ability to serve as a transcriptional coactivator for T-cell factor (TCF) proteins, the downstream transcriptional regulators of the Wnt pathway.
Keywords: colorectal cancer, peptide engineering, targeted therapy
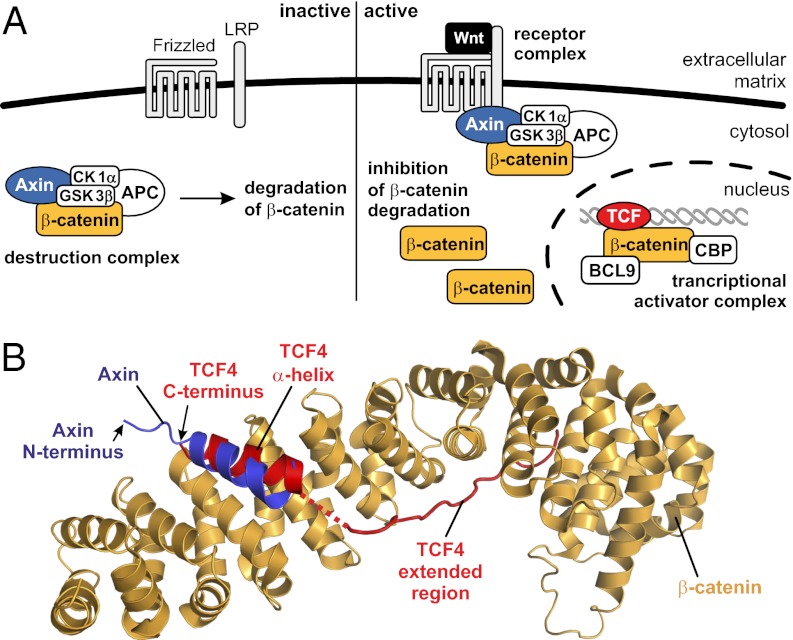
The canonical Wnt (Wingless and INT-1) signal transduction cascade regulates the expression of numerous genes involved in cell differentiation, proliferation, and survival (1). Wnt is a diffusible ligand protein that initiates the Wnt signaling cascade through engagement with a transmembrane complex comprising Frizzled and the low-density lipoprotein-related receptor (LRP) complex. The events that ensue from this engagement are critically dependent upon the protein β-catenin, which serves as a hub for transmission of the Wnt signal from the plasma membrane to the nucleus. In the absence of Wnt, β-catenin is recruited into a multicomponent “destruction complex” formed inter alia by the proteins adenomatous polyposis coli (APC), Axin, casein kinase 1α (CK1α), and glycogen synthase kinase 3β (GSK3β) (Fig. 1A). In this destruction complex, β-catenin undergoes phosphorylation by CK1α and GSK3β, which is followed by ubiquitination and proteasomal degradation of β-catenin (1). This constitutive degradation of β-catenin in the absence of a Wnt signal results in low steady-state levels of the protein. Engagement of Wnt with Frizzled/LRP causes Axin to translocate to the transmembrane receptor complex, thereby inhibiting the destruction complex (1). Consequently, β-catenin accumulates in the cell and undergoes translocation to the nucleus, where it serves to recruit powerful coactivators (1) such as CREB-binding protein (CBP) and B-cell lymphoma 9 protein (BCL9) to DNA-bound transcriptional regulators of the T-cell factor (TCF)/lymphoid enhancer binding factor (LEF) family (Fig. 1A). In addition, β-catenin acts as a structural component of cell–cell adherens junctions, a function that is considered to be independent of its involvement in canonical Wnt signaling. In these cell junctions, β-catenin is tightly bound to phosphorylated cadherins (2).
Fig. 1.
Wnt/β-catenin signaling. (A) The canonical Wnt signaling pathway with its major components in the inactive (Left) and active (Right) state. BCL9 and CBP are transcriptional coactivators. (B) Superimposed crystal structures of the β-catenin armadillo domain bound to the CBDs of Axin (PDB ID 1QZ7) (18) and TCF4 (PDB ID 2GL7) (17).
Activated Wnt signaling serves a critical role in many tissue types during embryonic development but ordinarily becomes quiescent after development in most tissues. In adults, activated Wnt signaling continues to serve a role in stem cell maintenance and tissue regeneration, being essential, for example, in perpetual regeneration of the intestinal lining. Inappropriate and chronic activation of Wnt signaling is strongly implicated in the initiation and progression of several types of cancer (1) but is most strikingly manifest in sporadic colorectal cancers, with ∼90% of all cases being associated with high-level constitutive activation of Wnt signaling (3). In most cases, constitutive Wnt activation seems to result from a reduced ability of the destruction complex to recruit and degrade β-catenin, owing to mutations in components of the complex, especially APC and Axin, and in β-catenin itself (3, 4). The Wnt pathway is thus one of several embryonic pathways, also including Notch and Hedgehog, that are co-opted in certain transformed cells to support tumor growth and survival (5). Activation of these embryonic pathways is generally associated with increased stem cell-like behavior in cancers and thus is linked with a poor prognostic outcome (5). Consequently, there exists particular interest in specific inhibitors of these embryonic pathways in general, and of the Wnt pathway in particular.
Published efforts to inhibit Wnt-dependent signaling have for the most part involved phenotypic screening of small-molecule libraries (6–14). The most active compounds reported thus far either inhibit tankyrase, an Axin destabilizer (IWR-1 and XAV939) (7, 8), or activate CK1α (pyrvinium) (9); these affect Wnt signaling primarily by stimulating the activity of the β-catenin destruction complex. Although these efforts have provided valuable tool compounds, the therapeutic utility of tankyrase inhibitors and CK1α activators is likely to be limited by the fact that most disease-causing mutations are either in APC or in pathway components downstream of the destruction complex (3). Additionally, tankyrase and CK1α are involved in non–Wnt-dependent cellular processes (8, 9), increasing the risk of undesirable off-pathway effects. Other inhibitors of the Wnt signaling pathway have been reported to decrease the transcriptional activity of the downstream TCF/LEF transcriptional activator complex and to reduce the growth of cultured human colon cancer cells (11–14). One of these compounds, NC043, has been shown not to interact directly with the transactivation complex (11) but rather to act by a yet-uncharacterized indirect mechanism. Another such compound, ICG-001, targets the general coactivator CBP (12) and therefore is unlikely to be Wnt pathway-specific. In three cases, molecules have been identified that disrupt the β-catenin/TCF interaction in vitro (13–15), but the molecular target(s) and therefore the exact mechanisms of these inhibitors have not been conclusively identified, rendering difficult the optimization of these relatively weak hit compounds. Thus, there remains significant impetus for the discovery of compounds that directly target key functional interaction sites on β-catenin via a rigorously defined molecular mechanism.
Assessing various options for a design-oriented approach toward inhibition of the β-catenin pathway, we opted for several reasons to focus on the interaction between β-catenin and TCF transcription factor. First, the β-catenin/TCF interaction is well established as a key downstream nodal point of the Wnt pathway (16–18), and its importance in tumor cell growth maintenance has been highlighted by coexpression of dominant negative versions of TCF as well as by the sensitizing effect of small interfering RNAs that down-regulate TCF protein levels (19). Second, the nature of the interaction between β-catenin and peptide ligands, including the β-catenin binding domains (CBD) of TCF proteins, has been characterized structurally (16–18). In β-catenin/TCF structures, the CBD comprises a C-terminal α-helical segment and an extended structure, connected by a disordered loop (16, 17). These wrap themselves around the midriff of the β-catenin armadillo repeat domain (Fig. 1B).
Inhibition of protein–protein interactions (PPIs) by cell-permeable molecules is considered particularly challenging, especially when the protein–protein interface is highly extended, as it is with β-catenin and TCF; thus it is unsurprising that efforts to identify direct-acting small-molecule antagonists of the β-catenin/TCF interaction have not met with success. On the other hand, recent successes in inhibiting even extended PPIs using the hydrocarbon-stapled α-helix technology (20–27) suggested the possibility of using this approach toward β-catenin/TCF antagonism; this notion was strengthened by the presence of the requisite α-helical interaction domain in TCF. Helix stapling entails the introduction of a synthetic all-hydrocarbon brace across one face of an α-helical peptide, which in numerous cases has been shown to stabilize the bioactive α-helical conformation, to increase binding affinity for the target, to decrease the rate of proteolytic degradation, and most importantly, to effect robust cell penetration by endocytic vesicle trafficking (20–29). Importantly, stapled peptides have demonstrated robust efficacy in animal models of human disease, including T-cell acute lymphoblastic leukemia, by direct targeting of the Notch transactivation complex (24) and BCL2-driven lymphoma (27). Here we report the combined use of stapled peptide technology and directed evolution to identify stapled peptides that directly bind the TCF-docking site on β-catenin, thereby antagonizing transcriptional activation via the Wnt signaling pathway. We provide definitive structural and biochemical evidence for the molecular mechanism of these compounds, and we demonstrate that they interfere selectively with Wnt/β-catenin signaling in cells and in a pathway-specific manner reduce the viability of Wnt-addicted cancer cells.
Results
Stapled α-Helical Peptides Targeting β-Catenin.
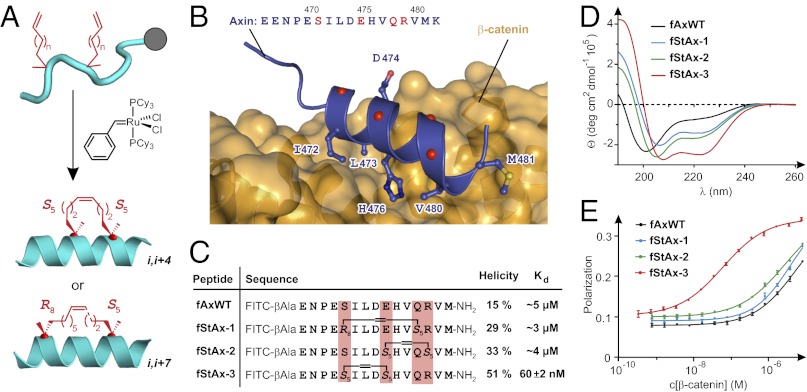
Because our goal was to inhibit TCF binding to β-catenin, we first considered the CBD of TCF4 as a candidate for production of stapled peptides. The extended portion of this peptide (Fig. 1B) represents a potential liability, because such unstructured regions are typically susceptible to proteolytic cleavage; hence, we first assessed the importance of the extended region for binding to β-catenin. Using fluorescence polarization (FP) assays, we found that a fluorescein-labeled peptide comprising only the α-helical region of the TCF4 CBD (Fig. 1B) (17) exhibited no detectable interaction with β-catenin (Kd > 10 μM; Fig. S1B), whereas binding was readily observed with a peptide containing both the α-helical and extended regions (Kd = 0.26 μM). Having determined that the extended region thus seems to be essential for recognition of β-catenin by TCF4 CBD, we sought alternative peptide frameworks for production of stapled peptides. We noted that the CBD of Axin is composed exclusively of α-helical structure, and the binding site for the Axin CBD on β-catenin overlaps substantially with that of the TCF4 α-helix, although the two helices bind in reverse orientation (Fig. 1B). We therefore evaluated Axin-derived peptides for binding to β-catenin, again using FP. Although the fluorescein-labeled 15-mer Axin CBD peptide exhibited only moderate affinity for β-catenin (Kd ∼5 μM; Fig. S1B), this was sufficient to move forward with this framework, given the expected affinity boost that typically results from staple incorporation.
An all-hydrocarbon staple is incorporated into a peptide by introduction of two α-methyl,α-alkenyl amino acids (Fig. S1C) during chain extension via solid-phase peptide synthesis, followed by closure of the macrocyclic bridge using ruthenium-mediated ring-closing olefin metathesis (Fig. 2A) (30–32). Inspection of the crystal structure of the Axin/β-catenin complex (18) revealed four positions in the Axin CBD (Fig. 2B, red) that are not involved in β-catenin binding; these were selected as potential staple incorporation sites. From this set of prospective stapling positions, those having the preferred relative spacings of i,i+4 and i,i+7 were then selected (31). On the basis of these design criteria, stapled Axin (StAx) CBD peptides StAx-1, -2, and -3 (Fig. 2C) were designed (Fig. S2 and Table S1). Fluorescein-labeled versions of these stapled peptides (fStAx-1, -2, and -3) were synthesized and evaluated for α-helicity and binding to β-catenin. As judged by circular dichroism spectra (Fig. 2D), the α-helical character of all three stapled peptides increased relative to that of the fluorescein-labeled wild-type Axin (fAxWT), with StAx-3 showing the greatest α-helicity (51%). The affinities of the first-generation StAx peptides to β-catenin were then determined by FP (Fig. 2E). Whereas fStAx-1 and -2 showed no affinity enhancement upon incorporation of the staple, fStAx-3 exhibited significantly enhanced affinity for β-catenin (Kd = 60 ± 2 nM) relative to fAxWT. The StAx-3 sequence was thus selected for further optimization.
Fig. 2.
Axin-derived stapled peptides. (A) Peptide stapling: Olefin-modified amino acids are incorporated at two positions in the peptide sequence at residues having the relative phasing i,i+4 or i,i+7 and are cross-linked by ruthenium-mediated olefin metathesis. (B) Crystal structure of β-catenin bound to Axin CBD (PDB ID 1QZ7) (18) with the side-chains of interacting residues shown explicitly and the α-carbons of noninteracting residues of the Axin α-helix shown as red spheres. (C) Sequences, helicities, and dissociation constants (Kd, mean ± SE) of Axin-derived FITC-labeled peptides. (D) Circular dichroism spectra of peptides. Details on calculation of α-helicity are provided in SI Methods. (E) FP assay of fluorescein-labeled peptides binding to β-catenin.
Affinity and Cell Permeability Optimization of StAx Peptides.
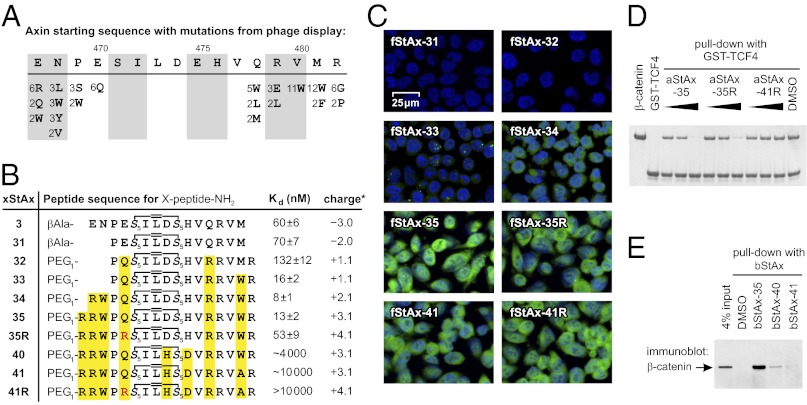
In a parallel series of experiments, we sought to affinity-optimize the Axin peptide sequence through the use of phage display technology (33). We therefore generated a library of phage-displayed peptides based on the Axin CBD but having different quadruplets of residues completely randomized (Fig. S3A). Three stringent selection cycles were performed, yielding 32 new sequences (Fig. S3B). Summarized in Fig. 3A are all of the variants observed at least twice in the selected sequences. Taking into account the frequencies of amino acid substitution at various positions in the phage-derived Axin CBD variants, we synthesized three consensus peptides bearing a fluorescent label and measured their affinity for β-catenin using FP. All three of these sequences show improved binding to β-catenin, with the magnitude of the increase ranging from eight- to 39-fold (Fig. S3 C and D). This information was then used in the design and evaluation of second-generation StAx peptides. Sequence inspection of the phage-optimized peptides (Fig. 3A) suggested that incorporation of a hydrophobic residue at position 468 and a Trp (or Phe) at positions 480 or 481 gave rise to the boost in affinity for β-catenin, hence these changes were chosen for incorporation into second-generation stapled peptides (Fig. 3B). At the same time we inspected the phage-derived peptides for patterns of residue utilization that were likely to promote or deter cell penetration. Although no hard-and-fast rules for cell penetration have yet been derived, removal of negatively charged residues and introduction of Arg generally enhances cell penetration by stapled peptides (26), as it does for other cell-penetrating peptides (34). We noted that the sequences of the phage display-derived peptides suggested at least tolerability of a Glu to Arg substitution at position 467 and Glu to Gln at position 470. These changes were therefore also incorporated into the second-generation stapled peptides.
Fig. 3.
Cell permeability and β-catenin binding of StAx peptides. (A) Summary of variants observed at least twice in the 32 selected phage sequences. Numbers indicate the frequency of appearance at a position (complete list in Fig. S3B). (B) Stapled peptide sequences including their Kd (mean ± SE) with β- catenin and overall charge (calculated for fStAx at pH 7.5 with Marvin 5.2.3, 2009, ChemAxon). (C) Analysis of DLD1 cellular uptake of fStAx peptides (7.5 μM, 24 h) by confocal fluorescence microscopy (overlaid images with blue: nuclear DAPI, green: fluorescein; details in Fig. S5). (D) In vitro competition of aStAx peptides (0.1, 0.5, 2.5 μM) with bead-immobilized GST-TCF4(1-52) for β-catenin (0.5 μM). (E) Pull-down assay with bStAx peptides in DLD1 lysates.
Shown in Fig. 3B are sequences and β-catenin–binding affinities for stapled peptides bearing various sequence alterations grafted onto the StAx-3 framework. Truncation of the peptide to eliminate Glu467 and Gln468 (fStAx-31) had little effect on binding. Introduction of Glu470 to Gln and Gln478 to Arg mutations in fStAx-32 moderately reduced affinity for β-catenin. Replacement of Met481 with Trp improved binding affinity by a factor of eight (fStAx-33). Another twofold increase in affinity was observed upon addition of Arg and Trp at positions 467 and 468 (fStAx-34), respectively. To increase the positive charge of the peptides, we further modified fStAx-34 by addition of arginine residues yielding peptides fStAx-35 and -35R. Competition FP assays and surface plasmon resonance experiments show reversibility of StAx binding to β-catenin (Fig. S4). As negative controls, we designed and evaluated fStAx-40, -41, and -41R, which show decreasing affinity for β-catenin with an increasing number of modifications (Fig. 3B).
Confocal fluorescence microscopy was used to evaluate cell penetration by fStAx peptides (Fig. 3C and Fig. S5). DLD1 colon cancer cells were incubated with 7.5 μM fluorescein-labeled fStAx peptides for 24 h, followed by fixation and DAPI nuclear staining. Negatively and slightly positively charged peptides did not show significant cellular uptake (fStAx-31 to -33), whereas peptides with an overall charge of more than +2 (fStAx-34 to -41R) did efficiently penetrate cells. In particular, StAx-35 and -35R, as well as the corresponding negative control peptides fStAx-41 and -41R, showed high levels of intracellular fluorescence distributed throughout the cytosol and nucleus (green in Fig. 3C).
The efficiency of the most highly cell-penetrant StAx peptides to compete with TCF4 for binding of β-catenin was investigated using an in vitro pull-down assay. Briefly, the GST-tagged CBD of TCF4(1-52) was immobilized on glutathione-labeled agarose beads and used to precipitate β-catenin (Fig. 3D). In the presence of N-terminally acetylated stapled peptides aStAx-35 and -35R, the binding of β-catenin to GST-TCF4(1-52) was inhibited competitively but was unaffected by the presence of the negative control peptide aStAx-41R. To determine whether the stapled peptides bind endogenous β-catenin, pull-down assays were performed in DLD1 cellular lysates. DLD1 is a Wnt-driven colon cancer cell line with elevated levels of β-catenin. Biotinylated versions of stapled peptides bStAx-35, -40, and -41 were incubated with the lysate and subsequently treated with streptavidin-conjugated beads to immobilize the peptides. For bStAx-35, Western blot probing with β-catenin–specific antibodies verified the formation of a bStAx-35/β-catenin complex (Fig. 3E). Treatment with control peptides bStAx40 and -41 resulted in only faint signals for β-catenin.
Structural Characterization of StAx-35/β-Catenin Interaction.
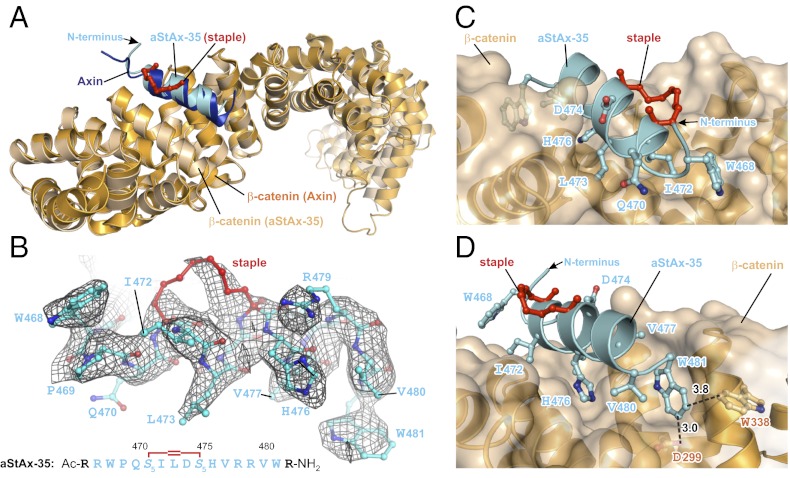
To understand the molecular basis of the interaction between the StAx peptides and β-catenin we determined the crystal structure of N-terminally acetylated aStAx-35 in complex with residues 134–665 of β-catenin at 3.0-Å resolution (Fig. 4 and Table S2). A Fo-Fc difference map calculated using an unliganded β-catenin structure (PDB ID 1QZ7) (18) as a phasing model revealed unambiguously the presence of aStAx-35 in the structure and its binding site on β-catenin (Fig. 4 A and B). The Cα superposition of residues 194–409 between β-catenin in the β-catenin/Axin and the β-catenin/aStAx-35 structures (rmsd = 0.47 Å) revealed a nearly identical overlay between the backbone of the wild-type Axin CBD peptide with the aStAx-35 stapled peptide (Cα rmsd = 0.38 Å; overlay in Fig. 4A). The wild-type residues of aStAx-35 interact with β-catenin in the same way as Axin itself, and the mutated residues seem to gain additional favorable interactions not made by the wild-type Axin CBD. In particular, the Trp residues at positions 468 and 481 that increased the binding affinity for β-catenin (Fig. 3B) seem to act as molecular “clasps” that bracket the ends of the peptide and engage recognition pockets of the surface of β-catenin (Fig. 4 C and D). Trp481 forms an edge-to-face interaction with Trp338 of β-catenin (shortest C-C distance = 3.8 Å) and also seems to contribute a σ-hydrogen bond between the edge of Trp481 and the carboxyl oxygen of Asp299 in β-catenin (O-C distance = 3.0 Å). These two interactions between Trp481 of aStAx-35 and β-catenin are absent in the Axin peptide, which has a methionine at position 481, and is presumably the origin of the eightfold increase in affinity that accompanies this amino acid substitution. As mentioned above, arginines were added to aStAx-35 at the N- and C-terminal positions 466 and 482, respectively, as well as at positions 467 and 478 to increase cell permeability. The terminal amino acids Arg476 and Arg482 are disordered in the crystal structure, and Arg467 and Arg478 do not contact β-catenin. The hydrocarbon atoms of the staple, most of which are well-ordered (Fig. 4 C and D), are oriented away from the interaction face of aStAx-35 and β-catenin and do not contribute directly to the protein/peptide interaction.
Fig. 4.
Crystal structure of the β-catenin/aStAx-35 complex. (A) Cα superposition of the X-ray structures of β-catenin bound to the Axin-CBD (PDB ID 1QZ7) (18) and aStAx-35 (rmsd = 0.47 Å). (B) Fo-Fc difference map (grid) calculated using β-catenin alone from the Axin-liganded structure (PDB ID 1QZ7) (18) as a phasing model and the density for the final structure of the β-catenin/aStAx-35 complex. The Axin numbering system was retained throughout for the stapled peptides. (C and D) Two different views of the aStAx-35/β-catenin interaction, with selected interacting side-chains shown explicitly. Distances in angstroms to several nearby residues are shown (coordinates of the β-catenin/aStAx-35 complex structure have been deposited under PDB ID 4DJS).
Inhibition of β-Catenin–Mediated Transcriptional Activities.
A reporter gene assay based on Wnt3a stimulation of HeLa cells was chosen to explore the effects of the StAx peptides on transcriptional activity. Cells were transfected with two constructs, one in which 10 tandem repeats of a TCF4 binding element with a minimum promoter was located upstream of the firefly luciferase cDNA (TOPflash), and the other construct containing a noninducible Renilla luciferase cDNA for normalization. In the presence of the ligand Wnt3a, HeLa cells were treated with fStAx peptides for 24 h, followed by luciferase activity measurement. From the panel of peptides tested, fStAx-35 and -35R showed the most potent inhibition of β-catenin/TCF4-driven firefly luciferase activity (Fig. 5A). At a concentration of 15 μM, fStAx-35 and -35R were roughly three times more active in inhibiting luciferase activity than fStAx-34. Although StAx34 exhibits a higher affinity for β-catenin (Fig. 3B), the cellular uptake of this peptide is reduced in comparison with fStAx-35 and fStAx-35R (Fig. 3C). The luciferase activity of cells treated with active fStAx-35 and -35R shows dose dependency, whereas control peptide fStAx-41 does not affect reporter activity (Fig. 5B).
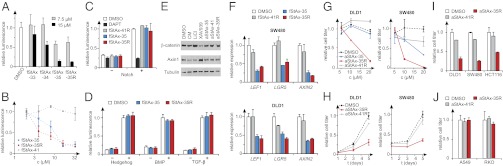
Fig. 5.
StAx peptides inhibit β-catenin–mediated transcriptional activity. (A) fStAx peptides inhibit TOPflash luciferase reporter activity in Wnt3a-stimulated HeLa cells. (B) fStAx-35 and -35R suppress luciferase activity in a dose-dependent manner. Negative control fStAx-41 is inactive. (C and D) fStAx peptides do not affect Notch, Hedgehog, BMP, and TGF-β signaling as examined in corresponding luciferase reporter assays in cells treated with 15 μM fStAx (DAPT). (E) β-Catenin and Axin1 protein levels in SW480 cell line were revealed by Western blot. Unlike the tankyrase inhibitor XAV939 (5 μM) or GSK3-β inhibitor LiCl (10 mM), aStAx peptides (10 μM) did not affect protein levels of Axin1 or β-catenin (CM: Wnt3a conditioned medium). (F) fStAx peptides (10 μM) inhibit the mRNA level of Wnt/β-catenin target genes in DLD1 and SW480 cells. Relative mRNA level was normalized with the housekeeping gene β-actin (all treatments for 24 h with 10% serum). (G) Proliferation of DLD1 and SW480 cells was blocked by aStAx-35R in a dose-dependent manner (5-d treatments). (H) Proliferation tendency of DLD1 and SW480 cells was significantly repressed after treatment with 10 μM aStAx-35R for 5 d. (I and J) aStAx-35R showed inhibitory effects on cell proliferation only in β-catenin–dependent cancer cell lines DLD1, SW480, and HCT116 but not on β-catenin–independent cell lines A549 and RKO (all treatments with 10 μM aStAx for 5 d, cell titer determined via cellular ATP level). Data points, mean ± SD.
To validate the specificity of StAx peptides, we examined their activity in reporter assays having luciferase expression driven by promoter elements responsive to signaling pathways other than Wnt, namely Notch, Hedgehog, BMP, and TGF-β. Whereas the Notch-driven reporter showed strong activation with NOTCH1 overexpression (Fig. 5C), which was diminished by treatment of the Notch pathway antagonist DAPT (N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycinet-butyl ester) (35), no such diminution was seen upon corresponding treatment with 15 μM fStAx peptides. Similarly, fStAx peptides had no statistically significant effect on either basal or ligand-stimulated reporter gene transcription downstream of the Hedgehog, BMP, or TGF-β signaling pathways (Fig. 5D). These data demonstrate that transcriptional antagonism by the StAx peptides is specific for the Wnt/β-catenin signaling pathway.
Wnt pathway perturbants that act upstream of β-catenin can alter levels of the β-catenin protein. To determine whether StAx peptides modulate β-catenin levels, we used Western blotting with an anti–β-catenin antibody to examine total β-catenin protein levels, and also levels of the Axin1 protein, in the presence and absence of StAx peptides (Fig. 5E). Consistent with previous reports (7–9), we observed stabilization of β-catenin in presence of LiCl, which is known to inhibit phosphorylation of β-catenin by GSK3β. Again as previously reported, treatment of cells with the small-molecule tankyrase inhibitor XAV939, a known Axin-stabilizing agent (8), resulted in decreased β-catenin and elevated Axin levels. On the other hand, treatment with 10 μM aStAx peptides did not significantly alter total levels of either β-catenin or Axin, consistent with the StAx peptides acting by a mechanism downstream of the pathway components involved in regulating levels of both proteins. In addition, we investigated the ability of StAx peptides to interfere with β-catenin/E-cadherin dimerization, which is essential for cell integrity and independent of Wnt signaling. Pull-down experiments show that StAx-35R does not inhibit the binding of β-catenin to phosphorylated E-cadherin in vitro (Fig. S6B), which is in agreement with the reported high stability of this complex (2). The most likely mode of action for the StAx peptides entails direct interference of β-catenin binding to TCF transcription factors in the nucleus.
Next we examined the effect of StAx peptides on mRNA expression levels of target genes previously shown to be transcriptionally up-regulated by Wnt pathway activation. Two colorectal cancer cell lines known to have elevated β-catenin levels, DLD1 and SW480, were treated with StAx peptides for 24 h, followed by total RNA extraction and quantitative RT-PCR of known β-catenin target genes LEF1, LGR5, and AXIN2 (1). In agreement with the findings of the TOPflash luciferase reporter assays described above, both fStAx-35 and -35R cause a substantial reduction in the mRNA levels of β-catenin/TCF target genes (Fig. 5F).
Reduced Viability of Wnt-Dependent Cancer Cells.
Previous work has shown that inhibition of Wnt/β-catenin signaling can decrease the proliferation of Wnt-dependent cancer cell lines (7–9). We therefore tested the StAx peptides for their effects on the proliferation of cancer cells harboring genetic changes that result in dependence on β-catenin for growth and survival. The colorectal cancer cell lines DLD1 and SW480 harbor deletions of APC, whereas HCT116 harbors both APC deletion and a mutation in β-catenin that blocks ubiquitination; these lines were chosen on the basis of their known dependence on β-catenin for growth and survival. Treatment of DLD1 and SW480 cells with increasing concentrations (5–20 μM) of aStAx-35R caused a significant decrease in cellular ATP levels, compared with DMSO and the negative control peptide aStAx-41R (Fig. 5G). Notably, time course experiments revealed efficient inhibition of cell proliferation after 5 d of treatment with 10 μM aStAx-35R (Fig. 5H). aStAx35-R has similar effects on the proliferation of DLD1 cells as does the tankyrase inhibitor XAV939 (Fig. S6C). On the basis of the selectivity of active StAx peptides for the inhibition of the Wnt signaling pathway, we expected the growth inhibitory effects of aStAx-35R to be selective for Wnt-dependent cancer cells. To explore the selectivity of aStAx-35R, we compared its effect on Wnt-dependent cancer cell lines such as DLD1, SW480, and HCT116, with cell lines that are known to be independent of sustained Wnt signaling (RKO and A549) (7, 8). After a 5-d treatment with 10 μM aStAx-35R, we indeed observed selective inhibition of cellular proliferation ranging from 52–74% for the Wnt-dependent cell lines (Fig. 5I), whereas in RKO and A549 cancer cells proliferation was not affected (Fig. 5J).
Discussion
An emerging theme in cancer biology is the inappropriate constitutive activation of signaling pathways that are ordinarily turned on during embryonic development but that subsequently become quiescent in most adult tissues. These pathways—Hedgehog, Notch, and Wnt, among others—have aroused great interest of late, because they seem to confer stem cell-like properties or “stemness” on cancers and are associated with aggressive growth, propensity toward migration and invasiveness, and poor responsiveness to current treatments. Whereas agents that target the Hedgehog and Notch pathways are currently in clinical development, no such therapeutic option has emerged in the case of Wnt. The Wnt pathway was discovered by Nusse and Varmus for its association with virally induced mammary cancers in mice, but is now known to play a key role in establishment and maintenance of a wide range of cancers, the archetypal example being hereditary and spontaneous colon cancers. Oncogenic activation of Wnt signaling can ensue from a variety of primary molecular lesions, but nearly all share the common feature of increasing cellular levels of β-catenin by interfering with its constitutive degradation. β-Catenin serves as a signaling hub for the Wnt pathway, engaging in crucial protein–protein interactions with both negative and positive effectors of the Wnt pathway; the negative regulatory interactions with β-catenin are specifically disrupted in cancer, either through mutations in β-catenin or in its interacting partners. These findings have directed particular focus on β-catenin as the target of choice in Wnt-driven cancers, but attempts to discover selective high-affinity binders to β-catenin have not previously met with success. This is perhaps unsurprising in light of numerous X-ray structures of β-catenin bound to physiologic peptide ligands, which show extended binding interfaces some 60 Å in length (as in Fig. 1B) and devoid of pronounced hydrophobic pockets conducive to the binding of small molecules (6–14). In the present work, we have used the stapled peptide approach to obtain cell-permeable molecules rigorously shown to bind β-catenin directly and thereby block its ability to engage the transcriptional activator TCF4.
Our initial studies focused on the CBD from the downstream target TCF4, but we found that this motif required not only the α-helical portion but also an extended appendage to bind with high affinity, and we were concerned that the latter would represent a proteolytic liability. We noticed, however, that a nearly all-helical CBD peptide from Axin bound to the same site as TCF4. Through a systematic approach that entailed screening of candidate stapling positions, affinity optimization via phage display, and introduction of residues that promote cell penetration, we were able to discover stapled Axin-derived (StAx) peptides that function as direct β-catenin antagonists in vitro and in cultured cells. These peptides, but not closely related negative control molecules, interfere with transcriptional activation of canonical Wnt pathway genes but leave unaffected genes responsive to the Hedgehog, BMP, and TGF-β cellular signaling pathways. The most active stapled peptide, aStAx-35R, was found to induce growth inhibition of Wnt-dependent cancer cells without affecting the growth of cancer cells that do not rely on deregulated Wnt signaling.
Interestingly, although crystallographic analysis clearly demonstrates that StAx-35R binds at the same site as does Axin, the stapled peptide does not increase steady-state levels of β-catenin. This suggests that either the binding site for the stapled peptide is inaccessible in the cytoplasmic destruction complex or that binding of the stapled peptide to that complex is insufficient to disrupt it. Whereas no changes were seen at the level of the β-catenin protein, strong suppression was observed at the mRNA level for a panel of canonical Wnt-driven genes but not for genes downstream of other signaling pathways. This transcriptional suppression indicates that aStAx-35R binding is sufficient to antagonize the nuclear form of β-catenin, which is supported by in vitro competition experiments with the CBD of TCF4. Future efforts will aim to gain a more detailed understanding of the role of StAx peptides in this complex interaction network, to assess StAx peptides in animal models of Wnt-driven cancers, and to optimize the potency and pharmacologic properties of this new class of Wnt pathway-specific transcriptional antagonists.
Methods
Peptide synthesis and characterization, phage display selection and cell-based assays were performed as described in SI Methods. Details related to X-ray crystallography and structure determination are presented in SI Methods. The coordinates of the complex structure have been deposited under PDB 4DJS.
Supplementary Material
Acknowledgments
We thank M. Kolar and Y.-W. Kim for assistance with automated solid-phase peptide synthesis; the Broad Institute Chemical Biology Program for access to instrumentation; A. Koehler and C. Feau at the Broad Institute for their support of the Biacore experiments; C. Sevenich at Technical University Dortmund, Germany for performing high-resolution mass spectroscopy experiments; W. Xu at University of Washington, Seattle for kindly providing the β-catenin expression construct; and H. Clevers at Hubrecht Institute, The Netherlands for kindly providing TOPflash reporter construct. This research was supported by GlaxoSmithKline. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. T.N.G. is the recipient of a fellowship (LPDS 2009-2) from Deutsche Akademie der Naturforscher Leopoldina. J.T-H.Y. was supported by a grant from Susan G. Komen for the Cure (KG091054). R.E.M was supported by an American Association for Cancer Research Centennial Pre-doctoral Research Fellowship.
Footnotes
Conflict of interest statement: The stapled peptide technology used in this paper has been licensed to Aileron Therapeutics, in which G.L.V. has a financial interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4DJS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208396109/-/DCSupplemental.
References
- 1.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Huber AH, Weis WI. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell. 2001;105(3):391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- 3.Fearnhead NS, Wilding JL, Bodmer WF. Genetics of colorectal cancer: Hereditary aspects and overview of colorectal tumorigenesis. Br Med Bull. 2002;64:27–43. doi: 10.1093/bmb/64.1.27. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima M, et al. Reduced expression of Axin correlates with tumour progression of oesophageal squamous cell carcinoma. Br J Cancer. 2003;88(11):1734–1739. doi: 10.1038/sj.bjc.6600941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8(2):97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 6.Shan JF, Shi DL, Wang JM, Zheng J. Identification of a specific inhibitor of the dishevelled PDZ domain. Biochemistry. 2005;44(47):15495–15503. doi: 10.1021/bi0512602. [DOI] [PubMed] [Google Scholar]
- 7.Chen BZ, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang SMA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 9.Thorne CA, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol. 2010;6(11):829–836. doi: 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu DS, et al. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci USA. 2011;108(32):13253–13257. doi: 10.1073/pnas.1110431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Liu H, Wang S, Hao X, Li L. A diterpenoid derivative 15-oxospiramilactone inhibits Wnt/β-catenin signaling and colon cancer cell tumorigenesis. Cell Res. 2011;21:730–740. doi: 10.1038/cr.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emami KH, et al. A small molecule inhibitor of β-catenin/cyclic AMP response element-binding protein transcription [corrected] Proc Natl Acad Sci USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepourcelet M, et al. Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell. 2004;5(1):91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 14.Gonsalves FC, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci USA. 2011;108(15):5954–5963. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian W, et al. Structure-based discovery of a novel inhibitor targeting the β-catenin/Tcf4 interaction. Biochemistry. 2012;51(2):724–731. doi: 10.1021/bi201428h. [DOI] [PubMed] [Google Scholar]
- 16.Graham TA, Weaver C, Mao F, Kimelman D, Xu WQ. Crystal structure of a β-catenin/Tcf complex. Cell. 2000;103(6):885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 17.Sampietro J, et al. Crystal structure of a β-catenin/BCL9/Tcf4 complex. Mol Cell. 2006;24(2):293–300. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Xing Y, Clements WK, Kimelman D, Xu WQ. Crystal structure of a β-catenin/axin complex suggests a mechanism for the β-catenin destruction complex. Genes Dev. 2003;17(22):2753–2764. doi: 10.1101/gad.1142603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Wetering M, et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 20.Bird GH, et al. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc Natl Acad Sci USA. 2010;107(32):14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek S, et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc. 2012;134(1):103–106. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 22.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6(8):595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18(5):411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavathiotis E, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455(7216):1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129(9):2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305(5689):1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharya S, Zhang HT, Debnath AK, Cowburn D. Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid. J Biol Chem. 2008;283(24):16274–16278. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danial NN, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14(2):144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackwell HE, Grubbs RH. Highly efficient synthesis of covalently cross-linked peptide helices by ring-closing metathesis. Angew Chem Int Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 31.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 32.Kim YW, Grossmann TN, Verdine GL. Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat Protoc. 2011;6(6):761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 33.Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 34.Hansen M, Kilk K, Langel U. Predicting cell-penetrating peptides. Adv Drug Deliv Rev. 2008;60(4-5):572–579. doi: 10.1016/j.addr.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3(7):688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.