Abstract
Genetic variation between individuals has been extensively investigated, but differences between tissues within individuals are far less understood. It is commonly assumed that all healthy cells that arise from the same zygote possess the same genomic content, with a few known exceptions in the immune system and germ line. However, a growing body of evidence shows that genomic variation exists between differentiated tissues. We investigated the scope of somatic genomic variation between tissues within humans. Analysis of copy number variation by high-resolution array-comparative genomic hybridization in diverse tissues from six unrelated subjects reveals a significant number of intraindividual genomic changes between tissues. Many (79%) of these events affect genes. Our results have important consequences for understanding normal genetic and phenotypic variation within individuals, and they have significant implications for both the etiology of genetic diseases such as cancer and for immortalized cell lines that might be used in research and therapeutics.
Keywords: somatic mosiacism, genomic rearrangement
Genetic variation between individuals has been extensively explored (1, 2), but far less effort has been made to ascertain somatic genetic variation in healthy individuals. This information is important for understanding how somatic mutations arise de novo that lead to both phenotypic variation and somatic genetic human diseases such as cancer. There is presently considerable interest in generation of induced pluripotent stem cells (iPS cells) from the cells of adult tissues because these cells have the potential to be used for therapeutic purposes. Evidence of genomic alterations has been found in human embryonic stem cells (3) and human iPS cells (4–7). Recent findings suggest that immortalized cell lines (8) contain significant copy number differences, although it is unclear whether these variations arise in vitro during cell culturing or originate in vivo in somatic cell populations.
On the basis of results from the 1,000 Genomes Project and other analyses, it is estimated that there are upward of 2,500 structural variations (duplications, insertions, and inversions) between any two individuals, 700–1,000 of which are greater than 1 kb (9, 10). The 1,000 Genomes Project analyses estimate upward of 3 million single nucleotide polymorphisms between any two humans (2). Sequence analysis of parent–offspring trios and quartets has revealed a mutation rate corresponding to ∼70 de novo single nucleotide variations that occur between parents and offspring, presumably through germline mutations (11, 12). Structural variations between parents and offspring were not measured.
Somatic mosaicism is the presence of genetically distinct populations of somatic cells within an organism and even within the same tissue. Somatic rearrangements in various cancer cell types are well documented, and more than 30 Mendelian diseases have been associated with somatic mosaicism (13). Somatic mosaicism is well known to occur in normal germline cells as the process of meiotic recombination and in cells of the immune system within Ig and T-cell receptor genes as a result of V(D)J recombination that provides diversity in immune responses. Other studies have revealed age-related structural variation in human blood cells (14), aneuploidy and retrotransposition in the human brain (15, 16), and aneuploidy in human preimplantation embryos (17). Evidence of copy number variation (CNV) in monozygotic twins (18) also reveals the presence of somatic mosaicism in tissues arising from the same zygote.
High-resolution genomic arrays enable the monitoring of genetic variation with an accuracy not previously applied to somatic tissues. Somatic mosaicism has been studied in cultured cells (8), but only one study has documented copy number variation between unfixed human tissues within individuals using BAC arrays for array-comparative genomic hybridization (aCGH) (19). These arrays produce low-resolution data, and they have high error rates, leaving uncertainty. Although deep whole-genome sequencing provides the highest resolution and the ability to resolve rearrangements to the base pair, mapping sequences from mosaic samples remains a challenge using available sequencing technologies. aCGH using arrays that can resolve rearrangements as small as ∼2 kb provides a sensitive and well-established technology for this investigation and can detect mosaic events in tissues.
Results
Identification of Copy Number Variations (CNVs) in Somatic Tissues.
The presence of somatic CNVs was investigated using organ tissues from subjects obtained during routine autopsy. In total, tissues from six individuals (age range, 45–85 y) not known to be affected by any disease with a genetic component (e.g., hereditary disorders or cancer) were analyzed using Nimblegen 2.1M oligonucleotide whole-genome arrays (20). Between three and 11 tissues were tested for each of the six subjects. The tissue that yielded the most abundant and high-quality DNA for each subject was selected as the reference. The remaining tissues, referred to as test tissues, were each hybridized against the reference for each subject. DNA from the test tissues was labeled with Cy3 fluorescent dye and hybridized to DNA from the reference tissue labeled with Cy5 fluorescent dye. For each tissue comparison, “dye swap” experiments were also performed in which the reference DNA was labeled with Cy3 and the test DNA was labeled with Cy5. We required that an event be observed in both “dye swap” experiments to be included in the list of total somatic CNVs (Fig. 1A and Table S1). As a control, the reference DNA of one subject was labeled with Cy3 and hybridized to the same DNA labeled with Cy5. Positive events called using Nexus Copy Number software were detected when different tissues types were hybridized, and the number varied with the threshold used (Table S2). CNVs above the threshold chosen for calling somatic CNVs were not detected when the reference DNA was hybridized to itself.
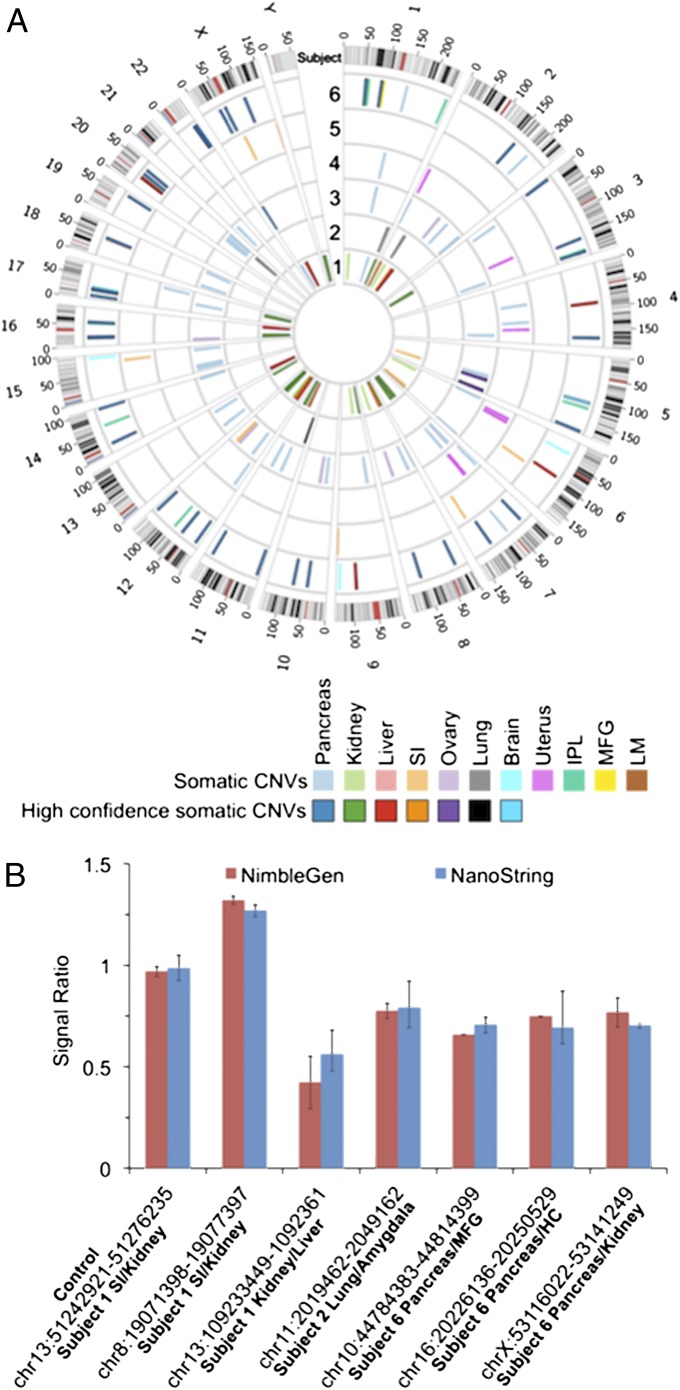
Fig. 1.
(A) Locations of somatic CNVs for each of six subjects. (B) Ratios of selected somatic CNVs by NimbleGen aCGH and NanoString validation. A control region with no CNV between somatic tissues is also included. HC, hippocampus; IPL, inferior parietal lobe; LM, leptomeninges; MFG, middle frontal gyrus; SI, small intestine.
To set a threshold for calling CNVs, we analyzed a number of CNVs in various tissues using an initial set of 50 NanoString probes. A threshold was established that minimized the false discovery rate (10%; Materials and Methods). Two hundred twenty-nine CNVs passed the threshold. CNVs from telomeric and centromeric regions were removed, because these regions are prone to cause cross-hybridization signals on aCGH. CNVs that appeared in the same location in multiple test tissues from the same individual were ascribed to the reference tissue. This resulted in a set of 178 candidate somatic CNVs. Of the 178 regions, 168 were amenable to NanoString probe design and subjected to a second round of validation. Of the 168 regions tested by NanoString technology, 73 were validated across six people and 36 tissue comparisons (Fig. 1A and Table S1). We refer to this list of 73 events as the high-confidence set.
The frequency of high-confidence events varied from 0 to 45 per individual, with most events detected within the two individuals with the most tissues tested. Reference tissues often yielded the greatest numbers of CNVs per individual. The increased number of tests performed with the reference tissues presumably increased detection of CNVs in those samples. It is plausible that all 178 events that were reproducibly detected by the arrays are bona fide events but that many were not confirmed using the NanoString technology owing to inadequate probe design or they were at a level below the stringent threshold used in our validation process. Regardless, these results indicate that there are a large number of somatic CNVs in the tissues of adults.
CNVs Occur in Many Tissue Types and Can Be Large.
Somatic CNVs were detected by aCGH in a variety of tissues (Fig. 1A, Table S1, and Table S3). The greatest number of somatic CNVs was detected in the pancreas of subject 6 (43 events), followed by those observed in pancreas of subject 3 (35 events) and kidney of subject 1 (21 events). Events in brain tissues were detected by aCGH in the leptomeningeal tissue (two events) of subject 1, as well as in tissues of the inferior parietal lobe (10 events) and middle frontal gyrus (one event) of subject 6. There were five events in subject 6 that appeared in more than one brain tissue and likely affect the entire brain. Events in the small intestine tissues were observed in subjects 1 and 5 (seven and six events, respectively). Somatic CNVs were detected by aCGH in all of the liver tissues tested (four subjects). Tissues of the reproductive system were obtained for the two female subjects analyzed. Ten ovary-specific events were detected in subject 3, and eight uterus-specific events were detected in subject 4. Examples of CNVs discovered by NimbleGen and validated by NanoString are shown in Fig. 1B and Fig. 2. The validated events in Fig. 1B demonstrate that somatic CNVs have signals less than 1.5 (a 3:2 ratio for a duplication) or greater than 0.5 (a 1:2 ratio for a deletion). In part, this is to be expected because the tissues and organs contain a substantial fraction of nonparenchymal cells from blood vessels and connective tissue. However, the signals suggest that the events may not be homogeneous throughout the parenchymal cells of the tissue.
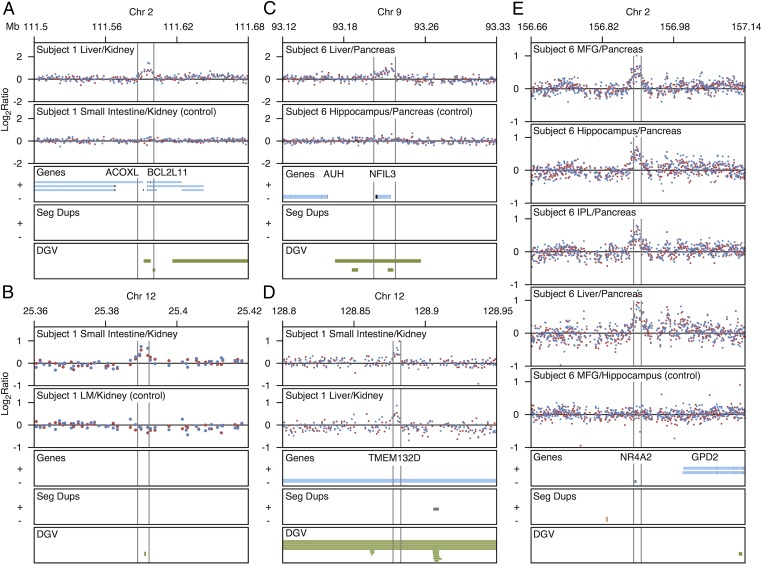
Fig. 2.
aCGH signal for somatic CNVs. (A) Increased signal in liver tissue when hybridized against kidney reference tissue of subject 1 corresponding to a duplication of chromosome 2 DNA in liver. (B) Increased signal in small intestine tissue when hybridized against kidney reference tissue of subject 1 corresponding to a duplication of chromosome 12 DNA in small intestine. (C) Increased signal in liver tissue when hybridized against pancreas reference tissue of subject 6 corresponding to a duplication of chromosome 9 DNA in liver. (D) Increased signal in small intestine and liver tissues when hybridized against kidney reference tissue of subject 1 corresponding to a deletion in the kidney reference tissue on chromosome 12. (E) Increased signal in MFG, hippocampus, IPL, and liver tissues hybridized against pancreas tissue in subject 6 corresponding to a deletion in the pancreas reference tissue on chromosome 2. Controls show no signal change between hybridized tissues. DGV, database of genomic variants; IPL, inferior parietal lobe; LM, leptomeninges; MFG, middle frontal gyrus; Seg Dups, segmental duplications. Blue and red dots represent dye-swap experiments.
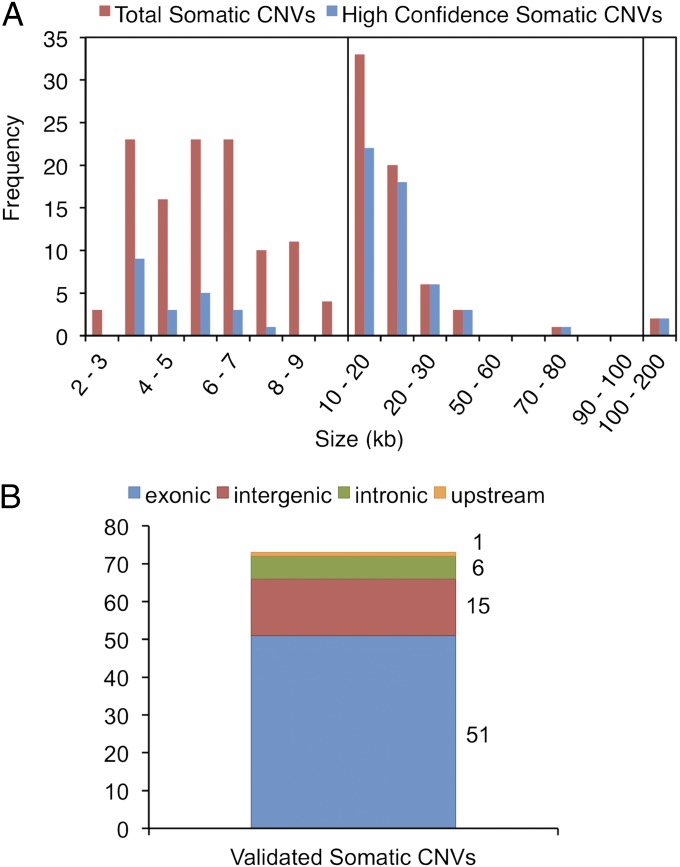
The size distribution of somatic CNVs was analyzed (Fig. 3A). Somatic CNVs with sizes smaller than 10 kb were binned into 1-kb intervals, somatic CNVs with sizes between 10 kb and 100 kb were binned into 10-kb intervals, and somatic CNVs with sizes larger than 100 kb were binned into 100-kb intervals. The sizes of the total somatic CNVs discovered by aCGH ranged from 2.0 kb to 184 kb, and those of the NanoString-validated set ranged from 3.2 kb to 184 kb. Thus, larger events were more likely to be validated by NanoString: 30 of 32 CNVs (93.8%) larger than 20 kb were validated, whereas only 43 of 146 CNVs (29.5%) smaller than 20 kb were validated. It is possible that the smaller CNVs were either false positive, or the validation probes were not optimal for the more limited target regions.
Fig. 3.
Analysis of somatic CNVs. (A) Frequency vs. size of all somatic CNVs discovered by aCGH and frequency vs. size of all high-confidence somatic CNVs validated by secondary methods. (B) Genomic elements affected by validated high-confidence CNVs.
Many Somatic CNVs Affect Genes Involved in Regulation.
We determined the number of high-confidence CNVs that affect genes and found 58 (79%) intersect genes, including introns, exons, or upstream sequence; 51 of these (70% of the total) directly affect exons (Fig. 3B). Fig. 2 shows examples of CNVs that affect the ACOXL, BCL2L11, TMEM132D, NFIL3, and NR4A2 genes. Fig. 2A shows a ∼13.7-kb increase in signal in DNA from liver tissue when hybridized against DNA from reference kidney tissue, suggesting a duplication in liver of a region overlapping both the BCL2L11 gene and the ACOXL gene. BCL2L11 mRNA is expressed in several human tissues, including liver (21). Fig. 2C shows an increase in signal of a ∼21.2-kb region on chromosome 9 of subject 6 in liver tissue DNA when hybridized against pancreas reference tissue DNA, suggesting a duplication of a region encompassing the NFIL3 gene in liver tissue. Expression of NFIL3 was previously reported in human liver tissue (22). Fig. 2 D and E show examples of somatic CNVs observed in the DNA of reference tissues. Fig. 2E shows an increase in signal for DNA from tissues of three brain regions and liver when hybridized against pancreas tissue DNA of subject 6 in a region encompassing the NR4A2 gene. Because this event is observed in all hybridizations against the pancreas reference tissue, the event is likely a deletion in the pancreas. More examples are shown in Fig. S1. These results indicate that most somatic CNVs affect genes. Some of these genes have previously been reported to be expressed in the affected tissues.
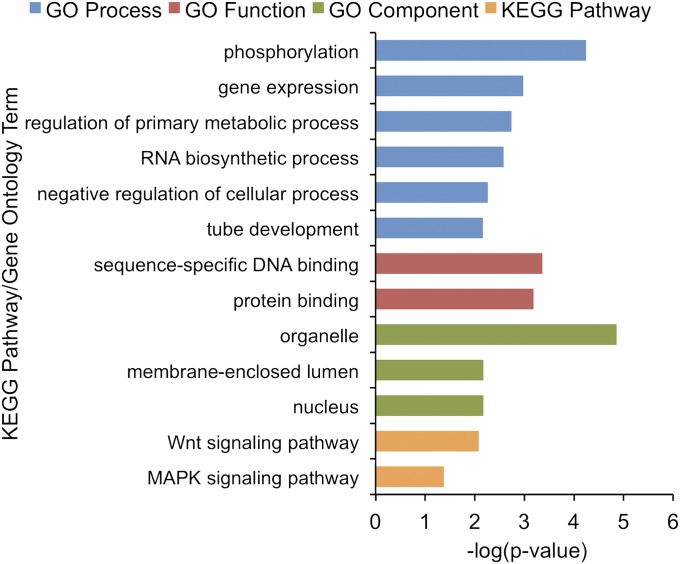
Investigation of the types of genes affected by high-confidence somatic CNVs was performed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (23, 24) (Fig. 4 and Table S4). GO analysis revealed modest enrichments for genes within somatic CNVs in ∼30 GO categories. Many of the GO categories relate to regulation of cellular processes such as regulation of phosphorylation, regulation of primary metabolic processes, and regulation of gene expression. Sequence-specific DNA binding and protein binding were also enriched in genes within somatic CNVs. KEGG pathway analysis revealed modest enrichments in the Wnt signaling pathway and the MAPK signaling pathway. Thus, many of the CNVs are likely to affect regulatory processes in the cell.
Fig. 4.
Selected GO terms and KEGG pathways enriched in genes within high-confidence somatic CNVs.
Further analysis was performed on genes within the somatic CNVs discovered to investigate whether any of the affected genes within the same subject or tissue are known to interact. Deletion events affecting the MAP2K3 gene on chromosome 17 and the SMAD7 gene on chromosome 18 were observed in pancreas tissue of subject 6. MAP2K3 and SMAD7 were previously shown to be expressed in pancreas tissue (25), and SMAD7 has been shown to be overexpressed in pancreatic cancer (26). The SMAD7 and MAP2K3 genes were previously reported (27) to interact as part of the TGF-β–dependent activation of p38 MAPK pathway inducing apoptosis in prostate cancer cells. Thus, in at least one case the CNVs affect interacting components.
Several Somatic CNVs Occur in the Same Location in Multiple Individuals.
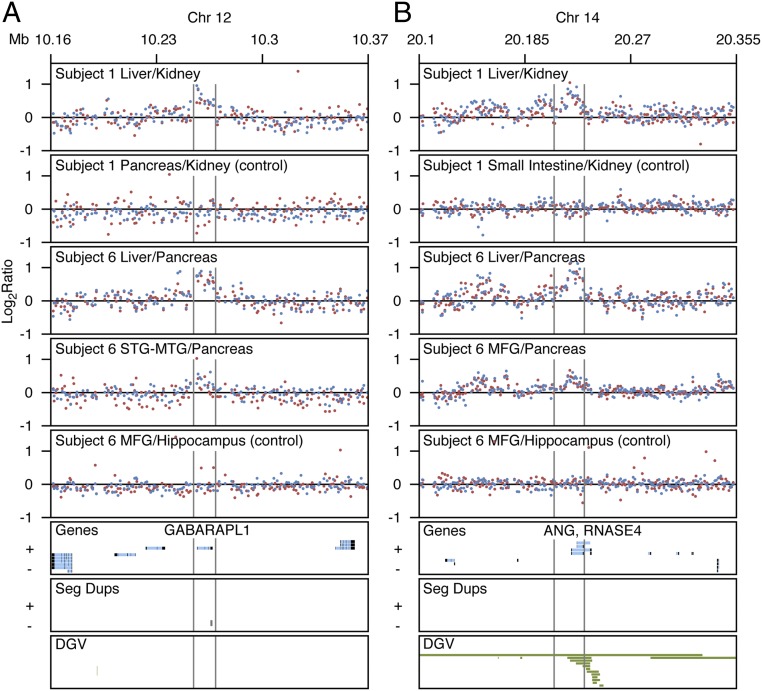
Nearly all events were unique; only seven regions overlap between two individuals, three of which were validated by NanoString. Fig. 5 A and B show regions on chromosomes 12 and 14, respectively, where CNVs were observed in both subjects 1 and 6. Fig. 5 A and B show increased signal in DNA from liver tissue when hybridized against DNA from kidney tissue of subject 1. An increased signal is observed for subject 6 in the same regions in DNA from liver tissue and tissue from brain regions when hybridized against DNA from reference pancreas tissue. Increased signal in all of the test tissues of subject 6 correspond to deletions in the reference pancreas tissue for both chromosomal regions. The events on chromosome 12 occur in a region encompassing the GABARAPL1 gene (Fig. 5A). The events on chromosome 14 overlap both the ANG gene and the RNASE4 gene (Fig. 5B). ANG and RNASE4 were previously shown to be expressed in pancreas and liver (28, 29). Thus, a few CNVs (4%) lie in the same regions and contain genes.
Fig. 5.
aCGH signal for somatic CNVs that appear in more than one subject. (A) Increased signal in liver tissue hybridized against kidney tissue of subject 1 corresponding to a duplication of chromosome 12 DNA in liver. Increased signal in liver tissue and superior temporal gyrus–middle temporal gyrus (STG-MTG) tissue hybridized against pancreas tissue of subject 6 corresponding to a deletion in the pancreas reference tissue on chromosome 12. (B) Increased signal in liver tissue hybridized against kidney tissue of subject 1 corresponding to a duplication of chromosome 14 DNA in liver. Increased signal in liver tissue and MFG tissue hybridized against pancreas tissue of subject 6 corresponding to a deletion in pancreas reference tissue on chromosome 14. Controls show no signal change between hybridized tissues. Blue and red dots represent dye-swap experiments.
Somatic CNV Breakpoints Analysis.
Analysis of the breakpoint regions of 73 high-confidence somatic CNVs was undertaken to assess the frequency of repetitive elements near somatic CNV breakpoints. aCGH does not provide base pair resolution that is necessary for determining exact CNV formation mechanisms (e.g., nonallelic homologous recombination, nonhomologous end-joining, and transpositions of mobile genomic elements). However, repetitive sequence is known to be integral in many of the CNV formation mechanisms. NimbleGen 2.1M oligonucleotide microarrays can achieve breakpoint resolution to ∼2 kb based on probe spacing of ∼1 kb. Therefore, a 2-kb window was analyzed around each breakpoint for the presence of six classes of repetitive sequence: L1, L2, Alu, LTR, segmental duplications, and microsatellites (Table 1). To test for significant enrichment or depletion for each class of repetitive element, P values were calculated by performing permutations of 10,000 sets of random intervals. Within each set of random intervals the number and size of the intervals was kept equal to those of the validated somatic CNVs being analyzed. The locations of the intervals were randomized within the regions probed by the NimbleGen 2.1M microarray. The number of somatic CNVs that intersect a repetitive element class was compared with the average number of random intervals that intersect the element to calculate an enrichment/depletion ratio. Thirty-nine of 73 somatic CNVs have a breakpoint that intersects a microsatellite, an enrichment of ∼1.29 over the genomic background (P < 3.8e−2). Microsatellites have been suggested to play a role in chromosomal rearrangement (30), and a colocalization of microsatellites with CNVs was previously reported (31). Twenty-five of 73 somatic CNVs have a breakpoint that overlaps an L1, a significant depletion of ∼0.53 compared with the genomic background (P < 1.2e−7). Twenty-nine of 73 somatic CNVs have a breakpoint that intersects an LTR, a significant depletion of ∼0.77 compared with random intervals (P < 4.8e−2). Segmental duplications, Alu, and L2 were not significantly enriched or depleted near somatic CNV breakpoints compared with average random intervals.
Table 1.
Analysis for enrichment or depletion of several classes of repetitive elements at somatic CNV breakpoints
| Repeat type | No. of high-confidence somatic CNVs that intersect repeat type (out of 73) | Size range of high-confidence somatic CNVs that intersect repeat type (kb) | Enrichment | P |
| Microsatellite | 39 | 3.23–184 | 1.291 | 3.8e−2 |
| LTR | 29 | 3.23–73.6 | 0.773 | 4.8e−2 |
| L1 | 25 | 3.47–39.2 | 0.534 | 1.2e−7 |
| L2 | 32 | 3.23–184 | 1.150 | 3.1e−1 |
| Alu | 55 | 3.23–184 | 1.056 | 4.5e−1 |
| Segmental duplication | 5 | 5.33–22.8 | 0.743 | 4.8e−1 |
In summary, these results show that somatic CNVs occur in humans in a variety of tissues. Although the sample size is small, the number of events detected was often higher in dividing tissues (e.g., liver, small intestine, and pancreas) relative to nondividing tissues (e.g., brain).
Discussion
This study confirms that humans have extensive genetic variation in somatic tissues. aCGH and the validation method used can detect heterogeneous cell populations within tissue samples. Many of the events detected have aCGH signals corresponding to less than a full-copy duplication or deletion, corroborating that there are indeed heterogeneous cell populations within tissues. To detect these events, they must be present in a substantial fraction of the parenchymal cells. In proliferating tissues, such as in the cases of p53 mutations in UV-irradiated skin (32) and some of the CNVs seen in normal blood cells (14), it seems probable that the somatic mutation conferred a growth advantage to the affected cells. In organs that undergo little or no proliferation in the adult, one possibility is that the observed CNV may have given a growth or differentiation advantage to the affected precursor cells during development. It is also likely that many more CNVs exist in small numbers of parenchymal cells but do not give any growth advantage to the cells.
These results have important implications for the detection of disease formation and treatment. A stepwise theory of cancer has been proposed to explain the origin of tumor populations (33–36). This theory posits that genetic instability in precancerous cells leads to an accumulation of mutations that eventually result in cancer. Our results suggest that CNVs may occur commonly during somatic cell growth and division. In some cases, these might result in predilection for diseases such as cancer. Detection of such events in cells of healthy individuals may have prognostic value for the risk of malignancy. These results also indicate that choosing the relevant tissue for comparison of normal and disease tissues is important. Use of tissues from distant lineages may find events that are not relevant to formation or maintenance of the disease state and that might simply be “background” events in another tissue.
In developmental biology, there is great interest in understanding how common progenitor cells differentiate into distinct tissue types. Our results show that somatic tissues can genetically vary. Although our sample size is small, several of the tissues with the highest numbers of observed events are dividing tissues (e.g., liver and intestine). The pancreas, liver, and small intestine are all derived from the endoderm germ layer. Studies of differentiation have revealed that the ventral endoderm can give rise to each of these tissues, depending on signaling from various pathways (37–39). We observed seven events that occurred in the same locations in two individuals, suggesting that there are potential hotspots for somatic genomic variation. These types of events were observed in several different tissues, including pancreas, liver, and small intestine. It remains to be seen whether the somatic genetic differences observed here play a role in the differentiation process.
Finally, this work is particularly important for therapies that use iPS cells. iPS cells are typically derived from somatic tissues and may contain somatically altered DNA. Indeed, CNVs have been observed in iPS cells relative to the starting material from which they were derived (4–7). Our findings indicate that many of these events (and perhaps all) are not induced during iPS cell formation but perhaps exist in somatic cells in vivo. Although genetic alterations are generally viewed as negative, somatic variation could be beneficial. For example, in tissues that frequently encounter pathogens, CNVs that eliminate viral receptors might enhance host survival. Somatic genomic variation may have wide-reaching implications for diverse aspects of human biology and health, making it an important area of future research.
Materials and Methods
aCGH.
Tissues that were identified as noncancerous were obtained from routine autopsy. DNA was extracted from tissues using the Qiagen DNeasy Blood & Tissue Kit (Qiagen). For each subject, the tissue that yielded the greatest amount of DNA was selected as the reference tissue. Each of the other tissues from a subject was compared against the reference tissue. aCGH was performed using NimbleGen (Roche NimbleGen) 2.1M microarrays according to the NimbleGen protocol. Dye swap replicates were carried out for each tissue comparison to control for dye-specific bias. Arrays were scanned using the Roche MS 200 Microarray Scanner. Images were analyzed using NimbleScan 2.6 software. CNV calls were made with Nexus Copy Number version 6.0 (BioDiscovery) using the Fast Adaptive States Segmentation Technique 2 (FASST2) segmentation algorithm, a hidden Markov model-based approach.
Validation.
NanoString probes were designed to 50 regions of possible copy number change. Half of the regions had two probes designed, whereas the other half had three probes designed. Hybridization, sample prep, and scanning were performed according to the NanoString protocol. Data analysis was performed using the nCounter CNV Collector tool. Copy number was determined by averaging over two to three probes per region. To validate the NimbleGen CNV calls, test/reference NanoString ratios were compared with NimbleGen ratios. For ratios below 1 (i.e., deletions), the ratios were inverted for a direct comparison. Events with NanoString (validation) ratios of 1.25 or greater were considered to be valid CNVs. A false-positive rate of 10% was used as a cutoff, which corresponded to a NimbleGen ratio of 1.3 for subjects 1–5 and 1.35 for subject 6 (Figs. S2 and S3). All thresholding analyses were performed using the ROCR package for the R statistical computing package. These ratios were used as the thresholds for calling CNVs from the NimbleGen data for each tissue comparison using the FASST segmentation algorithm of the Nexus Copy Number (BioDiscovery) software. A second round of NanoString probes was designed to the candidate CNVs that had not been previously validated. One probe was used for each of 168 regions. Candidate events with NanoString ratios of 1.25 or greater were considered validated.
Note Added In Proof.
Our speculation that many of these events are not induced during iPS cell formation but perhaps exist in somatic cells in vivo received support from complementary data in a recent paper (40).
Supplementary Material
Acknowledgments
We thank Alexander Vortmeyer and colleagues at the Yale Pathology Department for providing tissue samples. This work was supported by National Institutes of Health Grants 5R01MH094740 (to M.P.S. and A.E.U.) and 5P50HG002357 (to M.P.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40159).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1213736109/-/DCSupplemental.
References
- 1.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durbin RM, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funk WD, et al. Evaluating the genomic and sequence integrity of human ES cell lines; comparison to normal genomes. Stem Cell Res (Amst) 2012;8:154–164. doi: 10.1016/j.scr.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 5.Hussein SM, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 6.Laurent LC, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martins-Taylor K, et al. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488–491. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- 8.Mkrtchyan H, et al. Early embryonic chromosome instability results in stable mosaic pattern in human tissues. PLoS ONE. 2010;5:e9591. doi: 10.1371/journal.pone.0009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mills RE, et al. 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conrad DF, et al. 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada T, Candotti F. Somatic mosaicism in primary immune deficiencies. Curr Opin Allergy Clin Immunol. 2008;8:510–514. doi: 10.1097/ACI.0b013e328314b651. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg LA, et al. Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet. 2012;90:217–228. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yurov YB, et al. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem. 2005;53(3):385–390. doi: 10.1369/jhc.4A6430.2005. [DOI] [PubMed] [Google Scholar]
- 16.Baillie JK, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]
- 18.Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piotrowski A, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 20.Nuwaysir EF, et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaviraghi M, et al. Identification of a candidate alternative promoter region of the human Bcl2L11 (Bim) gene. BMC Mol Biol. 2008;9:56. doi: 10.1186/1471-2199-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, et al. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan D, Prodduturi N, Zhang B. WebGestalt2: An updated and expanded version of the Web-based Gene Set Analysis Toolkit. BMC Bioinformatics. 2010;11(Suppl 4) [Google Scholar]
- 25.Marselli L, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93:1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleeff J, et al. The TGF-beta signaling inhibitor Smad7 enhances tumorigenicity in pancreatic cancer. Oncogene. 1999;18:5363–5372. doi: 10.1038/sj.onc.1202909. [DOI] [PubMed] [Google Scholar]
- 27.Edlund S, et al. Transforming growth factor-beta1 (TGF-beta)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 2003;14:529–544. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg HF, Dyer KD. Human ribonuclease 4 (RNase 4): Coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res. 1995;23:4290–4295. doi: 10.1093/nar/23.21.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Futami J, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997;16:413–419. doi: 10.1089/dna.1997.16.413. [DOI] [PubMed] [Google Scholar]
- 30.Ugarković D, Plohl M. Variation in satellite DNA profiles—causes and effects. EMBO J. 2002;21:5955–5959. doi: 10.1093/emboj/cdf612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim PM, et al. Analysis of copy number variants and segmental duplications in the human genome: Evidence for a change in the process of formation in recent evolutionary history. Genome Res. 2008;18:1865–1874. doi: 10.1101/gr.081422.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein AM, Brash DE, Jones PH, Simons BD. Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc Natl Acad Sci USA. 2010;107:270–275. doi: 10.1073/pnas.0909738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 34.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 35.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 36.Jackson AL, Loeb LA. On the origin of multiple mutations in human cancers. Semin Cancer Biol. 1998;8:421–429. doi: 10.1006/scbi.1998.0113. [DOI] [PubMed] [Google Scholar]
- 37.Deutsch G, Jung J, Zheng M, Lóra J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- 38.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung WS, Shin CH, Stainier DYR. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–748. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino A, Rosenberg Belmaker L, Szekely A, Wilson M, Kocabas A, Calixto NE, Grigorenko EL, Huttner A, Chawarska K, Weissman S, Urban AE, Gerstein M, Vaccarino FM. Somatic copy-number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. doi: 10.1038/nature11629. in press. DOI 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.