Abstract
TGFβ activation and signaling have been extensively studied in experimental models of allergen-induced asthma as potential therapeutic targets during chronic or acute phases of the disease. Outcomes of experimental manipulation of TGFβ activity have been variable, in part due to use of different model systems. Using an ovalbumin (OVA)-induced mouse model of asthma, we here show that innate variation within TGFβ1 genetic modifier loci, Tgfbm2 and Tgfbm3, alters disease susceptibility. Specifically, Tgfbm2129 and Tgfbm3C57 synergize to reverse accentuated airway hyperresponsiveness (AHR) caused by low TGFβ1 levels in Tgfb1+/− mice of the NIH/OlaHsd strain. Moreover, epistatic interaction between Tgfbm2129 and Tgfbm3C57 uncouples the inflammatory response to ovalbumin from those of airway remodeling and airway hyperresponsiveness, illustrating independent genetic control of these responses. We conclude that differential inheritance of genetic variants of Tgfbm genes alters biological responses to reduced TGFβ1 signaling in an experimental asthma model. TGFβ antagonists for treatment of lung diseases might therefore give diverse outcomes, dependent on genetic variation.
Asthma is an allergic disease of the airways affecting more than 5% of the US population. It is characterized by airway hyperresponsiveness (AHR), inflammatory infiltration, increased mucus production, elevated serum IgE levels, and airway remodeling (1). Asthma can present within a wide range of disease severity, from mild and intermittent to severe, persistent, and drug refractory. It is considered a multifactorial disorder in which complex interplay between environmental and genetic factors determines disease risk and severity. Here we investigated the contribution of genetic factors that have previously been shown to interact with transforming growth factor β (TGFβ) in vivo, to disease severity in a mouse model of asthmatic response.
Genetic variants of the human TGFB1 gene are associated with asthma severity (2–5) and TGFβ is synthesized by, and has effects on, several cell types of the lung in response to an asthmatic stimulus. Thus, the TGFβ signaling pathway has been considered a potential therapeutic target in lung disease (6). It is a potent suppressor of inflammation, illustrated by lethal T-cell–mediated multifocal inflammation in Tgfb1−/− mice (7, 8). It also regulates epithelial cell growth and differentiation and stimulates smooth muscle and myofibroblast differentiation and extracellular matrix deposition (6). TGFβ appears protective in acute models of asthmatic pathology, seen both genetically and pharmacologically (9–12). Conversely, excess active TGFβ can exacerbate chronic asthma pathology by induction of fibrosis (13, 14). It can also stimulate pulmonary inflammation and accumulation and contraction of smooth muscle through induction of TH17 cells (15) and effects on intraepithelial mast cells (16), leading to airway obstruction and decreased lung function.
We have reported genetic loci, Tgfbm2 and Tgfbm3, which dramatically alter phenotypic outcome of low TGFβ1 levels in mice, with effects on both vascular development and skin tumor susceptibility (17–20). We also showed that, as for human TGFB1, the mouse Tgfb1 gene is polymorphic, with allelic variants that drive different expression levels in diverse mouse species, consequently conferring strain-specific variation in tumor susceptibility (20). Interestingly, the biological outcome of Tgfb1 genetic variation in terms of tumor risk is dependent on interaction with an unlinked genetic locus, Skts15 (20), illustrating the power of epistasis in masking single gene effects and determining disease risk. Significantly, Skts15 colocalizes with Tgfbm3 on the genome (19). This locus is thus synonymous with Skts15 and a potent modifier of two distinct TGFβ-dependent phenotypes.
In the current study, we demonstrate that different components of the asthmatic response to the allergen, ovalbumin (OVA), are dependent on mouse strain background. More specifically, we show that potentiation of AHR by loss of a single Tgfb1 allele is dependent on synergistic interaction between variant alleles of the two TGFβ1 modifier loci, Tgfbm2 and Tgfbm3. Moreover, we demonstrate uncoupling of the inflammatory vs. the AHR response to an asthmatic stimulus, mediated by these two genetic variants.
Results
Tgfb1 Haploinsufficiency Potentiates AHR in a Mouse-Strain–Specific Manner.
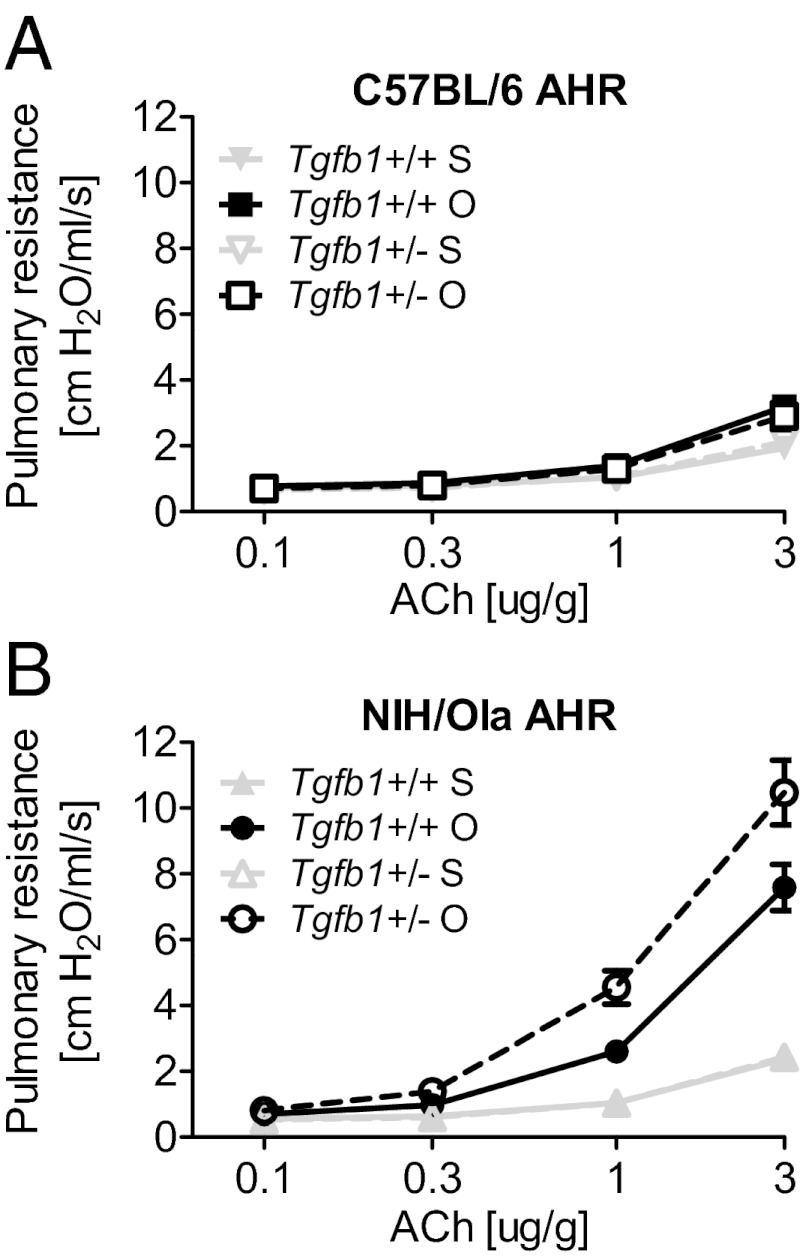
Several reports have suggested that TGFβ1 is protective against allergen-induced lung pathology (9–12). We compared wild-type with Tgfb1+/− mice on two different strains, NIH/OlaHSD (NIH) and C57BL/6NTac, and investigated their physiological and cellular responses to acute exposure to the allergen OVA after a 3-wk period of OVA sensitization. As assessed by acute AHR, C57 wild-type mice were relatively resistant to the asthmatic challenge compared with NIH wild-type mice (Fig. 1 A and B). Interestingly, we showed no effect of Tgfb1 gene dosage on the C57 genetic background, such that there were identical cellular and physiological responses to the asthmatic stimulus regardless of Tgfb1 genotype (Fig. 1A). However, we found strain differences with respect to Tgfb1 gene dosage on asthmatic response; loss of one Tgfb1 allele exacerbated AHR in NIH (Fig. 1B) but not C57 mice (Fig. 1A). We conclude that the effect of reduced TGFβ1 levels on asthmatic pathology was dependent on mouse strain background.
Fig. 1.
Tgfb1 haploinsufficiency potentiates AHR in a mouse-strain–specific manner. Respiratory resistance in response to escalating doses of ACh in mice sensitized and challenged with OVA (O) or saline (S) in (A) C57.Tgfb1+/+ and C57.Tgfb1+/− mice and (B) NIH.Tgfb1+/+ and NIH.Tgfb1+/−. Data shown as mean ± SEM for each group (n = 10). Linear regression model comparing treatment (P = 3 × 10−11) and genotype (P = 0.017).
Tgfbm2129 and Tgfbm3C57 Synergize to Reduce AHR in NIH-Tgfb1+/− Mice.
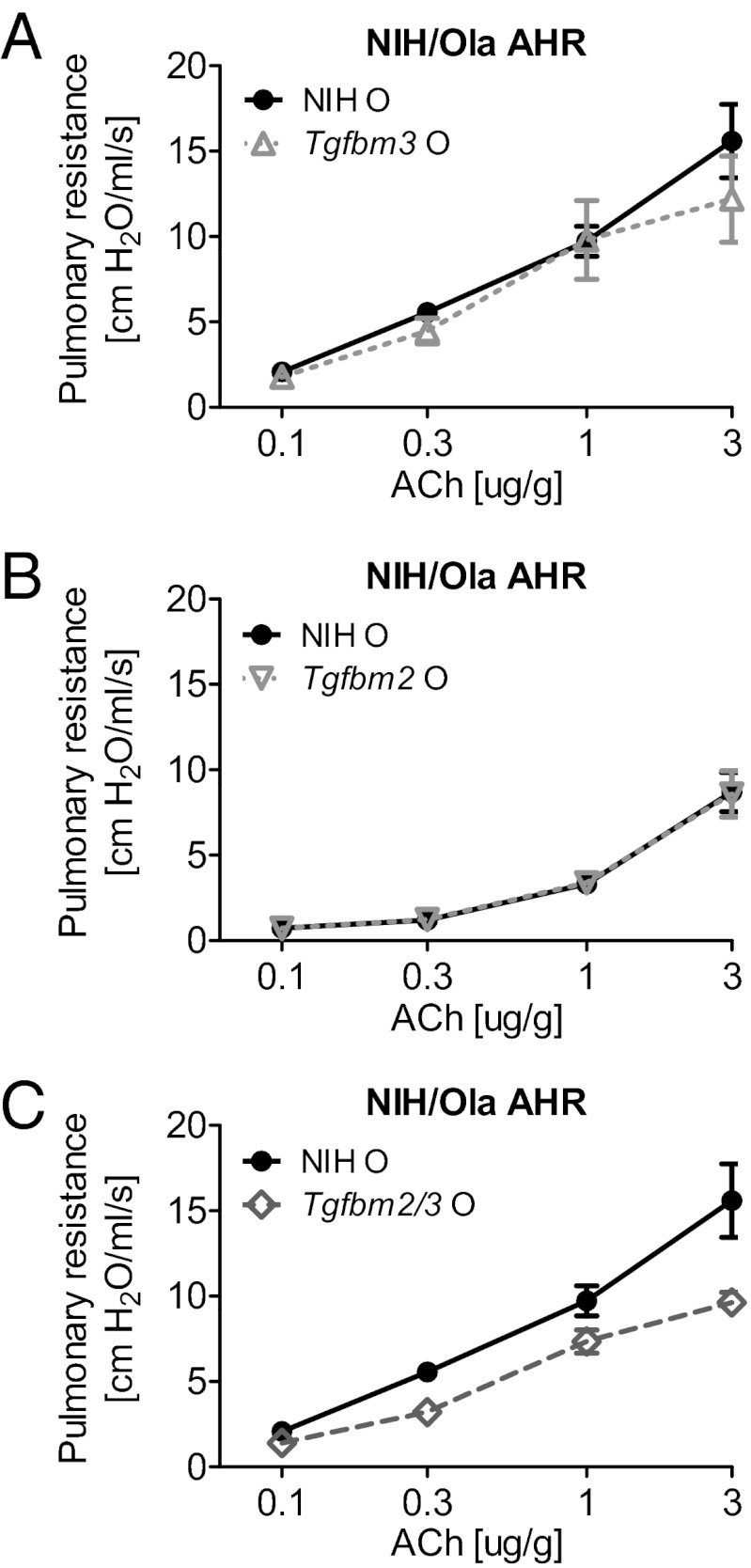
In two independent mouse models of pathology, namely tumor susceptibility (20) and vascular dysplasia (21), we previously demonstrated that the phenotypic outcome of Tgfb1 gene dosage is mouse strain dependent and strongly influenced by endogenous variation within Tgfbm2 and Tgfbm3 (20, 21). To examine the influence of these genetic loci on pulmonary responses to OVA challenge, we used the previously reported NIH.129-Tgfbm2 congenic mice that are >99% NIH except for a 1-Mb 129-derived interval around Tgfbm2 on distal chromosome 1 (Fig. S1A) (22). We also generated NIH.B6-Tgfbm3 congenic mice that harbor a 15-Mb interval of the C57 genome on proximal chromosome 12 around Tgfbm3 (Fig. S1B and Materials and Methods). Subsequently, we used the most asthma-susceptible strain in the current study, namely, NIH Tgfb1+/−, to compare the effects of inheritance of either or both modifier loci on responses to the asthmatic stimulus. Specifically, we compared four strains of NIH-Tgfb1+/− mice, namely, parental NIH, and NIH carrying either Tgfbm2129/129 (Tgfbm2) or Tgfbm3C57/C57 (Tgfbm3) or both variant modifier loci, Tgfbm2129/129 and Tgfbm3C57/C57 (Tgfbm2/3) (Fig. S1C).
We hypothesized that the C57 allele of Tgfbm3 would protect from OVA-induced AHR, because mice on the C57 genetic background were relatively resistant to OVA challenge. However, homozygosity for the C57 allele of Tgfbm3 had no effect on AHR in NIH-Tgfb1+/− mice (Fig. 2A). Similarly, 129Ola homozygosity at Tgfbm2, when acting alone, had no significant effect on AHR in NIH-Tgfb1+/− mice (Fig. 2B). In contrast, NIH-Tgfb1+/− mice bred to be homozygous variant for both loci, Tgfbm2/3, were significantly more resistant to acetylcholine (ACh)-induced AHR (Fig. 2C). This synergistic interaction between Tgfbm2 and Tgfbm3 was reproduced in two further independent experiments (Fig. S2). Thus, neither genetic modifier locus alone confers resistance to AHR, but the two loci synergize to protect the animal from an OVA-induced asthmatic airway response.
Fig. 2.
Tgfbm2 and Tgfbm3 synergize to reduce AHR in NIH-Tgfb1+/− mice. Respiratory resistance was measured in response to escalating doses of ACh in Tgfb1+/− mice sensitized and challenged with OVA (O) or saline (S). Respiratory curves of OVA-stimulated lungs are displayed for (A) NIH vs. NIH.Tgfbm3, (B) NIH vs. NIH.Tgfbm2, and (C) NIH vs. Tgfbm2/3 mice. Data shown as mean ± SEM from each group (n = 10). Linear regression model comparing OVA-treated NIH to OVA-treated Tgfbm2/3 mice (P = 0.002).
Tgfbm2 and Tgfbm3 Interaction Compensates for the Effect of Tgfb1 Haploinsufficiency on AHR.
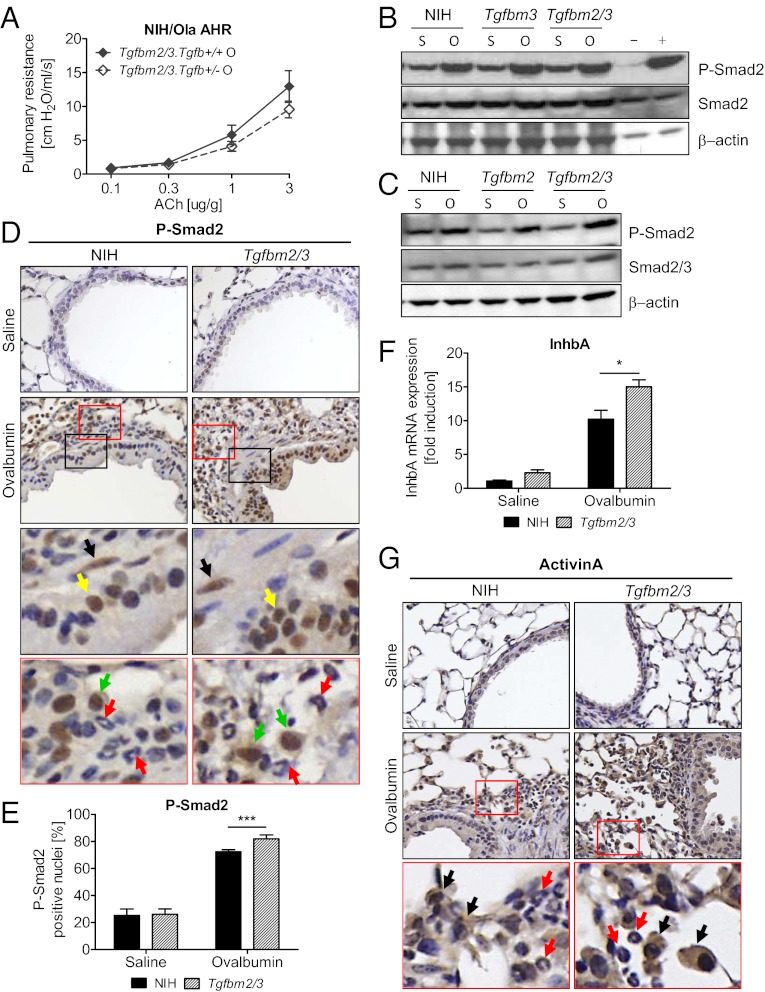
Because Tgfb1 haploinsufficiency potentiated OVA-induced AHR in NIH mice, and synergy between Tgfbm2 and Tgfbm3 attenuated AHR, we postulated that Tgfbm2 and Tgfbm3 might interact epistatically to compensate for the reduced Tgfb1 gene dosage effects seen in Tgfb1+/− mice. We predicted that the sensitizing effect of Tgfb1 haploinsufficiency on AHR would be masked in Tgfbm2/3 mice. As expected, whereas loss of a Tgfb1 allele potentiated the asthmatic response to OVA on the NIH background (Fig. 1B), on the Tgfbm2/3 background, Tgfb1 hemizygosity had no potentiating effect on AHR (Fig. 3A). Indeed, loss of a single Tgfb1 allele on the Tgfbm2/3 background even showed a trend toward protection from asthma.
Fig. 3.
Tgfbm2 and Tgfbm3 together compensate for Tgfb1 haploinsufficiency in Tgfbm2/3 mice. (A) Respiratory resistance measured in response to escalating doses of ACh after challenge with OVA, in Tgfb1+/− and wild-type mice on a Tgfbm2/3 strain background. Data shown as mean ± SEM from each group (n = 10). (B and C) Western blot analysis using indicated antibodies was done on protein lysates from lung tissue after saline (S) or OVA (O) challenge. Of 18 independent pairwise comparisons made between lung tissues, 11 showed a higher elevation in P-Smad2 in Tgfbm2/3 compared with NIH mice. (D) Immunohistochemistry for P-Smad2 with positive nuclear staining of bronchiolar epithelial cells of parental NIH and Tgfbm2/3 lungs after OVA or saline exposure. Enlargement of stained section (red and black box) demonstrates positive staining of bronchiolar epithelial cells (yellow arrow), macrophages (green arrow), and some smooth muscle cells (black arrow) but not eosinophils (red arrow). (E) NIH ImageJ was used to quantify positively stained nuclei for P-Smad2. Quantification of P-Smad2 staining used five fields of view (n = 5–6 mice per group, 5 sections per lung). (F) Inhba mRNA expression analysis of parental NIH and Tgfbm2/3 lungs after either saline or OVA challenge. Results are represented compared with GAPDH Ct value. Data shown as mean ± SEM from each group (n = 5). P values were determined using Student t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). (G) Immunohistochemistry for activinA with positive cytoplasmic staining of bronchiolar epithelial cells, macrophages (black arrow), and some smooth muscle cells but not eosinophils (red arrow) of parental NIH and Tgfbm2/3 lungs after OVA or saline exposure.
Tgfbm2/3 Mice Have a Greater P-Smad2 Response to OVA Challenge than NIH Mice.
As interaction between Tgfbm2 and Tgfbm3 appears to compensate functionally for reduced Tgfb1 gene dosage, we postulated that these mouse strains might differ in TGFβ signaling levels. To address this issue, we examined pulmonary P-Smad2 level as a marker of active TGFβ signaling in the various mouse strains. Western blot analysis demonstrated similar and invariable levels of total Smad2/3 regardless of mouse strain or treatment agent (saline vs. OVA). In contrast, all strains showed a significant elevation in P-Smad2 levels after OVA challenge, and this increase appeared marginally greater in the Tgfbm2/3 than in the NIH mice (Fig. 3 B and C). Immunohistochemical analysis showed that P-Smad2 was localized predominantly within bronchiolar epithelial cells and macrophages, with the occasional smooth muscle cell nucleus staining for P-Smad2 (Fig. 3D). Consistent with Western data, immunohistochemical analysis revealed that Tgfbm2/3 mice exhibited a significant increase in the percentage of P-Smad2 positive bronchiolar epithelial cells (Fig. 3E). It is possible that some of the differential increase in P-Smad2 staining seen by Western analysis was due to infiltrating macrophages. However, any greater elevation in macrophages in double congenic vs. NIH mice was not statistically significant, making it difficult to draw unequivocal conclusions (see Tgfbm2/3 Mice Exhibit Enhanced Airway Inflammation and Eosinophilia Compared with Parental NIH Mice).
Microarray analysis (see Molecular Analysis of Differential Asthmatic Responses in NIH vs. Tgfbm2/3 Mice) showed no significant difference in expression of Tgfb1, Tgfb2, or Tgfb3, or their canonical receptors between the two mouse strains. We therefore postulated that the observed increase in bronchiolar P-Smad2 signaling may be driven by other members of the TGFβ superfamily. Interrogation of microarray data (see Molecular Analysis of Differential Asthmatic Responses in NIH vs. Tgfbm2/3 Mice) for differentially expressed TGFβ signaling genes (Dataset S1A), suggested higher OVA-induction of Inhba in Tgfbm2/3 than NIH mice. Inhba encodes activinA, another Smad2-activating ligand. This finding was validated by qRT-PCR, confirming significantly higher induction of Inhba gene expression in lungs of Tgfbm2/3 (15.02-fold) vs. NIH mice (10.22-fold) (Fig. 3E). Immunohistochemical staining for activinA showed a very similar distribution to that of P-Smad2, with most activinA localized to bronchial epithelium and macrophages (Fig. 3F).
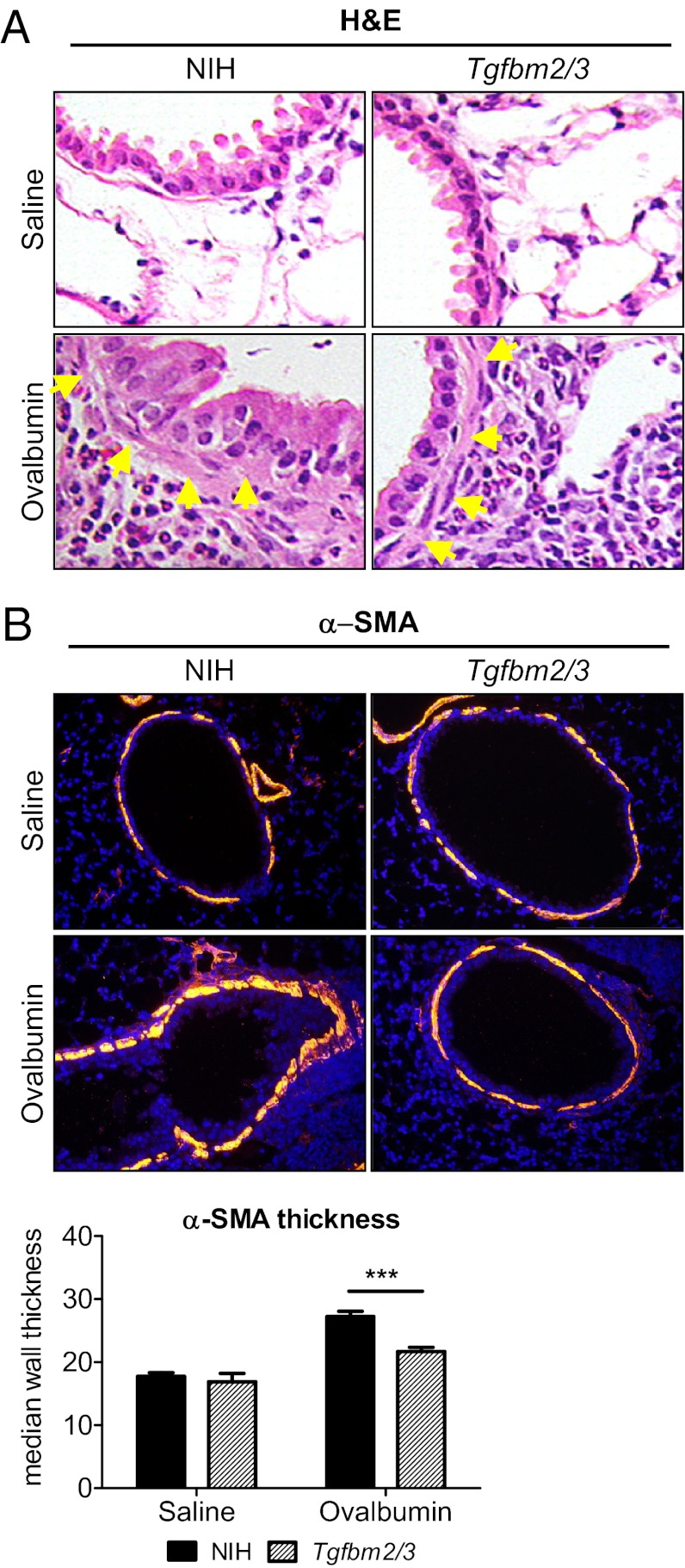
Enhanced Muscularization of Bronchiolar Submucosa in NIH vs. Tgfbm2/3 Mice.
H&E staining of the lungs suggested an apparent increase in subbronchiolar smooth muscle between OVA-treated NIH and Tgfbm2/3 mice (Fig. 4A). To further investigate these changes, anti–α-smooth muscle actin (α-SMA) immunofluorescence staining was undertaken on paraffin sections to identify myofibroblasts and smooth muscle cells (Fig. 4B). In both saline control groups, the submucosal smooth muscle layer of the bronchioles was discontinuous, with no difference between NIH and Tgfbm2/3 mice in the percent investment of bronchiolar epithelium with smooth muscle (Fig. S3). However, quantification of bronchiolar smooth muscle thickness demonstrated that both NIH and Tgfbm2/3 mice showed a thickening of the smooth muscle layer in response to OVA challenge, and this effect was significantly greater in the NIH than in the Tgfbm2/3 mice (Fig. 4B).
Fig. 4.
Thickened peribronchiolar smooth muscle layer in NIH lungs after OVA challenge. (A) H&E staining of heterozygous NIH and Tgfbm2/3 lung sections (5 µm thickness). Yellow arrows indicate smooth muscle layer. (20× fold magnification) (B) Anti α-SMA immunofluorescence staining of NIH and Tgfbm2/3 lungs after treatment with OVA or saline. Quantification of α-SMA thickness around the bronchioli of parental NIH and Tgfbm2/3 lungs after treatment with OVA or saline. NIH ImageJ was used to determine α-SMA thickness. Quantification used five fields of view per sample, 10 independent measurements per bronchiole. P values were determined using Student t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Tgfbm2/3 Mice Exhibit Enhanced Airway Inflammation and Eosinophilia Compared with Parental NIH Mice.
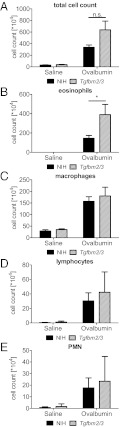
Because asthma is considered an inflammatory disease, and the Tgfbm2/3 mice were relatively protected from OVA-induced AHR, we hypothesized that this strain would show a reduced pulmonary inflammatory infiltrate compared with NIH mice after OVA challenge. On the contrary, although both mouse strains showed a robust increase in total bronchoalveolar lavage (BAL) cell counts after OVA challenge (Fig. 5A) there was a larger influx of inflammatory cells into the lungs of Tgfbm2/3 compared with NIH mice (Fig. 5A). These findings were most evident when comparing eosinophil cell numbers (Fig. 5B; NIH: 145 ± 27.6; vs. Tgfbm2/3: 390 ± 105.8; P = 0.03). Comparison of infiltrating numbers of macrophages, lymphocytes, or non-eosinophil polymorphonuclear cells did not differ significantly between the mouse strains (Fig. 5 C–E). Thus, the Tgfbm2/3 mice paradoxically showed greater pulmonary eosinophilia than parental NIH, despite the relative resistance of the NIH strain in terms of AHR.
Fig. 5.
Double congenic NIH-Tgfbm2/3 mice exhibit a stronger eosinophilic response to OVA than NIH. (A–E) BAL fluid from NIH and Tgfbm2/3 Tgfb1+/− mice was analyzed for (A) total cell count, (B) eosinophil cell number, (C) macrophage number, (D) lymphocyte number, and (E) polymorphonuclear neutrophil (PMN) cell number after saline or OVA challenge. Data shown as mean ± SEM from each group (n = 10).
Robust TH2-Mediated Inflammatory Response in both NIH and Tgfbm2/3 Mice.
Because the Tgfbm2/3 mice had more pronounced eosinophilia than NIH mice, we expected the cytokine profiles of these two strains to differ considerably. IgE-dependent mast cell activation induces the TH2 allergic response, via release of TH2-associated cytokines IL-4, IL-5, and IL-13. To investigate the cellular mechanisms of action of the TGF-β modifier loci in enhancing eosinophila but protection from AHR, we examined these parameters. Despite robust OVA-induced increases in serum IgE levels, as well as elevated transcript levels encoding TH2-associated cytokines IL-4, IL-5 and IL-13 in both mouse strains, there were no significant differences in the magnitude of these differentials between the two mouse strains (Fig. S4). Additional parameters examined included gene expression of osteopontin (SPP1), interferon g, PAI-1, and FoxP3 as markers of asthmatic response and of TH1 or Treg involvement, but no significant differences between mouse strains were found (Fig. S4).
Molecular Analysis of Differential Asthmatic Responses in NIH vs. Tgfbm2/3 Mice.
To investigate the possible underlying molecular causes leading to greater AHR in NIH than Tgfbm2/3 we undertook microarray gene expression analysis on lungs from both mouse strains after challenge with either saline or OVA. Expression analysis revealed that Tgfbm2/3 mice, despite relative resistance in terms of AHR, had more extensive OVA-induced gene expression changes than NIH mice. We postulated that this was due to the greater pulmonary influx of inflammatory cells in this mouse strain. Indeed, gene annotation analysis of differentially expressed transcripts between OVA-treated NIH and Tgfbm2/3 mice indicated enrichment of genes in the categories, inflammation, macrophage, cytokines, and immune regulation (Dataset S2A). Analysis of specific genes confirmed that these were part of the asthmatic response signature, including up-regulation of Sprr2a, Sprr1a, Mucin5ac, and Il6, and down-regulation of Hnmt (Dataset S3A). In contrast, transcripts differentially expressed between the two strains after saline exposure (Dataset S3B) were enriched for the classes: protein synthesis, ribonucleoproteins, ribosomal genes, cell cycle, and posttranslational modifications, namely, acetylation and methylation (Dataset S2B). This suggests that polymorphic variants of Tgfbm2 and/or Tgfbm3 control fundamental differences in the biosynthetic machinery of the cell even in the absence of OVA exposure.
Because the extensive OVA-induced inflammatory response in Tgfbm2/3 mice might mask gene expression differences responsible for the higher OVA-induced AHR in NIH, we mined the dataset for transcripts exhibiting a significantly larger differential in gene expression between saline and OVA treatment (ΔS–O) in NIH mice vs. Tgfbm2/3 mice (Dataset S1B). As a control, we undertook the converse analysis, comparing genes that had a significantly higher saline-to-OVA differential (ΔS–O) in Tgfbm2/3 than NIH mice (Dataset S1A). As expected, transcripts showing significantly larger ΔS–O differentials in Tgfbm2/3 than NIH mice were enriched for inflammatory signatures including chemokines and chemokine receptors, as well as markers of epithelial keratinization (Dataset S4A). In contrast, genes exhibiting greater ΔS–O differentials in NIH than Tgfbm2/3 mice were enriched for protein biosynthesis and posttranslational modification, including phosphoproteins, acetylation, and methylation, as well as nucleotide metabolism and chromosomal proteins (Dataset S4B). Once again this indicates a fundamental difference in the overall cellular biosynthetic machinery, not only between baseline characteristics of NIH vs. Tgfbm2/3 lungs (Dataset S2B), but also in the pulmonary responses of NIH to OVA (Dataset S4B).
Discussion
Because TGFβ1 levels are elevated and implicated in the pathology of both acute and chronic asthma models, and this signaling pathway is a potential therapeutic target for reducing excessive airway remodeling and pulmonary fibrosis, many studies have addressed the role of this growth factor in experimental asthma. Manipulation of TGFβ activity either genetically or pharmacologically has had variable outcomes. In a model of chronic OVA-induced asthma, therapeutic anti-TGFβ antibody dosing reduced peribronchiolar extracellular matrix, airway smooth muscle cells, and mucus production without stimulating airway inflammation or TH2 cytokine production (11). In contrast, in a house dust-mite–induced model, anti-TGFβ antibodies had no effect on airway remodeling but exacerbated eosinophilia resulting in potentiated AHR (23). More recently, it was shown that TGFβ produced by bone marrow stromal cells that home to the lung during an asthmatic episode, is implicated in reducing the pulmonary allergic inflammatory response (24), which is consistent with other reports that TGFβ plays a protective role in the asthmatic response (9, 10, 25, 26).
Here we have shown strain-specific differences in the magnitude of responses of C57 and NIH mice to an asthmatic stimulus. Both mouse strains showed elevated AHR to an OVA challenge, yet the C57 strain was relatively resistant compared with NIH. More importantly, whereas in NIH mice innate TGFβ1 was protective from OVA-induced AHR, on the C57 genetic background, loss of a single Tgfb1 allele had no effect on AHR. Thus, the functional response to reduced TGFβ1 levels in a murine asthma model is determined by genetic background. Contrasting with our findings, others found that Tgfb1 haploinsufficiency potentiates AHR in C57 mice (10). However, it is possible that the strain used in the earlier study was not a pure inbred C57 background, but carried genetic contaminants from other sources that might influence AHR (22). In the current study, Tgfb1+/− mice had been backcrossed through >20 generations to C57 and found to be pure by whole genome scan 500K SNP array (Jax mouse diversity genotyping array).
In an attempt to identify genetic variants that might regulate the differential responses to reduced TGFβ1 signaling on AHR, we hypothesized that the Tgfb1 genetic modifier loci, Tgfbm2 and Tgfbm3 might play a role. When we used NIH mice into which we had bred variant Tgfbm2 and/or Tgfbm3, we found that neither locus alone affected AHR response but the two synergized to significantly reduce AHR. These two variant loci functionally compensated for reduced TGFβ1 levels in NIH-Tgfb1+/− mice, in that mice carrying both variant loci no longer showed increased AHR in response to loss of a Tgfb1 allele. Thus, dependent on genetic variation within Tgfbm2 and Tgfbm3, the asthma-protective effect of TGFβ1 may be completely suppressed.
Another finding of the current study is the uncoupling between regulation of eosinophilia, airway remodeling, and AHR during an asthmatic episode, which also depends on Tgfbm2 and Tgfbm3. Recent studies on human asthma have demonstrated clinical stratification of asthmatics into TH2 low and TH2 high populations (27, 28), implicating a noneosinophilic mechanism of disease pathology in TH2 low asthmatics. However, the molecular etiology of asthma in the TH2 low population has yet to be elucidated. In mice, variation in genetic background has previously been found to affect different components of the asthmatic response in different manners. For example, in an acute model of OVA-induced asthma, C57 and BALB/c mice both developed similar extents of eosinophilic inflammation. However, the BalbC strain shows a significantly larger elevation in AHR and peribronchiolar smooth muscle expansion compared with C57 (29). Moreover, in C57 mice, the asthmatic response is totally dependent on eosinophils, as C57 ΔdblGATA1 knockout mice that lack eosinophils are completely resistant to OVA-induced AHR, whereas BALB/c ΔdblGATA1 knockouts still show elevated AHR in response to OVA (30). In the current study, we found that synergy between Tgfbm2 and Tgfbm3 regulates the eosinophilic and AHR responses to OVA challenge but these two physiological responses are regulated in oppositional directions by this genetic interaction, resulting in reduced AHR in the presence of increased eosinophilia in NIH mice homozygous for Tgfbm2 and Tgfbm3.
Tgfbm2 and Tgfbm3 might functionally compensate for reduced TGFβ1 of Tgfb1+/− mice by stimulating TGFβ signaling downstream of the ligand, therefore reducing AHR. Indeed, mice carrying both variant modifier loci exhibited higher levels of OVA-induced pulmonary P-Smad2 than mice harboring the NIH alleles. ActivinA may contribute to this effect, because it was the only Smad2-activating ligand showing a greater induction in Tgfbm2/3 than NIH mice. Both P-Smad2 and activinA were predominantly localized to bronchiolar epithelium and alveolar macrophages, with some staining in smooth muscle cells but none in eosinophils. Intriguingly, activinA has been reported, like myostatin, to suppress muscle mass (31) and may therefore contribute to the lack of bronchiolar smooth muscle expansion and consequent reduced AHR in Tgfbm2/3 compared with NIH mice.
Two outstanding issues are the identity of the Tgfbm2 and Tgfbm3 causative genetic variants that compensate for TGFβ1 and elicit differential asthmatic responses. It is now widely accepted that genetic modifier loci that influence multifactorial diseases frequently constitute clusters of functionally related polymorphic genes. This has been observed in mouse models of lupus (32) and cancer (33), making it difficult to dissect the underlying molecular mechanisms. With respect to Tgfbm2, Cenpf harbors an amino acid polymorphism between mouse strains at an evolutionarily conserved residue (18) (Fig. S1A). Cenpf has been proposed to regulate muscle determination (34, 35), although its major function is regulation of the centrosome and cytokinesis. Conceptually, distinct Cenpf isoforms might drive variable proliferation and/or differentiation in distinct tissue subsets.
To address the issue further, we undertook microarray gene expression analysis of NIH vs. Tgfbm2/3 mice. Consistent with the near genetic identity of NIH and Tgfbm2/3 mice, microarray analysis revealed no large differentials in gene expression between the two strains. Both mouse strains showed induction of an inflammatory signature after OVA treatment, albeit to differing extents. Because Tgfbm2/3 mice exhibited significantly more inflammation than NIH, this confounded interrogation of microarray data for genes that might contribute to the higher AHR in NIH vs. Tgfbm2/3. Around 500 genes showed greater differential gene expression (ΔS–O) in NIH than in Tgfbm2/3 mice. Intriguingly, this set of genes was enriched in proteins involved in protein biosynthesis and posttranslational modifications, including acetylation, phosphorylation, and methylation. In this respect, it is provocative that several genes encoding regulators of DNA or chromatin modification are localized in the Tgfbm3 region, including Asxl2, a polycomb protein; Dnmt3a, DNA methyl transferase 3a; DNAJC27 and Ddx1; as well as a regulator of translation and message stability, Pum2 (Fig. S1B). In particular, it was recently demonstrated that DNA methyltransferase 3a acts directly on the Il-13 gene within TH2 cells to down-regulate IL-13 expression, and hence allergic airway inflammation in mice (36). It is certainly possible that genetic variation at Dnmt3a may contribute to the phenotypic differences conferred by Tgfbm3, acting in concert with Tgfbm2. Microarray gene expression analysis of whole lung may be insufficiently sensitive to detect differentials in gene expression within minor cell type subsets of the lung, such as bone marrow stromal cells (24) or TH2 subsets (36), that nevertheless have a large impact on the outcome of the allergic response.
To summarize, the major conclusion from this study is that the biological responses to reduced TGFβ1 signaling in an experimental asthma model are dependent not only on the model (acute vs. chronic), or the cell types affected (smooth muscle vs. inflammatory vs. epithelial), but on the differential inheritance of endogenous genetic variants of wild-type modifier genes. This effect is pronounced, because differential inheritance of Tgfbm2 and Tgfbm3 can reverse the biological outcome of reduced Tgfb1 levels. These findings present a cautionary note for the use of TGFβ agonists or antagonists for the treatment of acute or chronic lung diseases, indicating a more personalized approach to the use of such agents. However, further dissection of the genes and molecules involved in modifying the outcome of reduced TGFβ activity may shed light on alternative pathways that could be targeted for treatment of lung diseases.
Materials and Methods
Animals.
The Tgfb1 null allele used in this study was Tgfb1Tm1n (7), referred to as Tgfb1+/− throughout the text and figures. These mice were bred as previously described (19). NIH.C57-Tgfbm3 congenic mice were generated by backcrossing B6NIHF1 mice through more than seven generations to recipient NIH mice. At each generation, selection was made for C57 genetic markers spanning from the centromere of chromosome 12 to exon 3 of the Lipn1 gene. NIH.129-Tgfbm2 congenic mice were the result of biological coselection with the Tgfb1 null allele of 129 genomic DNA at Tgfbm2 through multiple backcross generations to NIH mice (22). The Tgfbm2129 region spans from exons 57–60 of Ush2a to exons 2–6 of Ptpn14.
All experiments involved 10-wk-old male animals in groups of 10 mice per arm except for experiment 4, where 8 animals were used in the OVA arm and 5 animals were used in the saline control group. All animal husbandry practices were in full compliance with the recommendations published in Guide for the Care and Use of Laboratory Animals (37). All proposed protocols have been approved by the University of California San Francisco Institutional Animal Care and Use Committee.
Analysis of MicroArray Dataset.
Affymetrix Mouse Gene 1.0 ST Array CEL files were preprocessed (background correction, quantile normalization, probeset summarization) using the aroma.affymetrix R package (38). Differential expression analysis using moderated t statistic with Benjamini–Hochberg false discovery rate (adjusted P value) correction was performed using the Limma R package (39).
Genes responding to treatment with adjusted P value of <0.05 were initially selected. To focus on genes with substantial differences in log fold change (FC) between NIH and Tgfbm2/3, the following two quantities, M = difference in log FC between NIH and Tgfbm2/3, and A = average log FC, were evaluated. Genes with absolute M/A > 0.33 were retained. These were segregated into a group with stronger NIH response, where [abs (logFC NIH/logFC Tgfbm2/3) > 1] and one with stronger Tgfbm2/3 response [abs (logFC NIH/logFC Tgfbm2/3) < 1].
Functional enrichments for the four gene lists were derived by uploading MicroArray ProbeID into DAVID using Affymetrix_Exon_Gene_ID as identifier and MoGene-1–0 as background setting. The output of SP_PIR keywords (SP_PIR = Swiss Prot Protein Information Resource) within the functional categories was determined with a P value threshold set to 0.05.
Supplementary Material
Acknowledgments
Microarray analysis was performed by the Gladstone Institute’s Microarray Core. Work was funded by National Institutes of Health Grants R01-HL078564 and R0-1GM60514 (to R.J.A.) and the Bouque Foundation (R.J.A.). F.F.C. was the recipient of fellowships from the Belgian American Educational Foundation and the American Heart Association.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.A.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205374109/-/DCSupplemental.
References
- 1.McFadden ER, Jr, Gilbert IA. Asthma. N Engl J Med. 1992;327:1928–1937. doi: 10.1056/NEJM199212313272708. [DOI] [PubMed] [Google Scholar]
- 2.Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFβ1 allele association with asthma severity. Hum Genet. 2001;109:623–627. doi: 10.1007/s00439-001-0617-y. [DOI] [PubMed] [Google Scholar]
- 3.Silverman ES, et al. Transforming growth factor-β1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med. 2004;169:214–219. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 4.Gentile DA, et al. Association between TNF-α and TGF-β genotypes in infants and parental history of allergic rhinitis and asthma. Hum Immunol. 2004;65:347–351. doi: 10.1016/j.humimm.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 5.Mak JC, et al. Analysis of TGF-β(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol. 2006;117:92–96. doi: 10.1016/j.jaci.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 6.Howell JE, McAnulty RJ. TGF-β: Its role in asthma and therapeutic potential. Curr Drug Targets. 2006;7:547–565. doi: 10.2174/138945006776818692. [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni AB, et al. Transforming growth factor β 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diebold RJ, et al. Early-onset multifocal inflammation in the transforming growth factor β 1-null mouse is lymphocyte mediated. Proc Natl Acad Sci USA. 1995;92:12215–12219. doi: 10.1073/pnas.92.26.12215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schramm C, et al. TGF-β regulates airway responses via T cells. J Immunol. 2003;170:1313–1319. doi: 10.4049/jimmunol.170.3.1313. [DOI] [PubMed] [Google Scholar]
- 10.Scherf W, Burdach S, Hansen G. Reduced expression of transforming growth factor β 1 exacerbates pathology in an experimental asthma model. Eur J Immunol. 2005;35:198–206. doi: 10.1002/eji.200425209. [DOI] [PubMed] [Google Scholar]
- 11.McMillan SJ, Xanthou G, Lloyd CM. Manipulation of allergen-induced airway remodeling by treatment with anti-TGF-β antibody: Effect on the Smad signaling pathway. J Immunol. 2005;174:5774–5780. doi: 10.4049/jimmunol.174.9.5774. [DOI] [PubMed] [Google Scholar]
- 12.Alcorn JF, et al. Transforming growth factor-β1 suppresses airway hyperresponsiveness in allergic airway disease. Am J Respir Crit Care Med. 2007;176:974–982. doi: 10.1164/rccm.200702-334OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-β1 induces prolonged severe fibrosis in rat lung. J Clin Invest. 1997;100:768–776. doi: 10.1172/JCI119590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kenyon NJ, Ward RW, McGrew G, Last JA. TGF-β1 causes airway fibrosis and increased collagen I and III mRNA in mice. Thorax. 2003;58:772–777. doi: 10.1136/thorax.58.9.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo M, et al. IL-17A produced by αβ T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. 2012;18:547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto K, et al. The αvβ6 integrin modulates airway hyperresponsiveness in mice by regulating intraepithelial mast cells. J Clin Invest. 2012;122:748–758. doi: 10.1172/JCI58815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonyadi M, et al. Mapping of a major genetic modifier of embryonic lethality in TGF β 1 knockout mice. Nat Genet. 1997;15:207–211. doi: 10.1038/ng0297-207. [DOI] [PubMed] [Google Scholar]
- 18.Tang Y, et al. Genetic modifiers interact with maternal determinants in vascular development of Tgfb1(-/-) mice. Hum Mol Genet. 2003;12:1579–1589. doi: 10.1093/hmg/ddg164. [DOI] [PubMed] [Google Scholar]
- 19.Tang Y, et al. Epistatic interactions between modifier genes confer strain-specific redundancy for Tgfb1 in developmental angiogenesis. Genomics. 2005;85:60–70. doi: 10.1016/j.ygeno.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Mao JH, et al. Genetic variants of Tgfb1 act as context-dependent modifiers of mouse skin tumor susceptibility. Proc Natl Acad Sci USA. 2006;103:8125–8130. doi: 10.1073/pnas.0602581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian F, et al. Reduction in Smad2/3 signaling enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer Res. 2003;63:8284–8292. [PubMed] [Google Scholar]
- 22.Benzinou M, et al. Mouse and human strategies identify PTPN14 as a modifier of angiogenesis and hereditary haemorrhagic telangiectasia. Nat Commun. 2012;3:616. doi: 10.1038/ncomms1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fattouh R, et al. Transforming growth factor-β regulates house dust mite-induced allergic airway inflammation but not airway remodeling. Am J Respir Crit Care Med. 2008;177:593–603. doi: 10.1164/rccm.200706-958OC. [DOI] [PubMed] [Google Scholar]
- 24.Nemeth K, et al. Bone marrow stromal cells use TGF-β to suppress allergic responses in a mouse model of ragweed-induced asthma. Proc Natl Acad Sci USA. 2010;107:5652–5657. doi: 10.1073/pnas.0910720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon ED, et al. A protective role for periostin and TGF-β in IgE-mediated allergy and airway hyperresponsiveness. Clin Exp Allergy. 2012;42:144–155. doi: 10.1111/j.1365-2222.2011.03840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Presser K, et al. Coexpression of TGF-β1 and IL-10 enables regulatory T cells to completely suppress airway hyperreactivity. J Immunol. 2008;181:7751–7758. doi: 10.4049/jimmunol.181.11.7751. [DOI] [PubMed] [Google Scholar]
- 27.Woodruff PG, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–395. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choy DF, et al. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. J Immunol. 2011;186:1861–1869. doi: 10.4049/jimmunol.1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Hove CL, et al. Comparison of acute inflammatory and chronic structural asthma-like responses between C57BL/6 and BALB/c mice. Int Arch Allergy Immunol. 2009;149:195–207. doi: 10.1159/000199715. [DOI] [PubMed] [Google Scholar]
- 30.Walsh ER, et al. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J Exp Med. 2008;205:1285–1292. doi: 10.1084/jem.20071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SJ, et al. Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 2010;24:1998–2008. doi: 10.1210/me.2010-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morel L, Blenman KR, Croker BP, Wakeland EK. The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci USA. 2001;98:1787–1792. doi: 10.1073/pnas.031336098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samuelson DJ, et al. Rat Mcs5a is a compound quantitative trait locus with orthologous human loci that associate with breast cancer risk. Proc Natl Acad Sci USA. 2007;104:6299–6304. doi: 10.1073/pnas.0701687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadimou E, Ménard C, Grey C, Pucéat M. Interplay between the retinoblastoma protein and LEK1 specifies stem cells toward the cardiac lineage. EMBO J. 2005;24:1750–1761. doi: 10.1038/sj.emboj.7600652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pabón-Peña LM, Goodwin RL, Cise LJ, Bader D. Analysis of CMF1 reveals a bone morphogenetic protein-independent component of the cardiomyogenic pathway. J Biol Chem. 2000;275:21453–21459. doi: 10.1074/jbc.M000518200. [DOI] [PubMed] [Google Scholar]
- 36.Yu Q, et al. DNA methyltransferase 3a limits the expression of interleukin-13 in T helper 2 cells and allergic airway inflammation. Proc Natl Acad Sci USA. 2012;109:541–546. doi: 10.1073/pnas.1103803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.National Research Council, Committee for the Update of the Guide for the Care and Use of Laboratory Animals, and Institute for Laboratory Animal Research . Guide for the Care and Use of Laboratory Animals. 8th Ed. Washington, DC: National Academies Press; 2011. [Google Scholar]
- 38.Bengtsson H, Simpson K, Bullard J, Hansen K. (2008) Aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory (Department of Statistics, Univ of California, Berkeley), Technical Report 745.
- 39.Smyth GK. 2005. Limma: Linear models for microarray data. Bioinformatics and Computational Biology Solutions Using R and Bioconductor, eds Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, Springer, New York), pp 397–420.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.