Abstract
Paramyxoviruses contain glycoprotein fusion machineries that mediate membrane merger for infection. The molecular framework and mechanistic principles governing receptor-induced triggering of the machinery remain unknown. Using measles virus (MeV) fusion complexes, we demonstrate that receptor binding to only one dimer of the tetrameric attachment protein (H) dimer-of-dimers induces fusion-protein (F) triggering; receptor binding and F triggering can be communicated across the dimer–dimer interface of H; and the physical integrity of the tetramer is maintained during fusion. The central MeV H ectodomain stalk region requires structural flexibility for activation of F, and alanine substitutions in this section, physical stress, or exposure of H to soluble ligands trigger conformational rearrangements in native H tetramers. Binding of soluble receptor to H is sufficient to initiate refolding of F, underscoring the physiological significance of this rearrangement of the H tetramer. These data outline a model of the triggering of the physiological MeV fusion machinery in which unilateral receptor binding to one dimer pair in the H tetramer is sufficient to induce a reorganization of H that affects the conformation of the central stalk section, severing interactions between H and the F trimer and activating refolding of F.
Keywords: paramyxovirus entry, protein refolding, virus envelope glycoproteins
Enveloped viruses display highly specialized glycoprotein fusion machineries, which, when activated, undergo a series of conformational changes ultimately resulting in membrane merger, fusion pore formation, and infection. All pathogens of the Paramyxovirinae subfamily depend on the concerted action of two glycoprotein complexes for infection; the attachment protein (H) binds to the cellular receptor and then activates refolding of the fusion protein (F), which facilitates membrane merger (1). Both proteins are thought to interact specifically in hetero-oligomeric fusion complexes (2–7).
Structural and biochemical studies have advanced our insight into conformational changes in F that are required for fusion (1). In contrast, basic questions about the molecular framework that defines productive receptor binding and the mechanism that links receptor binding to F triggering remain unaddressed: i.e., what is the minimal productive receptor:attachment protein stoichiometry; is receptor immobilization in the target membrane required for triggering of the paramyxovirus fusion machinery; does receptor binding affect the conformation of the attachment protein oligomer, and, if so, is this reorganization of the H tetramer instrumental for F triggering?
Measles virus (MeV), a representative of the Morbillivirus genus within the Paramyxovirinae, is a paramyxovirus archetype of high clinical significance. We have demonstrated that a homotetramer or higher-order multimer constitutes the physiological oligomer of the MeV H protein (8). Subsequent crystallization of the isolated globular head domains of MeV H complexed with soluble signaling lymphocyte activation molecule (SLAM/CD150) receptor has corroborated this view (9). Like all other Paramyxovirinae attachment proteins, the head domain of each MeV H monomer harbors receptor-binding sites (RBS) and assumes the classical β-barrel fold of sialidases, although the H protein lacks neuraminidase activity (9–12). A long stalk domain connects the head of the H protein to the transmembrane domain and short luminal tail. The binding sites for all three reported MeV receptors, CD46, SLAM, and nectin-4 (9, 11, 13), are located in an overlapping area of the head domain (14). MeV H and F complexes are thought to preassemble intracellularly (15), and discrete H stalk residues have been implicated in mediating F protein binding and triggering. Through biochemical analyses of full-length native MeV H fusion complexes, we have identified residues in the central section of the stalk domain (position 111–118) that, when mutated, prevent physical association of H and F. Position 98 in the H stalk, slightly more membrane proximal, was shown to be required for efficient triggering of F (16). Having developed an MeV H bimolecular complementation (H-BiC) assay, we furthermore demonstrated that the RBS, F-interacting, and F-triggering functionalities are truly distinct. Coexpression of H mutants with functional defects in these domains restores fusion-support activity through transcomplementation (8, 17).
The structure of the attachment protein stalk domains remains to be solved in its entirety, but the crystal structures of soluble Newcastle disease virus (NDV) hemagglutinin-neuraminidase (HN) attachment protein head and partial stalk domains show a four-helix bundle (4HB) organization of the stalk (18). This arrangement was corroborated by the structure of the parainfluenza virus type 5 (PIV5) HN stalk (19), suggesting that a 4HB stalk arrangement is conserved among Paramyxovirinae attachment proteins.
Crystal structures of free and receptor-bound isolated H head domains in monomeric, dimeric, or tetrameric configurations have revealed that the fold of individual head monomers and the organization of the monomer–monomer interface in covalently linked H dimers remain largely unchanged upon receptor binding (9–12). By contrast, tetrameric ectodomain fragments of both H- (9) and related HN-type (18, 20) attachment proteins crystallized in different spatial organizations. For H, these structures were speculated to represent pre- and postreceptor bound/F-triggering conformations. However, the physiological relevance of individual conformations of purified H ectodomain fragments and the question of whether receptor binding induces a reorganization of the attachment protein remain unaddressed.
Building on available structural and functional information and using an array of newly established functional assays, the present study identifies fundamental determinants that link H receptor engagement to F triggering. Cysteine engineering combined with the H-BiC assay defined the molecular framework of H tetramer–receptor interactions that are necessary for fusion triggering. Insertion of disulfide bonds at strategic positions probed the physiological relevance of existing structural models of the H tetramer, and biochemical and functional assays explored the effect of soluble and membrane-embedded receptor on the H tetramer and associated F trimer organization under physiological conditions.
Results
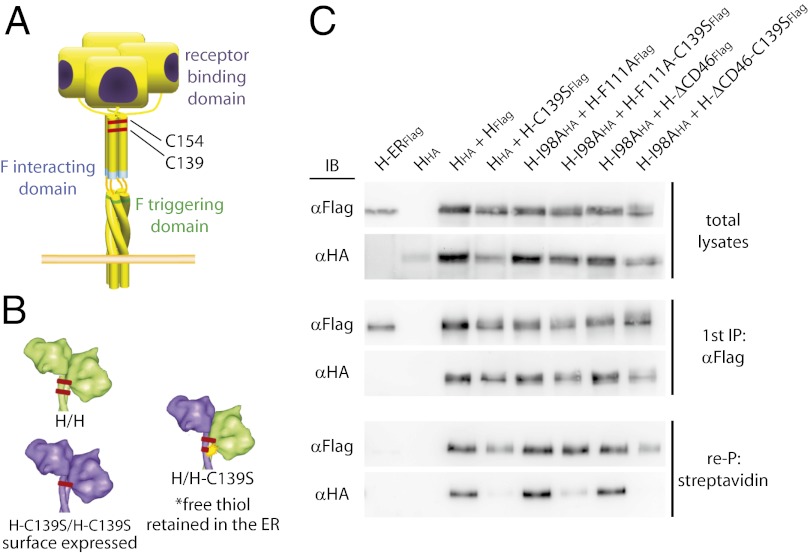
We have shown previously that bioactivity of MeV H mutants with discrete functional defects (illustrated in Fig. 1A) can be restored through protein transcomplementation after coexpression (8). This H-BiC assay provides a platform to explore individual H functionalities in the context of physiological, membrane-embedded fusion complexes. However, the approach is limited by an inherent inability to differentiate between complementation on a heterodimer or [which presumably would be more informative based on recent structural information (9)] on a homodimer/heterotetramer level. To explore transcomplementation across the dimer–dimer interface in H homodimer/heterotetramers complementation pairs, we hypothesized that the two natural disulfide bonds at H stalk positions 139 and 154, which mediate covalent H dimerization, could be used in a combination of H-BiC with disulfide bond engineering (Fig. 1B).
Fig. 1.
Cysteine engineering to assess selective H transcomplementation on a homodimer/heterotetramer level. (A) Schematic of the MeV H tetramer; the locations of independent complementation groups and naturally present disulfide bonds are indicated. (B) Illustration of the cysteine-engineering strategy. (C) Coimmunoprecipitation of epitope-tagged surface-exposed H dimers confirms that only H homodimers with paired thiol moieties reach the cell surface. Surface proteins were biotinylated, and total cell lysates were subjected to α-Flag immunoprecipitation (first IP), followed by reprecipitation (re-P) of the plasma membrane fraction with immobilized streptavidin. A previously described H variant carrying an endoplasmic reticulum-retention signal (H-ERFlag (15)] served as specificity control for the streptavidin reprecipitation. Immunoblots were developed with α-Flag or α-HA antibodies, respectively.
MeV H Heterodimers with Unpaired Stalk Thiols Are Intracellularly Retained.
Previously, we have demonstrated that substituting either cysteine 139 or cysteine 154 with serine retains H bioactivity (21). We therefore anticipated that coexpression of standard H with H-C139S will result in the formation of H-H and (H-C139S)-(H-C139S) homodimers that are fully intracellularly transport competent because of the pairing of all reactive thiol moieties present in the ectodomains. In contrast, H-(H-C139S) heterodimers are expected to be intracellularly retained by endoplasmic reticulum (ER)-resident isomerases that target unpaired, reactive thiol groups (22). H variants harboring HA or Flag epitope tags for distinction were generated to test this hypothesis conceptually in coimmunoprecipitation experiments of cell-surface–exposed and total H material.
HHA-HFLAG complexes were present in both whole-cell extracts and the plasma membrane fractions. However, plasma membrane levels of HHA-(H-C139SFLAG) heterodimers were greatly reduced (Fig. 1C), demonstrating that disulfide engineering is suitable to control the composition of H tetramers in surface-exposed, functional fusion complexes. Equivalent results were obtained with H variants featuring the C139S substitution in addition to one of the signature mutations of the different complementation groups, i.e., F111A, which affects F interaction (6); ΔCD46, which affects receptor binding (23, 24); or I98A, which affects F triggering; Fig. 1C (16).
Unilateral Receptor Engagement by a Single Dimer of the H Tetramer Is Sufficient for F Triggering.
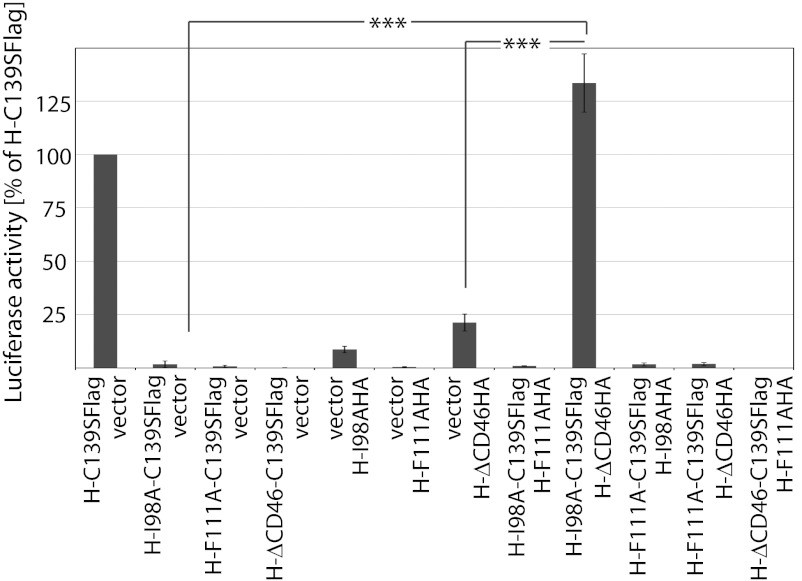
To explore whether transcomplementation of the individual H functionalities can be achieved in a homodimer/heterotetramer setting, we coexpressed H complementation variants pairwise in all combinations with MeV F and quantified bioactivity using a luciferase reporter-based cell-to-cell fusion assay. Efficient complementation, similar to that seen with the H-C139S reference, was observed upon combination of H-I98A-C139S with H-ΔCD46 (Fig. 2 and Fig. S1). In contrast, F-triggering activity was not restored when we paired F-interaction–deficient H-F111A homodimers with homodimers of either of the other two complementation groups. Naturally, the C139S substitution itself did not restore the triggering competence of any of the H mutants when expressed with F alone for control.
Fig. 2.
Unilateral interaction of a receptor with a single covalently linked H dimer is sufficient to initiate F triggering. Results of a quantitative firefly luciferase-based cell-to-cell fusion assay assessing H transcomplementation are shown; data represent averages of at least four experiments ± SEM; ***P < 0.001.
These results indicate that unilateral receptor binding to only one of the covalently linked dimer pairs in the attachment protein tetramer is sufficient for F triggering. Unlike efficient compensation of the H-F111A substitution in a heterodimer setting (8), however, F-binding–deficient H-F111A homodimers cannot be complemented in trans.
Distinct Conformations of Attachment Protein Tetramers.
Two distinct scenarios could account for this lack of trans-complementation: Heterotetramers containing H-F111A homodimers could form physically but lack functionality, suggesting that each covalent dimer pair must contain at least one F-binding–competent H monomer. Alternatively, H-F111A homodimers may be unable to associate into heterotetramers with either H-I98A or H-ΔCD46 homodimers, implying that they assume a conformation that is competent for intracellular transport but conformationally distinct from either H-I98A or H-ΔCD46.
To differentiate between these possibilities, we generated a set of electrophoretically distinct H variants by adding single-chain antibody moieties as size tags to the H C terminus as reported previously (25). Adding single or tandem copies of the single-chain antibody resulted in HXL and HXXL constructs, respectively, with an estimated increase of ∼25 kDa per single-chain antibody copy, largely unchanged bioactivities (Table S1), but clearly distinguishable electrophoretic mobility (Fig. S2A Upper). Electrophoretic separation of surface-expressed material after coexpression of standard and size-tagged H constructs under denaturing, nonreducing conditions underscored that heterodimers form readily but that heterodimers harboring an unpaired thiol moiety at position 139 are not competent for intracellular transport (Fig. S2A Lower), confirming that the added single-chain domain does not affect the homodimer engineering strategy.
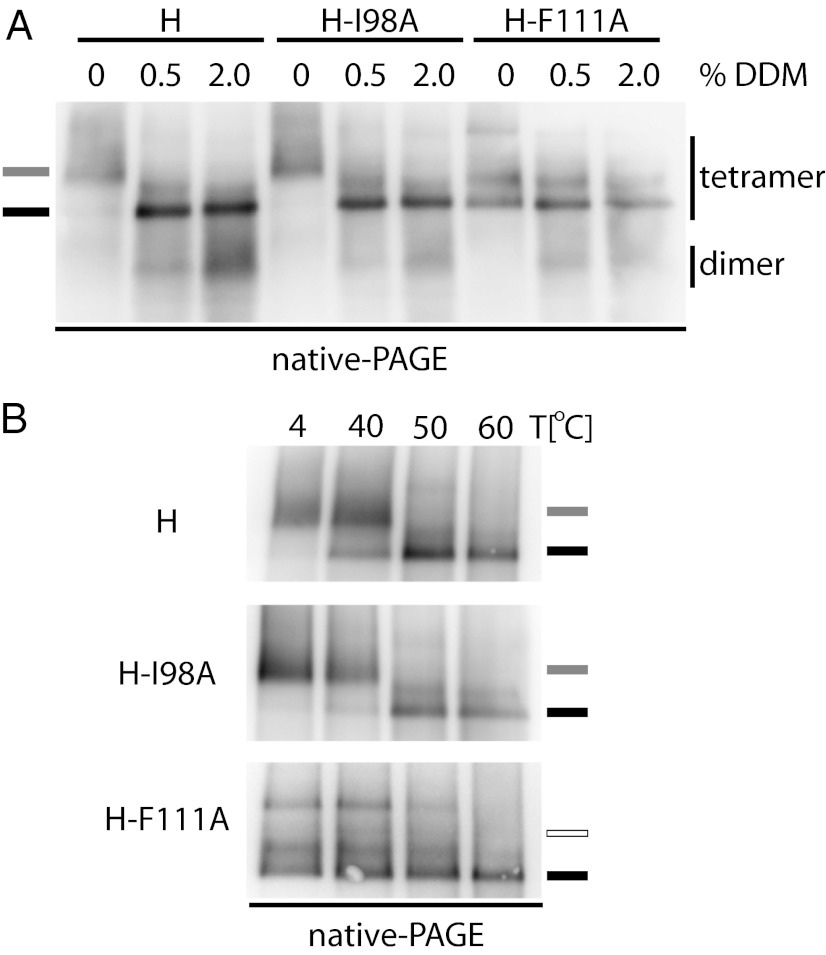
We next subjected the size-tagged HXXL constructs to digitonin extraction followed by native-PAGE, which fractionates the intact H tetramers according to mass, shape, and surface charge (8, 26). When standard and size-tagged variants of otherwise unmodified H, H-I98A, and H-ΔCD46, all with and without the C139S substitution, were coexpressed, tetramers of intermediate mobility representing mixed homodimer/heterotetramers complexes became readily appreciable in addition to untagged and size-tagged homotetramers (Fig. 3A). However, H homodimers harboring the F111A mutation failed to engage in mixed tetramers with standard H, H-I98A, or H-ΔCD46-dimers (Fig. 3A), suggesting that H-F111A tetramers have a distinct conformation. Fig. S2B summarizes the complementation and oligomerization phenotypes of all H constructs analyzed in an activity matrix.
Fig. 3.
H-F111A homodimers are structurally distinct from homodimers of unmodified H or other H-complementation groups. (A) Native-PAGE analysis of digitonin-extracted H tetramers reveals that H-F111A homodimers are unable to heterotetramerize with any other H homodimer species. To distinguish better untagged and size-tagged H oligomers in native-PAGE, size-increased H constructs contained a tandem copy of the tag HXXL. The migration positions of size-tagged H homo- and heterotetramers are shown. (B) H homotetramers harboring stalk mutations that prevent interaction with F (F111A, L114A, I118A, or 110-114A) show a distinct migration pattern in native gels. The predominant H tetramer migration profiles of standard H, H-I98A, and H-ΔCD46 (gray marker) and H-F111A (black marker) are shown.
Under native-PAGE conditions, conformational rearrangements that result in a change of protein shape and/or surface charge typically are reflected by an altered mobility pattern (27, 28). Indeed, direct comparison of standard H and representatives of the different H complementation groups without additional size tags revealed two distinct banding patterns: tetramers of all H mutants that are confirmed to abrogate physical interaction with F [i.e., H-F111A, H-L114A, H-I118A, and an H-110–114A quintuple mutant (6)] were found predominantly in a fraction of higher electrophoretic mobility than those of standard H, F-triggering–, or receptor-binding–defective H variants (Fig. 3B Upper). A small fraction of H-F111A, H-L114A, and H-I118A found at higher molecular weight may represent higher-order oligomers or protein aggregates. Standard reducing and denaturing SDS/PAGE of the same samples confirmed that the underlying H monomers are all of equivalent molecular weight (Fig. 3B Lower). Because the overall protein pI is unaffected by the amino acid substitutions introduced at H positions 111, 114, or 118, the altered native-PAGE mobility pattern indicates a different conformation of the F-binding–deficient H tetramers that is distinct from that of standard H, H-I98A, and H-ΔCD46.
Induced Rearrangement of the H Tetramer into an H-F111A–Like Conformation.
To test the physiological relevance of the different H tetramer organizations, we examined whether standard H tetramers can rearrange into an H-F111A–like conformation. We first explored the effect of increasing detergent stringency or temperature shock on the organization of H tetramers. n-Dodecyl β-d-maltoside (DDM) is, like digitonin, a nonionic detergent used for native-PAGE (26), but in our experience it applies higher scrutiny to larger protein complexes such as the MeV H tetramer (8). Adding increasing amounts of DDM to digitonin-extracted H tetramers reliably converted the standard H and H-I98A tetramer mobility pattern to the H-F111A profile (Fig. 4A). Consistent with our previous observations (8), H tetramers partially disintegrated into the covalently linked dimers at higher DDM concentrations.
Fig. 4.
Physical stress induces transition of H tetramers to an H-F111A–like conformation. (A) Digitonin extracts of H tetramers were treated with increasing amounts of DDM and subjected to native-PAGE fractionation. The predominant tetramer migration profiles as observed in Fig. 3B are shown. (B) Digitonin extracts of H tetramers were subjected to 10-min heat treatment before native-PAGE analysis.
Heat exposure has been used to trigger refolding of purified paramyxovirus F proteins into the postfusion conformation (29). When we subjected the MeV H complexes in an analogous experimental approach to brief (10-min) heat treatment followed by native-PAGE, we noted predominant reorganization of standard H and H-I98A tetramers into an H-F111A–like configuration at or above 50 °C (Fig. 4B), comparable to the conformational shift observed in the presence of increasing DDM concentrations.
Proteinaceous Ligands Induce H Tetramer Reorganization.
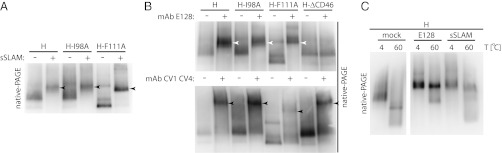
To assess further the relevance of detergent or heat-induced H tetramer rearrangements, we next exposed the panel of H variants to purified, soluble SLAM receptor (sSLAM) (Fig. 5A) (6) or a neutralizing monoclonal antibody (mAb E128) that recognizes an epitope located in the CD46 RBS (Fig. 5B Upper), followed by digitonin extraction and native-PAGE. After the addition of sSLAM or RBS-specific mAb, the electrophoretic mobility of H oligomers was reduced, indicating the formation of larger H tetramer-sSLAM and H tetramer-mAb complexes. Importantly, the mobility profile of all three H variants (standard H, H-I98A, and H-F111A) became similar when complexed with sSLAM or RBS-specific mAb. In contrast, the difference in mobility between H-F111A and standard H or H-I98A remained when a nonneutralizing α-MeV H mAb mixture was added for comparison (Fig. 5B Lower). sSLAM and the different mAbs displayed similar electrophoretic mobility in native-PAGE when fractionated in the absence of H complexes (Fig. S3A), and an H-ΔCD46 variant that is not recognized by mAb E128 (Fig. S3B) confirmed the specificity of the mAb E128–H interaction (Fig. 5B).
Fig. 5.
RBS-specific soluble ligands induce H tetramer reorganization. (A) Exposure of H-expressing cells to sSLAM before digitonin extraction and native-PAGE analysis returns H tetramer/sSLAM complexes of similar mobility (arrowheads). (B) Exposure of H-expressing cells to RBS-specific mAb E128 (competes with CD46 receptor binding) or nonneutralizing H-specific mAbs CV1 CV4. Although all E128/H tetramer complexes show indistinguishable migration profiles (white arrowheads), CV1 CV4/H-F111A tetramers retain distinct electrophoretic mobility (black arrowheads). (C) Consecutive exposure of H tetramers to heat and RBS-specific ligands. H material was extracted with digitonin and was kept at 4 °C (4) or was heated to 60 °C (60) for 10 min. Subsequently, samples were exposed to RBS-specific ligand (E128, sSLAM) or vehicle (mock).
To address whether gel shifts induced through physical stress (heat shock or stringent detergent extraction) and through the addition of protein ligands (sSLAM or α-RBS mAb) visualize equivalent rearrangements of the H tetramer, we examined the effect of heat shock and exposure to ligand in combination. If either procedure induced H tetramer rearrangements of the same molecular nature, we would expect to extract H complexes with mobility equal to that of complexes exposed to soluble ligand alone. This was indeed the case when gel shift assays were performed after consecutive exposure of standard H tetramers to heat and/or proteinaceous (sSLAM or mAb E128) ligands (Fig. 5C). These data indicate that standard H tetramers can rearrange into an H-F111A–like conformation and suggest that binding of proteinaceous ligands (either soluble receptor or RBS-specific mAbs) to H is sufficient to trigger this reorganization.
Upper H Stalk Sections Retain a Closed Arrangement During F Triggering.
Different conformations of H oligomers appear consistent with the two distinct tetrameric crystal structures that were reported recently for soluble MeV H head domains complexed with SLAM receptor (9). Of these, a predicted pre-F triggering conformation (form I) posits the H stalk domains in a 4HB arrangement, compatible with that observed in NDV and PIV5 HN stalk structures (18, 19). In contrast, a second structure (form II) that was suggested to represent a postreceptor-bound/F-triggering organization of the H tetramer features discrete H stalk dimers. The transition of H from form I to form II was predicted to coincide with the separation of the membrane-distal stalk sections (9).
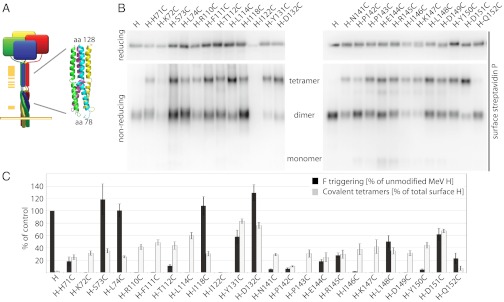
Fig. 6A provides a homology model of the central MeV H stalk section that we generated based on the PIV5 HN coordinates. The model positions residues 98, involved in F triggering (16), and 111 flanking a predicted transition from a twisted to a straight 4HB arrangement. Based on the inability of H-F111A tetramers to interact with F, we considered that the higher H-F111A–like mobility may represent a form II-like structural organization. To test the relevance of form II experimentally, we inserted cysteine substitutions at selected positions along the length of the H stalk. The resulting H variants were assayed first for intracellular transport competence (Table S1) and the formation of covalently linked tetramers assessed through electrophoretic fractionation under reducing and nonreducing conditions (Fig. 6B). Two of the constructs, H-K72C and H-I122C, were not surface expressed, presumably because of the presence of unpaired thiol moieties in the stalk. Of the remaining, intracellular transport-competent mutants, four H variants (H-L114C, H-Y131C, H-D132C, and H-D151C) exist predominantly (>50%) as covalently linked tetramers (Fig. 6 B and C).
Fig. 6.
Membrane-distal (upper) H stalk sections maintain close physical proximity during F triggering. (A) Overview of the cysteine-scanning mutagenesis performed along the H stalk, represented by a yellow bar (Left) in the cartoon. (Inset) Homology model of the central section of the MeV H stalk, generated in Swiss-MODEL based on the coordinates of a PIV5 HN stalk fragment (PDB ID 3TSI). (B) Assessment of the covalent oligomerization status of cell-surface–exposed H cysteine mutants under reducing and nonreducing conditions. (C) Quantitation of relative amounts of surface-exposed H in covalent tetramers (based on densitometric analysis of gels as shown in B) and F-triggering activity; data represent average of four experiments ± SEM.
When we assessed the bioactivity of these constructs in quantitative fusion assays, no clear correlation between covalent H tetramerization and fusion-triggering activity was observed. The three H variants that showed the highest degree of covalent tetramers (H-Y131C, H-D132C, and H-D151C) were all capable of robust F triggering, ranging from ∼60–120% of that found for standard H (Fig. 6C). These data indicate that the molecular integrity of preformed H tetramers is conserved during F triggering and reveal the consistently maintained close proximity of the membrane-distal (upper) stalk sections in pre- and postreceptor-bound H complexes.
F Triggering Requires Structural Freedom in the Central Section of the H Stalk.
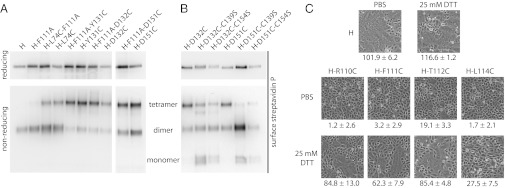
If the F111A mutation indeed induces a postreceptor-bound–like organization of H, this finding implies that the conformation of H-F111A tetramers should be distinct from that of crystal form II. To test this notion, we examined whether H-F111A variants remain capable of forming covalently linked tetramers when cysteine substitutions are added to stalk positions membrane proximal or distal of the F111A site (i.e., positions L74C, Y131C, D132C, or D151C). Comparative analysis of these constructs with the corresponding single mutants under reducing and nonreducing conditions demonstrated that the F111A exchange does not affect the degree of covalent H tetramerization (Fig. 7A). Of note, combining distal cysteine insertions with removal of the naturally existing disulfide bonds at stalk positions 139 or 154 abrogates essentially all covalent H tetramerization (Fig. 7B), indicating that the natural disulfide bonds at positions 139 and 154 introduce a rigid scaffold into the dimeric stalk pairs that is required for covalent tetramerization.
Fig. 7.
Structural flexibility of the central section of the H-stalk domain is required for F triggering. (A and B) Assessment of the covalent oligomerization status of selected H cysteine mutants. (C) Microphotographs of Vero cells coexpressing MeV H constructs with standard F protein. Before imaging, cells were exposed to mildly reducing conditions (25 mM DTT) or vehicle (PBS) for control. Numbers represent the amount of nuclei found in a syncytia/standardized field of view (each control field contains ∼100–120 cells). Averages of 75 fields of view ± SEM are shown.
In contrast to the high bioactivity of H tetramers with covalently rigidified upper-stalk sections, cysteine substitutions introduced into the central part of the stalk itself (residues 110, 111, 112, and 114) largely blocked F triggering (Figs. 6C and 7C). An exception was H-118C, which maintained efficient fusion support activity. Consistent with this finding, H-118C, but not H-118A, is able to associate efficiently with F (Fig. S4), underscoring the highly specific side-chain requirement in the H stalk section for productive F binding. However, when we partially released disulfide bonds of H-110C, H-111C, H-112C, and H-114C under mildly reducing conditions [25 mM dithiothreitol (DTT)], the bioactivity of all variants except H-114C was restored substantially, to ∼60–85% of that observed for standard H (Fig. 7C). This finding is consistent with our recent analysis of the homologous region of the related canine distemper virus H protein (30), suggesting that structural, possibly rotational, freedom between stalk monomers in a central, but not upper, stalk section is a conserved requirement for morbillivirus F triggering. Although additional disulfide bonds near stalk position 111 arrest H-F111C tetramers in a reactivatable, pre–F-triggering organization, the conformation of H-F111A tetramers may represent a distinct postreceptor-bound/F-triggering form of the tetramer.
Exposure of H Tetramers to Soluble Receptor Initiates F Trimer Refolding.
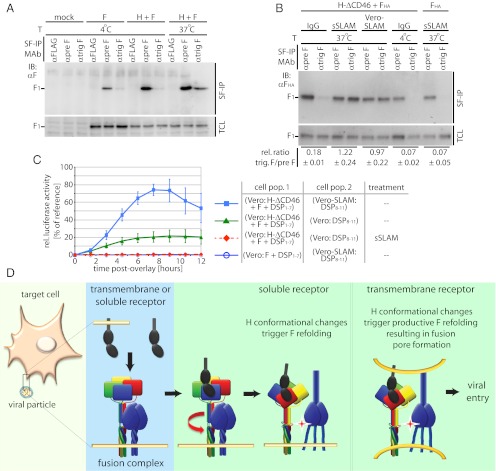
To test directly whether the H-F111A tetramer conformation represents a post-F-triggering form, we examined the effect of soluble receptor-induced H tetramer rearrangements on the initiation of F triggering in physiological, membrane-embedded fusion complexes. To detect the initiation of F trimer refolding, we used a pair of mAbs directed against the MeV F protein, mAb 186A (31) and, mAb 19GD (32), which we have found specifically detect a pretriggered (α-pre F) or a fusion-triggered (α-trig F) conformation of the F trimer, respectively (Fig. 8A).
Fig. 8.
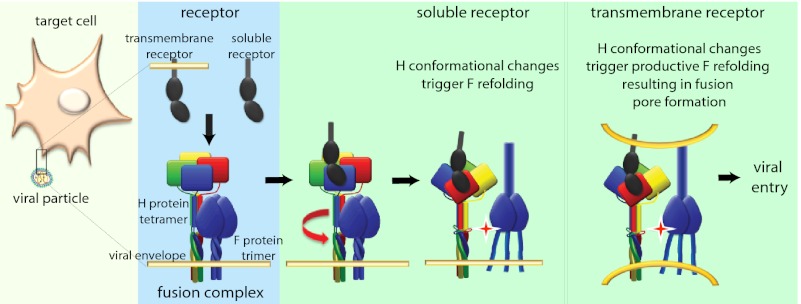
H tetramer conformational changes induced by soluble receptor are sufficient to initiate F refolding. (A) Surface immunoprecipitation (SF-IP) of Vero cells coexpressing MeV H and F. Precipitation of F material with a conformation-dependent mAb that recognizes a fusion-triggered F trimer (αtrig F) requires warming of samples to 37 °C, allowing F refolding to proceed. A mAb recognizing prefusion F (αpre F), and a control mAb (αFlag) were used for comparison. (B) Surface immunoprecipitation of MeV F protein with conformation-dependent αpre F and αtrig F mAbs as specified. Overlay of MeV SLAM receptor-positive cells (Vero-SLAM) or sSLAM is sufficient to initiate F refolding. Numbers represent densitometric ratios of triggered F/prefusion F, normalized for F material present in total cell lysates (TCL); averages of three experiments ± SEM are shown. (C) Initiation of F refolding by soluble receptor does not lead to fusion pore formation. Values were normalized for cells cotransfected with DSP1–7 and DSP8–11 plasmids (33) and represent averages of four to six experiments ± SEM. (D) Model of the MeV fusion machinery. Contact of a single H dimer present in functional fusion hetero-oligomers (light blue background; H tetramers are colored by monomer; F trimers are in dark blue) with membrane-embedded or soluble receptor leads to reorganization of the central H stalk section (red arrow); this reorganization most likely coincides with rearrangements of the H head domain. This H tetramer reorganization triggers the F refolding cascade. However, interaction of H with membrane-embedded receptor is required for productive membrane fusion.
To generate conditions void of high-affinity receptor stimulation of H, CD46-binding– incompetent H-ΔCD46 and F were coexpressed in Vero cells, which lack both SLAM and nectin-4 (9, 13) and therefore do not provide a receptor for H-ΔCD46. In this system, we observed strong reactivity of F trimers with the pretriggered F-specific mAb in cell-surface immunoprecipitation assays, whereas the corresponding α-triggered F-specific mAb showed little binding (Fig. 8B). Likewise, F expressed in the absence of H maintained full reactivity with the pretriggered F-specific mAb. These data indicate that prefusion MeV F trimers have a low rate of spontaneous refolding in the absence of H interaction with receptor and demonstrate that MeV F does not require physical contact with H to retain a metastable prefusion conformation.
When MeV glycoprotein-expressing cells were exposed to soluble SLAM, however, reactivity with the α-triggered F mAb increased substantially to levels comparable to those achieved through overlay with SLAM-positive Vero-SLAM cells, which served as a positive control for the assay (Fig. 8B). Corroborating previous reports that the I98A substitution in the H stalk impairs F triggering but not physical interaction between H and F complexes (8, 16), exposure of H-I98A tetramers to membrane-integral or soluble receptor in this experimental setting did not result in increased F reactivity with the α-triggered F mAb (Fig. S5A).
To explore whether the soluble SLAM-mediated initiation of F triggering is sufficient to drive the opening of fusion pores, we adapted a recently described real-time cell-content–mixing assay to the paramyxovirus system; this assay is based on the functional reconstitution of individually expressed N- and C-terminal halves of GFP/renilla luciferase dual-fusion proteins upon the formation of fusion pores (33). Monitoring the reconstitution of functional luciferase moieties in this setting revealed successful opening of fusion pores only in the presence of membrane-integral receptor (Fig. 8C). Low-level content mixing observed with the H-ΔCD46 and F combination in the absence of soluble SLAM is most likely caused by residual low-affinity docking of H-ΔCD46 to the CD46 receptor. Importantly, exposure of Vero cell populations expressing H-ΔCD46 and F to soluble SLAM did not lead to increased cell content mixing and syncytia formation in this assay or upon microscopic examination (Fig. S5B).
These studies demonstrate that binding of soluble receptor molecules to a native, membrane-embedded paramyxovirus attachment protein oligomer is sufficient to activate the F refolding pathway, suggesting that the ligand-induced changes in H electrophoretic mobility in native gels reflect an H tetramer reorganization that constitutes the central link between binding to a receptor and triggering of the F fusion machinery. Opening of fusion pores, however, requires the presence of membrane-embedded receptor.
Discussion
Paramyxoviruses depend on the concerted action of two envelope glycoprotein complexes to achieve membrane fusion and infection. In this study, we subjected full-length, membrane-integral MeV attachment protein complexes to an array of biochemical and functional assays to dissect the molecular mechanism that links receptor engagement by the H protein to F- triggering under physiological conditions.
From H transcomplementation assays (8) combined with cysteine-substitution–based homodimer engineering, we conclude, first, that an H monomer:receptor stoichiometry ≤2:1 initiates fusion and that unilateral receptor docking to only one of the covalently linked dimer pairs in the H tetramer is necessary and sufficient. Based on these results, we can exclude triggering models that assume separation of the tetrameric H head arrangement along the noncovalent dimer–dimer interface as a result of physical force generated through simultaneous binding of membrane-immobilized receptor molecules to each covalent dimer pair. However, productive complementation across cysteine-engineered F-triggering– or receptor-binding–defective H homodimers reveals functional cross-talk along the dimer–dimer interface.
The generation of triggering-competent covalently linked H tetramers through disulfide bond engineering into the upper (membrane-distal) stalk domain indicates that the molecular integrity of the tetramer itself remains unchanged during receptor binding and F triggering. Thus, efficient F triggering does not mandate complete separation of a tetrameric H stalk assembly, as proposed for an alternative H tetramer-SLAM cocrystal arrangement (form II in ref. 9). Rather, our study suggests that MeV H stalks form a single bundle, consistent with the recently described partial ectodomain structures of NDV (18) and PIV5 (19) HN proteins, and that F triggering does not mandate altering the overall integrity of this organization.
However, efficient F triggering requires structural flexibility between individual H monomers in the central section of the stalk bundle. Cysteine substitutions in this region (residues 110–114) completely abolish fusion-support activity, independent of whether the substitutions induce covalent tetramers or lead to an additional disulfide bond between monomers of an existing covalent H dimer. Considering our experience that unpaired stalk thiols result in intracellular retention of the protein, we assume that the engineered stalk cysteines are engaged in novel disulfide bonds. Confirmation for this notion comes from our observation that exposure of H-110C, H-111C, and H-112C complexes to reducing conditions partially restored fusion-support activity. This finding also demonstrates that the engineered disulfide bonds trap the H tetramer in a pre–F-triggering conformation. In contrast, disulfide bonds at position 114 inhibited fusion activation, even under reducing conditions. Because this variant also was fully intracellular transport competent, gross protein misfolding is unlikely. Alternatively, H-114C may assume a post–F-triggering–like conformation, or this substitution may impair interaction with F on a short-range basis.
Our successful previous engineering of N-linked glycans into consecutive positions (110-112) of the MeV H stalk highlights that each monomer enjoys a high degree of rotational freedom in this region (6). This flexibility is accentuated further by a modeling-predicted disturbance in the secondary structure of the monomers immediately upstream of residue 110. We postulate that local unraveling or rotational unwinding involving the central sections of each stalk monomer is required for F triggering (visualized schematically in Fig. 8D). Likewise, the stalk domains of the attachment proteins of many Paramyxovirinae subfamily members were implicated in F triggering (2, 16, 34). Our recent analysis of the related canine distemper virus H stalk domain (30) revealed a comparable requirement for structural freedom of the stalk center, suggesting a common theme of Paramyxovirinae attachment-protein–mediated fusion support.
The homodimer/heterotetramer complementation studies and native-PAGE assays prove that receptor binding induces biochemically appreciable conformational changes in the H tetramer. H homodimers harboring alanine substitutions in the central stalk section spontaneously assume an organization distinct from that of wild-type H dimers. They homotetramerize efficiently but are incapable of F binding (6) and are unable to associate into heterotetramers with standard H or any of the other complementation groups. However, exposure to physical stress, soluble receptor, or mAbs mimicking receptor binding through interaction with the RBS is sufficient to initiate the reorganization of standard H tetramers into an H-F111A-like conformation, suggesting that the latter may represent a final, postreceptor-binding/F-triggering structure. This hypothesis is substantiated by our demonstration that exposure to soluble receptor is sufficient to initiate F refolding in native, membrane-embedded paramyxovirus fusion complexes. The significance of this finding is threefold: It underscores the physiological significance of the H tetramer rearrangement observed in native-PAGE assays; it demonstrates that little external energy is required to trigger the Paramyxovirinae entry cascade; and it confirms that triggering is achieved readily at the plasma membrane under neutral pH conditions.
We note that, upon exposure to physical stress or soluble receptor, the F-triggering–defective H-I98A tetramers also undergo a conformational rearrangement resembling that of standard H tetramers. Previous studies have revealed a substantially higher F coimmunoprecipitation efficiency with H-I98A than with unmodified H; this finding was interpreted as indicating a stronger physical interaction of H-I98A F glycoprotein oligomer pairs (16). Our results may reflect that receptor binding to H-I98A is insufficient to induce the H tetramer rearrangements because of a stabilizing effect of tightly bound F. Alternatively, the H-I98A substitution could impair the separation of the two envelope glycoprotein complexes even after a receptor-induced H-I98A reorganization has occurred. Either scenario is compatible with our observation that exposure of H-I98A F complexes to membrane-integral or soluble receptor does not lead to the initiation of biochemically appreciable F-trimer refolding.
Because H-F111A could complement H-I98A or H-ΔCD46 functionally on a heterodimer level in previous complementation studies (8), we furthermore conclude that the H-F111A reorganization is not structurally dominant. For the related Nipah virus G attachment protein, rearrangements have been proposed based on exposure of an mAb epitope after receptor docking (35) and altered mAb reactivity after stalk mutagenesis (34). Because crystals of free and receptor-complexed G head domains show few structural differences (36), changes in the G oligomer organization likewise may lead to Nipah F triggering, suggesting that basic principles of fusion initiation indeed may be conserved across different Paramyxovirinae genera.
Last, additional conditions located downstream of receptor binding and the initial F triggering must be fulfilled for productive MeV F refolding and fusion pore formation. Conceivably, these requirements could comprise a demand for a defined proximity of donor and target membrane provided by the continued interaction of H with membrane-embedded receptor, the coordinated local assembly of multiple activated H/F complexes through receptor clustering in the target membrane, a requirement for additional physical force originating from H/receptor-driven local curvature in opposing donor and target membranes (37, 38), or, potentially a second contact between attachment and refolding F protein complexes (39) that is required to complete membrane merger but is not induced through the interaction of H with soluble receptor.
In conclusion, we postulate that the functional MeV membrane fusion machinery is comprised of preassembled H-F hetero-oligomers in a prereceptor-bound/F-triggering H tetramer and prefusion F trimer conformation (Fig. 8D). Upon ligand binding to at least one of the covalent dimer pairs in the H dimer-of-dimers, partial unwinding/unraveling of the central section of the H stalk domain ensues, most likely disrupting preexisting contacts between the stalk and the F trimer. Little energy is required for this step, because docking of soluble receptor moieties initiates the process efficiently. The resulting change in microenvironment at the former H–F contact zone may be sufficient for F refolding to commence. Unwinding of the stalk center most likely coincides with a rearrangement of the H head domain dimer–dimer interface. However, the overall physical integrity of the H tetramer and the tetrameric arrangement of the upper section of the H stalk remain intact during the process.
Methods
Cell Lines, Viral Stocks and Transfections.
Vero (African green monkey kidney epithelial) cells (CCL-81; ATCC) and Vero cells stably expressing human SLAM [Vero-SLAM cells (40)] were maintained in DMEM supplemented with 7.5% (vol/vol) FBS at 37 °C and 5% CO2. At every third passage, Vero-SLAM cells were kept under G418 selection. Lipofectamine 2000 (Invitrogen) was used for all transient transfection reactions. Modified vaccinia virus Ankara expressing T7 polymerase (41) was amplified in DF-1 (chicken embryo fibroblast) cells (CRL-12203; ATCC).
Site-Directed Mutagenesis and Epitope Tagging.
Site-directed mutagenesis was performed following the QuikChange protocol (Stratagene) using pCG-H (42) as template. In addition, mutant constructs were epitope tagged, resulting in a set of H variants that contained either an N-terminal HA tag or a triple FLAG tag (43). A single or double copy of a single-chain antibody directed against the carcinoembryonic antigen (CEA) (25) was added to the H C terminus in some experiments to impart a significant change in molecular weight of the protein. Changes were confirmed by DNA sequencing in all cases.
Flow Cytometry for Analysis of H Expression and Receptor-Binding Capacity.
Surface-expression levels of MeV H and the ability of MeV H variants to bind the SLAM receptor were monitored in a flow cytometer assay as previously described (6).
Quantitative Cell-to-Cell Fusion Assays.
Cell-to-cell fusion was assessed using an established luciferase reporter assay as previously described (6, 8). For qualitative assessment, transfected Vero cells were photographed at the indicated times following transfection at a magnification of 200×. To quantify the extent of cell-to-cell fusion after partial reduction of engineered H disulfide bonds through DTT treatment, the number of nuclei present in discrete cells and in syncytia was quantified in multiple, randomly selected fields of view (magnification: 200×). The number of nuclei present in syncytia was determined by the difference in the number of individual cells counted in the negative control (F-only population) and the treated samples.
Statistical Analysis.
To assess the statistical significance of differences between sample means, unpaired two-tailed t tests were applied using the Prism 5 (GraphPad) software package.
Envelope Glycoprotein Heterodimer Coimmunoprecipitation.
Vero cells were transfected with equal amounts of the differently tagged H variants as indicated. Thirty-six hours posttransfection, cell-surface proteins were biotinylated with 0.5 mg/mL sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (Thermo Scientific) for 20 min at 4 °C as previously described (6). Cleared supernatants were incubated with M2 mAb directed against the FLAG epitope (Sigma) at 4 °C. After precipitation with immobilized protein G at 4 °C, samples were washed and eluted in PBS, 2% SDS. The eluted material was diluted to 0.15% SDS with PBS, followed by adsorption of biotinylated protein material to immobilized streptavidin for 120 min at 4 °C. Bound material was eluted and denatured with urea buffer [200 mM Tris (pH 6.8), 8 M urea, 5% (wt/vol) SDS, 0.1 mM EDTA, 0.03% bromophenol blue, 1.5% (wt/vol) DTT] for 30 min at 50 °C, fractionated by SDS/PAGE, and blotted to PVD membranes. Immunoblots were decorated with α-FLAG (M2) and αHA (16b12) monoclonal antibodies, respectively, and were developed using an anti-mouse IgG light-chain conjugate and ChemiDoc XRS digital imaging system (Bio-Rad).
Detection of Envelope Glycoprotein Dimers on Cell Surface.
Vero cells were transfected with 4 µg per well of MeV H-encoding DNA total, 2 µg of a FLAG-tagged H variant, and 2 µg of FLAG-tagged H variants containing a single-chain antibody moiety (XL) as indicated. Thirty-six hours posttransfection, cell-surface proteins were biotinylated with 0.5 mg/mL sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate as detailed above. Precipitates were washed as outlined above, split into two equal fractions, and denatured in urea buffer under reducing [urea buffer containing 1.5% (wt/vol) DTT] or nonreducing (urea buffer without DTT) conditions. Reduced samples were fractionated on 10% (wt/vol) SDS-Tris/glycine gels. Nonreduced samples were analyzed on 3–8% (wt/vol) NuPAGE Tris/Acetate gradient gels (Invitrogen).
Attachment Protein Homodimer Heterotetramer Gel-Shift Analysis.
Vero cells were transfected with a total of 4 µg per well of MeV H-encoding DNA [1 µg encoding FLAG-tagged H variants and 3 µg encoding FLAG-tagged H featuring, in addition, a tandem copy of a single-chain α-CEA antibody (2XL)]. Thirty-six hours posttransfection, cells were subjected to native-PAGE analysis. Blots were visualized with α-FLAG M2 antibodies as described.
Cysteine Engineering and Assessment of Covalent H Tetramer Formation.
Vero cells were transfected with 4 µg per well of MeV H-encoding plasmid DNA harboring individual cysteine substitutions as specified. Thirty-six hours posttransfection, cell-surface–exposed proteins were biotinylated as described. After precipitation with immobilized streptavidin, bound material was divided into two equal fractions, denatured under reducing (urea-DTT) or nonreducing (urea without DTT) conditions, and fractionated on 10% (wt/vol) SDS-Tris/glycine or 3–8% (wt/vol) NuPAGE Tris-Acetate gradient gels (Invitrogen). For densitometric analysis of immunoblots, signal intensities were quantified using the QuantityOne software package (Bio-Rad). The extent of covalent H tetramer formation was quantified by calculating for each individual mutant the ratio of H tetramer to total H material.
Native-PAGE and Native Gel Shift.
Vero cells were transfected with 4 µg per well of plasmid DNA encoding different H constructs as specified. Thirty-six hours posttransfection, cells were washed with PBS, and protein was extracted using Native Tris Sample buffer [100 mM Tris⋅Cl, 10% glycerol, 0.0025% Bromophenol Blue (pH 8.6) with 0.1% digitonin] at 4 °C for 30 min. Samples were cleared (20,000 × g for 20 min) and fractionated on native-PAGE 3–12% (wt/vol) Bis-Tris gradient gels. PVDF blots were fixed with an 8% acetic acid wash for 10 min, followed by immunostaining and developing as outlined above.
For gel-shift experiments, antibodies (as specified) or affinity-purified soluble mouse Fc-SLAM receptor [sSLAM at 0.05 mg/mL final concentration (6)] were bound to H-protein–expressing cells on ice for 30 min before washing and digitonin extraction of proteins. In ex vivo binding experiments, digitonin extracts of H variants were prepared first and then were mixed with antibodies or sSLAM and incubated on ice for 30 min. In both cases, extracts were processed and subjected to native PAGE analysis as before.
Coimmunoprecipitation.
To assess physical association of MeV H and F proteins, coimmunoprecipitation was carried out as previously described (6, 8).
In Situ Assessment of F Triggering.
Vero cells were cotransfected with H-ΔCD46– and F-encoding plasmids, followed by incubation in the presence of fusion inhibitory peptide (FIP) as indicated to prevent any premature breakdown of the cell monolayer. Thirty-six hours posttransfection, cells were washed extensively to remove FIP and were subjected to surface immunoprecipitations by incubating intact monolayers in the presence of mAb [α-Flag M2 (Sigma) for control, α-pretrigger F(32), or α-triggered F(31); 1:750-dilution in DMEM each] at 4 °C to block conformational rearrangements of membrane-integral envelope glycoprotein complexes) or 37 °C to enable fusion for 1 h. Where specified, wells also received sSLAM or unspecific murine IgG (Sigma) at 0.05 mg/mL final concentration. Reference wells were overlaid with SLAM-positive Vero-SLAM cells before incubation at 37 °C. Subsequently, samples were incubated further for 1 h at 4 °C. Unbound antibody then was removed through extensive washing, cells were lysed in RIPA buffer, and cleared lysates were subjected to precipitation of immunocomplexes with immobilized protein G Sepharose and SDS/PAGE analysis as described above.
Dual Split-Protein Cell-Content–Mixing Assay.
Vero cells were transfected with plasmids encoding H-ΔCD46, F, and dual-split protein (DSP1–7) (33). In controls, H-encoding plasmid was omitted. A second population of Vero or Vero-SLAM cells was transfected with plasmids encoding SLAM and DSP8–11 (33) or DSP8–11 alone. As a reference for reconstitution of N-terminal (DSP1–7) and C-terminal (DSP8–11) GFP/renilla luciferase halves, controls were cotransfected with both DSP1–7 and DSP8–11 plasmids. Twenty-four hours posttransfection, cells were mixed at an equal ratio in the specified combinations and reseeded in black-walled 96-well plates; specified wells received sSLAM at 0.05 mg/mL final concentration. Cell-content mixing indicating fusion pore formation was monitored over a 12-h time window at 37 °C by following DSP1–7 and DSP8–11 reconstitution resulting in restored renilla luciferase activity. As substrate, EnduRen (Promega) was added according to the manufacturer’s instruction. Plates were examined at 90-min intervals in a BioTek Synergy H1 plate reader in luminescence top-count area scan mode. For each replicate, relative luciferase units were calculated through normalization of individual measurements for the maximum readout found in the DSP1–7/DSP8–11 double-transfected reference cell population. In some experiments, cell populations were reseeded into 12-well plates and incubated in the presence or absence of additional sSLAM (0.05 mg/mL final concentration), and fusion was assessed microscopically.
Supplementary Material
Acknowledgments
We thank B. Horvat and M. Ehnlund for providing mAbs directed against the MeV F protein and G. Melikyan, N. Kondo, and Z. Matsuda for providing split-luciferase reporter plasmids. This work was supported, in part, by National Research Service Award/National Institute of Allergy and Infectious Disease (NIAID) F32 AI085763 from the National Institutes of Health (NIH)/NIAID (to M.A.B.) and by Public Health Service Grant AI083402 from the NIH/NIAID (to R.K.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 17750 (volume 109, number 44).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210925109/-/DCSupplemental.
References
- 1.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng R, Mirza AM, Mahon PJ, Iorio RM. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch Virol Suppl. 1997;13:115–130. doi: 10.1007/978-3-7091-6534-8_12. [DOI] [PubMed] [Google Scholar]
- 3.Deng R, Wang Z, Mirza AM, Iorio RM. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology. 1995;209:457–469. doi: 10.1006/viro.1995.1278. [DOI] [PubMed] [Google Scholar]
- 4.Melanson VR, Iorio RM. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J Virol. 2006;80:623–633. doi: 10.1128/JVI.80.2.623-633.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanabayashi K, Compans RW. Functional interaction of paramyxovirus glycoproteins: Identification of a domain in Sendai virus HN which promotes cell fusion. J Virol. 1996;70:6112–6118. doi: 10.1128/jvi.70.9.6112-6118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paal T, et al. Probing the spatial organization of measles virus fusion complexes. J Virol. 2009;83:10480–10493. doi: 10.1128/JVI.01195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JK, et al. Functional interaction between paramyxovirus fusion and attachment proteins. J Biol Chem. 2008;283:16561–16572. doi: 10.1074/jbc.M801018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brindley MA, Plemper RK. Blue native PAGE and biomolecular complementation reveal a tetrameric or higher-order oligomer organization of the physiological measles virus attachment protein H. J Virol. 2010;84:12174–12184. doi: 10.1128/JVI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashiguchi T, et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat Struct Mol Biol. 2011;18:135–141. doi: 10.1038/nsmb.1969. [DOI] [PubMed] [Google Scholar]
- 10.Colf LA, Juo ZS, Garcia KC. Structure of the measles virus hemagglutinin. Nat Struct Mol Biol. 2007;14:1227–1228. doi: 10.1038/nsmb1342. [DOI] [PubMed] [Google Scholar]
- 11.Santiago C, Celma ML, Stehle T, Casasnovas JM. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat Struct Mol Biol. 2010;17:124–129. doi: 10.1038/nsmb.1726. [DOI] [PubMed] [Google Scholar]
- 12.Hashiguchi T, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyce RS, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;7:e1002240. doi: 10.1371/journal.ppat.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashiguchi T, Maenaka K, Yanagi Y. Measles virus hemagglutinin: Structural insights into cell entry and measles vaccine. Front Microbiol. 2011;2:247. doi: 10.3389/fmicb.2011.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plemper RK, Hammond AL, Cattaneo R. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J Biol Chem. 2001;276:44239–44246. doi: 10.1074/jbc.M105967200. [DOI] [PubMed] [Google Scholar]
- 16.Corey EA, Iorio RM. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J Virol. 2007;81:9900–9910. doi: 10.1128/JVI.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plemper RK, Brindley MA, Iorio RM. Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 2011;7:e1002058. doi: 10.1371/journal.ppat.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan P, et al. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc Natl Acad Sci USA. 2011;108:14920–14925. doi: 10.1073/pnas.1111691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose S, et al. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J Virol. 2011;85:12855–12866. doi: 10.1128/JVI.06350-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Plemper RK, Hammond AL, Cattaneo R. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J Virol. 2000;74:6485–6493. doi: 10.1128/jvi.74.14.6485-6493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feige MJ, Hendershot LM. Disulfide bonds in ER protein folding and homeostasis. Curr Opin Cell Biol. 2011;23:167–175. doi: 10.1016/j.ceb.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson JB, Scheiflinger F, Manchester M, Yilma T, Oldstone MB. Structural and functional studies of the measles virus hemagglutinin: Identification of a novel site required for CD46 interaction. Virology. 1999;256:142–151. doi: 10.1006/viro.1999.9644. [DOI] [PubMed] [Google Scholar]
- 24.Corey EA, Iorio RM. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology. 2009;383:1–5. doi: 10.1016/j.virol.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond AL, et al. Single-chain antibody displayed on a recombinant measles virus confers entry through the tumor-associated carcinoembryonic antigen. J Virol. 2001;75:2087–2096. doi: 10.1128/JVI.75.5.2087-2096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reisinger V, Eichacker LA. Solubilization of membrane protein complexes for blue native PAGE. J Proteomics. 2008;71:277–283. doi: 10.1016/j.jprot.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Corbin JD, et al. Metal ion stimulators of PDE5 cause similar conformational changes in the enzyme as does cGMP or sildenafil. Cell Signal. 2011;23:778–784. doi: 10.1016/j.cellsig.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Zettler J, Mootz HD. Biochemical evidence for conformational changes in the cross-talk between adenylation and peptidyl-carrier protein domains of nonribosomal peptide synthetases. FEBS J. 2010;277:1159–1171. doi: 10.1111/j.1742-4658.2009.07551.x. [DOI] [PubMed] [Google Scholar]
- 29.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci USA. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ader N, et al. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J Biol Chem. 2012;287:16324–16334. doi: 10.1074/jbc.M112.342493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malvoisin E, Wild F. Contribution of measles virus fusion protein in protective immunity: Anti-F monoclonal antibodies neutralize virus infectivity and protect mice against challenge. J Virol. 1990;64:5160–5162. doi: 10.1128/jvi.64.10.5160-5162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheshberadaran H, Chen SN, Norrby E. Monoclonal antibodies against five structural components of measles virus. I. Characterization of antigenic determinants on nine strains of measles virus. Virology. 1983;128:341–353. doi: 10.1016/0042-6822(83)90261-1. [DOI] [PubMed] [Google Scholar]
- 33.Kondo N, Miyauchi K, Matsuda Z. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr Protoc Cell Biol. 2011 doi: 10.1002/0471143030.cb2609s50. Chapter 26:26.9.1–26.9.9. [DOI] [PubMed] [Google Scholar]
- 34.Bishop KA, et al. Residues in the stalk domain of the hendra virus g glycoprotein modulate conformational changes associated with receptor binding. J Virol. 2008;82:11398–11409. doi: 10.1128/JVI.02654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguilar HC, et al. A novel receptor-induced activation site in the Nipah virus attachment glycoprotein (G) involved in triggering the fusion glycoprotein (F) J Biol Chem. 2009;284:1628–1635. doi: 10.1074/jbc.M807469200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu K, et al. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plemper RK. Cell entry of enveloped viruses. Curr Opin Virol. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melikyan GB. Membrane fusion mediated by human immunodeficiency virus envelope glycoprotein. Curr Top Membr. 2011;68:81–106. doi: 10.1016/B978-0-12-385891-7.00004-0. [DOI] [PubMed] [Google Scholar]
- 39.Porotto M, et al. Spring-loaded model revisited: Paramyxovirus fusion requires engagement of a receptor binding protein beyond initial triggering of the fusion protein. J Virol. 2011;85:12867–12880. doi: 10.1128/JVI.05873-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono N, et al. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol. 2001;75:4399–4401. doi: 10.1128/JVI.75.9.4399-4401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 42.Cathomen T, Naim HY, Cattaneo R. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol. 1998;72:1224–1234. doi: 10.1128/jvi.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Hernan R, Brizzard B. Multiple tandem epitope tagging for enhanced detection of protein expressed in mammalian cells. Mol Biotechnol. 2001;19:313–321. doi: 10.1385/MB:19:3:313. [DOI] [PubMed] [Google Scholar]