Abstract
Myofibril stability is required for normal muscle function and maintenance. Mutations that disrupt myofibril stability result in individuals who develop progressive muscle wasting, or muscular dystrophy, and premature mortality. Here we present our investigations of the Drosophila l(2)thin [l(2)tn] mutant. The “thin” phenotype exhibits features of the human muscular disease phenotype in that tn mutant larvae show progressive muscular degeneration. Loss-of-function and rescue experiments determined that l(2)tn is allelic to the tn locus [previously annotated as both CG15105 and another b-box affiliate (abba)]. tn encodes a TRIM (tripartite motif) containing protein highly expressed in skeletal muscle and is orthologous to the human limb-girdle muscular dystrophy type 2H disease gene Trim32. Thin protein is localized at the Z-disk in muscle, but l(2)tn mutants showed no genetic interaction with mutants affecting the Z-line–associated protein muscle LIM protein 84B. l(2)tn, along with loss-of-function mutants generated for tn, showed no relative mislocalization of the Z-disk proteins α-Actinin and muscle LIM protein 84B. In contrast, tn mutants had significant disorganization of the costameric orthologs β-integrin, Spectrin, Talin, and Vinculin, and we present the initial description for the costamere, a key muscle stability complex, in Drosophila. Our studies demonstrate that myofibrils progressively unbundle in flies that lack Thin function through progressive costamere breakdown. Due to the high conservation of these structures in animals, we demonstrate a previously unknown role for TRIM32 proteins in myofibril stability.
Keywords: muscle degeneration, TRIM protein, LGMD2H
Degenerative muscle disease results from mutations in genes that are diverse in function (1–3). Muscle degeneration in the cell can result from the force-dependent disruption of subcellular membranes and/or internal cellular architecture, which unbundles normal myofibril organization and prevents proper function of the myofibrils (1, 3). Progressive degenerative muscle disease, such as muscular dystrophy, worsens over time because myofibril elements are disrupted and are unable to be properly processed or replaced in muscle (1, 3). Widespread muscle degeneration impacts other organ systems, such as the respiratory and circulatory circuits, that rely on healthy muscle for proper function and support. Failure of these systems in individuals with degenerative muscle disorders often results in reduced life span after a protracted disease progression, limited mobility, and decreased quality of life (1, 3).
Many genes that are now known to be required in muscle tissue were originally discovered to be mutated in individuals with degenerative muscle disorders (1, 3). Investigations of such genes in both human and model systems have enabled researchers to build models of the conserved elements of muscle structure and function (1–13). Disruptions in sarcoglycan genes (α-, β-, γ-, or δ-sarcoglycan) prevent the proper production of these sarcolemmal transmembrane proteins (4–8). Consequent aberrant function (sarcoglycanopathy) in humans results in multiple forms of limb-girdle muscular dystrophy (LGMD), a heterogeneous disease characterized by progressive muscle wasting of the limb, trunk, shoulder, and pelvic muscles (9). Emery–Dreifuss muscular dystrophy (EMD) can result from mutations in either emerin or the gene LMNA (lamin A/C), which are proteins that interact with the nucleoskeleton and/or nuclear components to maintain nuclear envelope architecture (10). Mutations in Dystrophin, a protein central to muscle cell stability that functions as a linkage between actin filaments to the cell membrane, result in individuals with Duchenne or Becker muscular dystrophy (11–13). Identification of these disease genes, as well as an understanding of the roles that they play in normal healthy muscle, is central to diagnosis and the development of more effective therapies.
Mutations in the E3 ubiquitin ligase TRIM32 result in individuals with Bardet–Biedl syndrome, a pleiotropic human disease, or limb-girdle muscular dystrophy 2H (LGMD2H) (14–16). The mouse model was used to confirm that TRIM32 is highly expressed in muscle and accumulates during muscle remodeling (17, 18). Disruptions in murine TRIM32 were able to recapitulate the myopathies observed in human LGMD2H (17–19). Muscle studies conducted using the mouse model have shown the murine ortholog of Trim32 to be diverse in function as it has the ability to bind myosin and ubiquitinate actin and to interact directly with the dystrophin-associated complex member, dysbindin (17–19). Nevertheless, there is still much to learn concerning a mechanistic understanding of the nature of cellular-level breakdown involving the specific mutations in Trim32 that lead to myofibril disorganization and muscular dystrophy.
The genetically tractable Drosophila model has established itself as an efficient system for understanding human disease (20–22). In this report, we describe our analysis of a muscle degenerative mutant in Drosophila for which the affected gene is orthologous to TRIM32. Our analyses demonstrate that costamere structure is lost in the Drosophila mutants and suggest that compromised costamere function could contribute to the mechanism(s) of LGMD2H in humans.
Results
thin Is a Human Trim32 Ortholog.
The original l(2)thin [l(2)tn] mutation was described by Ball et al. (23) as a muscle degenerative mutant in Drosophila. To analyze the “thin” phenotype and further map the mutation, we crossed the l(2)tn allele to deficiency stocks either within or spanning the original mapped region of l(2)tn [2R: 55D-56B (1)]. We found that the l(2)tn chromosome was lethal in combination with Df(2R)Exel6068 but not with Df(2R)Exel6067 or Df(2R)BSC349, enabling us to narrow the location of the l(2)tn locus to a small section of chromosome II. This molecularly defined genomic region contained only eleven candidate genes (Fig. 1A). Among these genes was CG15105/another b-box affiliate (abba), an ortholog of human Trim32, a gene that is required for muscle maintenance and stability and is mutated in patients with LGMD2H. Comparisons of domain structure and amino acid sequence that confirm this orthology are shown in Fig. S1.
Fig. 1.
The l(2)tn mutation is located on the second chromosome and results in muscle degeneration. (A) The l(2)tn mutation was localized—by complementation analysis using deletions containing molecularly defined breakpoints—to the boxed area. This area contains 11 annotated genes. (B and C) Representative pupae from WT (w1118) and thin mutants (l(2)tn/Df). Compared with WT (B), l(2)tn mutants have narrower and elongated pupal cases (C). (D–I) Immunofluorescent stains of L3 larval pelts stained with anti-TM (green) and phalloidin to visualize F-actin (red). The basic patterning of the WT (D) larval musculature is similar in the l(2)tn mutants (E), but muscles appear more robust in WT animals. (F–I) High-magnification images of muscles show highly organized sarcomeres in WT (F and H) compared with l(2)tn mutants (G and I), which show thinner, looser muscles with a loss of cross-striation and irregular sarcolemma. Arrows indicate sarcomeres that have remained in register; arrowheads indicate gaps between myofibrils. (Scale bars, 1.3 mm for B and C; 300 μm for D and E; 50 μm for F and G; 12 μm for H and I.)
Compared with wild type (WT) (Fig. 1B), l(2)tn/Df animals produced pupae that were thin and long (Fig. 1C). In L3 larval muscles, in contrast to WT (Fig. 1D), “thin” mutant skeletal muscles were narrower, round in cross-section, and relatively loose at the attachment sites (Fig. 1E). At higher magnification, WT muscles were oval in cross-section and showed aligned sarcomeres within the tightly bundled myofibrils (Fig. 1 F and H). WT muscles also had evenly spaced, linearly organized nuclei aligned along the midline of each muscle (Fig. S2A). Gaps between the myofibrils were apparent in tn mutants, as the fibers appeared unbundled at higher magnification (Fig. 1 G and I). There was also irregular clustering of nuclei within the mutant muscle cells (Fig. S2B). The elongated pupal case results from the inability of the musculature to compact the animal at the onset of pupariation (23).
tn Is Allelic to abba and the Encoded Protein Is Localized to the Z-Disk in Muscle.
We next sought to determine if l(2)tn is allelic to the annotated abba gene. UAS-RNAi lines specific for abba were crossed to the muscle driver 24B-Gal4 to generate knockdown animals (24B > abbaRNAi). We also created abba deletion alleles (tn∆A and tn∆B) by standard Minos-mediated excision techniques (24) as well as a rescue allele (UAS-abba) for use in a double-mutant background (UAS-abba Df/tn∆A;24B). Both 24B > abbaRNAi and tn∆A/Df animals recapitulated the thin pupal phenotype (Fig. 2 A and B), whereas the rescue animals had partial-to-full restoration of the WT pupal phenotype (Fig. 2C). Whole mounts of L3 larval skeletal muscle for the tn∆A/Df and RNAi knockdown animals phenocopied the thinner, looser musculature observed in l(2)tn/Df animals (Fig. 2 D and E). UAS-abba Df/tn∆A;24B animals generally showed a WT myofibrillar organization (Fig. 2F). High-resolution images of muscle from both 24B > abbaRNAi and tn∆A/Df animals (Fig. 2 G, J, and K) revealed the same myofiber instability and unbundling as seen in l(2)tn/Df homozygotes (Fig. 1 G and I), whereas rescue muscles were relatively more stable and had sarcomeres appearing in register (Fig. 2L).
Fig. 2.
Knockdown of Thin function by RNAi or genetic deletion mirrors the degenerative muscle phenotypes observed in thin mutants. Representative pupal cases from 24B > abbaRNAi (A) or tnΔA/Df (B) individuals have the same elongated pupa phenotype observed in the original l(2)tn mutant. (C) (UAS-abba,Df/tn∆A;24B) are able to partially rescue the WT phenotype in a mutant background. (D–L) Immunofluorescent antibody stains of L3 muscle pelts stained with anti-TM (green) and phalloidin to visualize F-actin (red). Knockdown (D) and knockout (E) animals mimic the original l(2)tn phenotype by displaying thinner, looser muscles that appeared less robust than partial rescue (F) and WT animals. (G–L) Higher-magnification images of the individual myofibers in knockdown (G and J) and knockout (H and K) larvae show a general loss of sarcomere organization and cross-striation, whereas myofibrils of partial rescue animals are relatively well organized (I and L) and appear more stable than in the mutant. (M) Axial ratios for pupae of the indicated genotypes. The median value is indicated by the line through the box bounded by the upper quartile and lower quartile. Error bars indicate the upper and lower limits of the data points. The distribution of axial ratios revealed a consistently larger length/width ratio for tn mutants compared with w1118 and rescue animals. (N) Lethal-phase data indicate that the majority of the tn mutant offspring are embryonic and pupal lethal. Although some rescue animals were embryonic lethal, the majority of hatched larvae eclosed as adults. (Scale bars, 1.3 mm for A–C; 300 μm for D–F; 50 μm for G–I; 16 μm for J–M.)
We further characterized the thin phenotype quantitatively by measuring the axial ratios between groups of mutants and WT pupae (Fig. 2M). The length/width ratios of pupae between l(2)tn/Df, l(2)tn/tn∆A, l(2)tn/tn∆B, and 24B > abbaRNAi groups were substantially higher and displayed a greater degree of variability around the mean compared with WT and rescue (UAS-abba Df/tn∆A;24B) values. Lethal phase determination of l(2)tn allelic combinations revealed that mutant animals died at two stage-specific time points in the Drosophila life cycle. A large proportion of l(2)tn/Df, l(2)tn/tn∆A, l(2)tn/tn∆B, and 24B > abbaRNAi animals died in embryogenesis. Remarkably, either very low numbers or no mutant individuals were lost during the larval stages. Animals that survived through the larval stages died as pupae, and the rescue animals mostly eclosed as adults. Our lethality data suggested that the l(2)tn allele may be a hypomorph as more animals survived until the pupal stage. These data, together with the degenerative phenotypes observed in the muscles of all mutant L3 larvae, provide strong evidence that the original l(2)tn mutation affects the CG15105/abba gene, which we name tn.
Given that tn gene function is essential for myofiber stability, we next sought to determine the spatial and temporal accumulation of Thin protein during development. In embryos, tn mRNA and protein were expressed in the skeletal muscle precursors (Fig. 3 A and D) and remained high in a muscle-specific pattern until the completion of embryonic muscle development (Fig. 3 B and E). l(2)tn/Df animals showed a marked reduction in accumulation of tn mRNA, but the signal remained muscle specific (Fig. 3C). This muscle-specific, yet reduced, accumulation was also observed in l(2)tn/Df mutant embryos immunostained for Thin protein expression (Fig. 3F) and supports the idea that the l(2)tn allele is a hypomorphic allele. Through sequencing of cDNA for tn we did not observe any amino acid changes, suggesting that the original l(2)tn mutation might affect the regulatory regions of this gene.
Fig. 3.
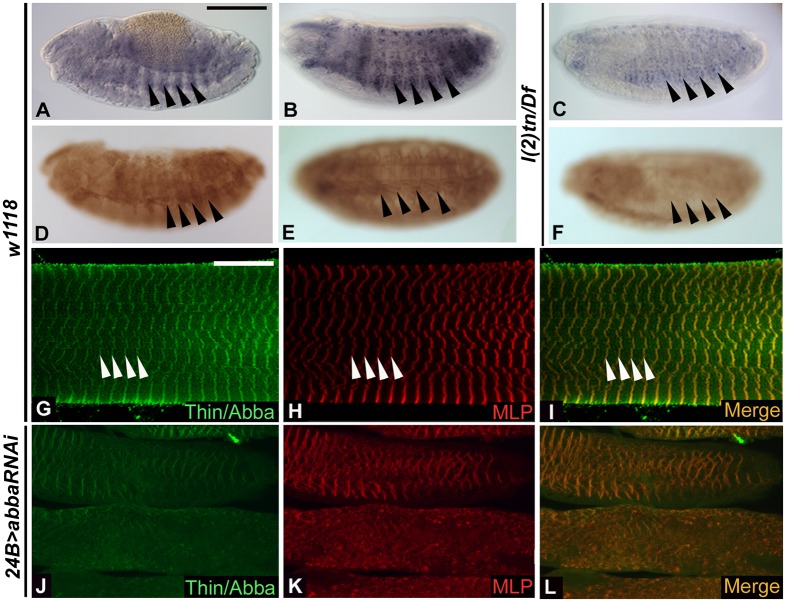
Thin is expressed in developing and mature muscle. (A–C) In situ hybridizations showing tn mRNA expression. tn mRNA is robustly expressed in stage 13 (A) and stage 16 (B) WT embryos. Reduction of tn levels is apparent in the l(2)tn/Df mutants (C). (D–L) Thin protein expression in developing muscle (D and E) and mature muscle (G and I). Thin protein accumulated in a muscle-specific pattern in stage 13 (D) and stage 16 (E) WT embryos, but was markedly reduced upon loss of tn function (F). (G) A WT preparation shows that Thin is present in L3 larval muscle (green) in a repeated pattern within the sarcomeres. Costaining for the Z-line protein MLP84B (H, red) reveals that Thin also is found at the Z-disk of the sarcomere (I). Knockdown of tn using RNAi results in a reduction of Thin accumulation at the Z-lines and a general decrease in muscle expression levels (J) compared with MLP84B, which is retained at the Z-lines (K and L). Black arrows (A–F) point to corresponding muscle segments in the developing embryos. White arrows (G–I) illustrate the corresponding Z-lines in the muscle of the L3 larva preparations. (Scale bars, 140 μm for A–F; 15 μm for G–L.)
Thin protein accumulation in WT animals was also detected in the mature muscles of L3 larvae and localized in a striated pattern across the muscle (Fig. 3G). Immunolocalization with the Z-disk–associated protein muscle LIM protein 84B (MLP84B) (Fig. 3H) revealed overlap between Thin and MLP84B at the Z-disks (Fig. 3I). Because there was a small amount of Thin accumulation in the l(2)tn/Df mutants, we examined 24B > abbaRNAi progeny to test the sensitivity and specificity of our anti-Thin antibodies (Fig. 3 J–L). Relative to WT, Thin accumulation was depleted at the Z-lines of the mutant muscles, and no other staining in muscle was present (Fig. 3 J and L). MLP84B remained present and localized within the disrupted myofibrils of 24B > abbaRNAi animals (Fig. 3 K and L). Together, these data demonstrate that the Thin protein is expressed within muscle tissue and that the encoded protein is closely associated with the muscle sarcomere, consistent with our demonstrated role for this protein in muscle maintenance.
tn Mutants Reveal No Genetic Interaction with mlp84B and sls.
The localization of Thin to the Z-disk raised the possibility that Thin associates with Z-line proteins already known to have roles in myofiber stability. To test this hypothesis, we performed genetic interaction analyses to determine if the l(2)tn allele enhanced the thin pupal phenotypes of mutations in mlp84B or the titin gene sallimus (Table S1). Mutants for tn, mlp84B, and salimus/D-titin (sls) alone each produced pupa with a “thin-like” phenotype, observable as higher axial ratios compared with WT (Fig. 4) (25). Both mlp84B and sls mutants exhibited a lesser thin phenotype compared with tn mutants (Fig. 4). Removal of one copy of mlp84B or sls in a l(2)tn/tn∆A background did not enhance the thin phenotype over l(2)tn/tn∆A individuals alone (Fig. 2M). An increased axial ratio was observed upon removal of one copy of sls in mlp84B homozygotes, consistent with the published genetic interactions between these two genes (25). Taken together, these results suggest that either there is no interaction between l(2)tn and sls and/or mlp84B, or the full thin phenotype observed in l(2)tn/tn∆A homozygotes and sls/mlp84B interaction crosses is the most extreme incarnation of the “thin” phenotype, leaving any genetic interaction unobservable in this phenotype.
Fig. 4.
tn does not genetically interact with mutations affecting the Z-disk components MLP84B or D-Titin. D-Titin, also known as sallimus (sls), is the fly ortholog of mammalian Titin. Box plots of the indicated genotypes show the distribution of axial ratios in mutant pupae. Pink shading represents the span of the box-plot ratios for control (w1118). Blue shading indicates the range of box-plot ratios for the original l(2)tn mutant and removal of mlp84B or sls in a tn mutant background. Df(3R)dsx2M is a deletion that removes Mlp84B.
α-Actinin, Actin, and MLP84B Remain Localized in tn Mutants, Whereas Myosin Heavy Chain and Tropomyosin Are Mislocalized.
Vertebrate TRIM32 can bind to the thick filament protein myosin and ubiquitinates actin (17). To determine if these and other sarcomeric components are altered upon loss of tn, we analyzed the expression and localization of key muscle proteins in our mutants. WT animals stained for α-Actinin or MLP84B showed specific localization to the Z-disk in well-organized sarcomeres (Fig. 5 A and B). Also, myosin heavy chain (MHC) and tropomyosin (TM) were found organized as cross-striations within the tightly bundled myofibrils of normal muscle (Fig. 5 C and D). α-Actinin, MLP84B, MHC, and TM were all present in both l(2)tn/Df and tn∆A/Df muscle. The key differences were in their relative subcellular localization within the muscle tissue compared with WT. Despite the widespread degeneration of myofibrillar organization in both l(2)tn/Df and tn∆A/Df animals, α-Actinin (Fig. 5 E and I) and MLP84B (Fig. 5 F and J) remained relatively well localized within the sarcomeres of the mutant muscle, with significant areas of cross-striations still observed. By contrast, MHC (Fig. 5 G and K) and TM (Fig. 5 H and L) were present, but mislocalized, in the mutants. Rather than retaining a periodic localization within the sarcomeres, both MHC and TM accumulated along the sarcolemmal membrane in globular structures. These results, combined with the observation that both MHC and TM accumulate normally in embryos mutant for tn, suggested that myofibril destabilization occurs in actively contracting muscles upon loss of Thin.
Fig. 5.
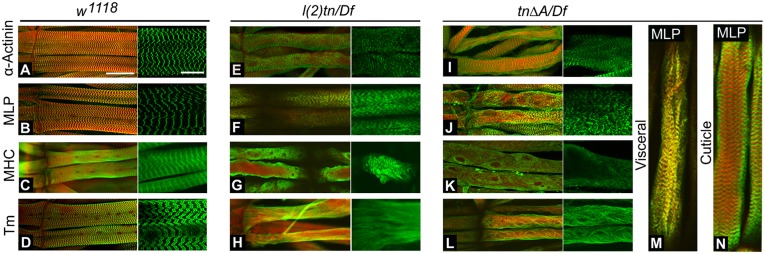
Loss of tn selectively affects the localization of known sarcomere proteins within the myofiber. (A–N) Confocal micrographs of the L3 larval musculature in WT (A–D) and tn mutants (E–N) stained with the indicated antibodies (green) and phalloidin to observe F-actin (red). In WT muscle, α-Actinin (A) and MLP84B (B) are Z-line associated. These proteins remain properly localized at the sarcomeres despite the loss of tn and disorganization of the myofibrils in l(2)tn/Df (E and F) or tn∆A/Df (I and J) mutants. WT stains exemplify the highly organized localization of the sarcomeric proteins MHC (C) and TM (D) in the myofiber. In the genotypes l(2)tn/Df and tn∆A/Df, MHC (G and K) and TM (H and L) are mislocalized, and in some preparations, the disassociated protein accumulates along the surface of the sarcolemma and occasionally forms globular structures. Visceral (M) and cuticle (N) images of muscle stained for MLP84B (green) and actin (red) show the directional disorganization of myofibrils. Muscle along the cuticle maintains a relatively well-organized cross-striated pattern (N) compared with the unbundling myofibrils along the visceral (M) side of the muscle. (Scale bars: full size, 30 μm; Inset, 12 μm.)
The degenerative muscle phenotype was further examined by comparing the relative state(s) of myofibrillar unbundling in confocal sections through the z axis in WT and mutant muscle preparations of the genotype tn∆A/Df (Fig. 5 M and N). Myofibrils of muscle cells became unbundled directionally: the muscle unbundling phenotype was more severe toward the visceral surface of the muscle (Fig. 5M), but appeared to retain a more WT appearance toward the most lateral (i.e., closest to the cuticle) region of the cell (Fig. 5N). We hypothesize that the loss of Thin in actively crawling larvae could be temporarily compensated for by a steady-pressure force directed from the cuticle. Muscle in older individuals appeared to have a greater degree of degeneration than that of younger individuals. This is consistent with the observations of Ball et al. (23) that suggest that the loss of tn results in progressive muscle degeneration that manifests in the unbundling of myofibrils as well as the dislodging of specific sarcomeric proteins. This evidence also supports the role of Thin as a stabilizing protein in the structural maintenance of muscle.
In vertebrates, the costamere is positioned at the Z-disk of the sarcomere and links the myofibrils to the overlying sarcolemma to provide sustained stability of the myofibrillar apparatus with the sarcolemma during muscle contraction. Although implied, the existence of costameres has not yet been demonstrated in flies (26, 27). Drosophila orthologs of the costameric proteins β-Integrin, Talin, Spectrin, and Vinculin exist and could play conserved roles in muscle stability and function (28). Given the nature of the physical breakdowns in the musculature of tn mutants, we next sought to determine the existence of the costamere in larval skeletal muscles.
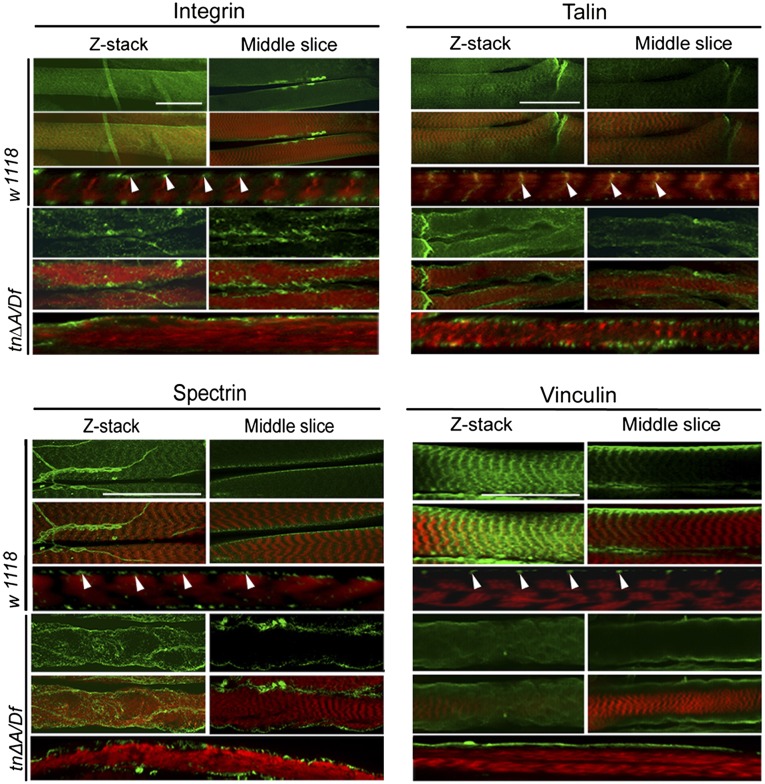
In WT larvae, β-integrin, Talin, Spectrin, and Vinculin were present in cross-striations overlying the Z-disk (Fig. 6). To confirm that these proteins were localized to a position in fly muscles similar to the position of the costamere in vertebrate muscles, confocal sections through the z axis were collected. These micrographs clearly showed that the components of the Vinculin–Talin–Integrin complex, along with Spectrin, exist in flies (Fig. 6). Furthermore, these proteins remain conserved as Z-line–associated structures between the myofibrils and the overlying sarcolemma. Scans through the center of the bundle of myofibrils showed that β-integrin, Talin, Spectrin, and Vinculin do not appear at the internal Z-lines (Fig. 6), as also seen for these proteins in vertebrate muscles. Thus, proteins that compose the vertebrate costamere have orthologs that accumulate in Drosophila larval muscles in a pattern highly reminiscent of the costamere. We conclude that costameres exist in Drosophila larval body-wall muscles and that the structure of the costamere shares many similarities with vertebrate costameres.
Fig. 6.
The costameric proteins Integrin, Talin, Spectrin, Vinculin are consistently mislocalized in mutants. Both WT and tn∆A/Df late-stage larvae were dissected and immunofluorescently labeled for the antibody (green) and for F-actin (red). Images were captured by confocal microscopy. Merged Z-stack scans show the surface of the muscles. A middle slice from these scans is also presented to illustrate the sarcolemmal accumulation of some proteins. WT animals share two localization features of costameric proteins: localization at the Z-disk and localization around the surface of the muscle, but not in the center. In tn∆A/Df mutants, these proteins are mislocalized from the Z-disk and often appear as clumps near the surface of the muscle. Arrowheads indicate the localization of costamere proteins along the surface of muscle. (Scale bars, 30 μm.)
We next tested if a loss of Thin has an impact on the stability of costamere-associated proteins in muscle tissue. The spatial organization of β-integrin, Talin, Spectrin, and Vinculin was examined in tn∆A/Df animals. The distribution and accumulation of β-integrin was normal in embryonic development in both mutant and WT embryos. (Fig. S3). However, in mutant larvae, we observed a similar phenotype for β-integrin, Talin, Spectrin, and Vinculin (Fig. 6), where these proteins no longer retained localization at the Z-disk and were randomly distributed along the membrane of the sarcolemma. In addition, noncostameric, trans-sarcolemmal proteins such as δ-sarcoglycan (Fig. S4) also showed widespread mislocalization due to tn loss of function. From this evidence, we conclude that Thin is required for maintaining proper costamere stability in muscle and that loss of Tn ultimately results in the degenerative muscle phenotype characteristic of tn mutants.
Discussion
This work demonstrates that Thin is a key structural protein in maintaining myofibrillar stability. Myofibrils in tn mutants progressively unbundled during development, most likely due to a loss of costamere integrity. Over time, this results in progressive muscle wasting. Gaps in the myofibrils of tn mutants may be analogous to gaps in the myofibrils of diseased muscle tissue from individuals with muscle myopathy that also displays myofibrillar disorganization and abnormal vacuole position (1). The myofibril defects that we observe in Drosophila are more extensive than those observed for TRIM32 deficits in mammals, suggesting that Thin has a greater functional role in muscle than do the vertebrate orthologs. Because it is likely that multiple mechanisms cooperate to produce the muscle breakdown that occurs in humans suffering from LGMD2H, the role of Thin in maintaining myofiber stability could be one contributing factor.
Antibodies generated during this study showed that Thin is expressed in muscle and localizes at the Z-line. This is consistent with the Z-line localization observed for murine TRIM32 (18). Given the high level of conservation from sequence to muscle phenotypes of both tn and Trim32 mutants, further studies using tn mutants will serve as a good model for understanding the molecular mechanisms underlying LGMD2H. Moreover, the apparent single Trim32 ortholog in flies allows for the direct examination of Thin function in myofibril stability without the potential complication of redundancy with other TRIM domain proteins.
We also illustrate in detail in Drosophila a detailed characterization of the muscle stability complex known in vertebrates as the costamere. Our observations place the costameric proteins β-integrin, Talin, Spectrin, and Vinculin at the Z-line on the periphery of the myofibers in flies. Loss of tn in flies leads to the disintegration of these core components and the overall breakdown of the costamere. Our phenotypic studies indicate that stability of the costameres is central to keeping sarcomeres in register and to preventing myofibrillar unbundling. Secondarily, muscle proteins such as MHC and TM are dependent on this complex for proper localization or stability.
Studies here assign a molecular role for a mutant first described over 20 y ago and named l(2)tn for its elongated, thin pupal phenotype (23). Our analysis of the l(2)tn allele shows that there is some residual transcript expression in the embryonic musculature compared with WT. Also, a substantially larger proportion of l(2)tn/Df mutants survive until the pupal stages compared with the earlier lethality observed in the tn deletion alleles or 24B > abbaRNAi knockdown. This indicates that l(2)tn is likely a hypomorphic allele and that there is a minimal requirement for Thin accumulation to maintain normal muscle stability. Interestingly, muscles form normally in tn knockout mutants, indicating that Thin is required for myofibril stability, but not required for initial myofibril formation.
Our data position Thin in close proximity to the costamere complex, and reveal it is associated with the Z-disk in more central muscle areas. Based on the relative positions of stable versus mislocalized Z-line–associated proteins in tn mutants, Thin function is likely to be critical to sustaining costameric associations between MLP84B and Vinculin or Spectrin, and it must also bind and stabilize/costabilize other proteins in the sarcomere. The Z-line proteins α-Actinin and MLP84B, along with actin, remained associated with the Z-disk in tn mutants. This observation indicates that their localization, other than general myofibril disassociation, was unaffected by the loss of Thin. Dislodged and disorganized accumulations of MHC and TM along the surface of the sarcolemma suggested that Thin could be a possible key anchor point of the myofibril architecture, and future studies aimed at defining binding partners for Thin will be informative in uncovering its molecular function.
The costameres form rings, positioned at the Z-lines of the outermost sarcomeres of myofibril bundles, just below the membrane (27, 28). These costamere rings provide physical support and stability as the muscle moves and contracts. Although the existence of these structures has been suggested in Drosophila muscle (26, 27), they had yet to be characterized. Loose, destabilized myofibrils in tn mutants are consistent with a phenotype of muscles that suffer from a lack of stabilizing pressure provided from intact and functional costameres.
How a loss of TRIM32 at the cellular level results in progressive muscle wasting in individuals with LGMD2H is unknown. Insight into TRIM32 is further complicated by its widespread expression and pleiotropic functions as observed in satellite cells as well as in nonmuscle tissues (29–31). Although separable from a specific role in muscle cells, loss of TRIM32 prevents satellite cells from proper maturation and proliferation, thus preventing muscle cell replacement and repair (30, 31). In vitro, the RING domain of TRIM32 has been shown to interact with myosin (17). This may explain why a loss of function in the fly ortholog results in such a severe phenotype for MHC accumulation, although it does not correlate with the general Z-disk localization of Thin protein. Consistent with the canonical function of RING finger motifs, TRIM32 possesses E3 ubiquitin ligase activity, where both actin and dysbindin are in vitro targets (17, 32, 33). Mutations that result in LGMD2H affect the C-terminal NHL repeats of TRIM32. Modeling of these repeats suggests that they fold into a β-propeller structure that may mediate protein–protein interactions or TRIM protein dimerization (34). Recapitulation of the causal mutations found in the NHL domains of LGMD2H patients demonstrate that TRIM32 can become destabilized and therefore lose its relative abundance in the cell (19, 34). TRIM32 participates in multiple cellular interactions and likely has a variety of roles in the cell.
We envision two additional roles for Thin/TRIM32 in muscle cells, which are not mutually exclusive. First, Thin may serve as a critical link to stabilize the structure of costameric proteins. The individual NHL domains that comprise the β-propeller structure could form a bridge by binding to Vinculin and/or Spectrin near the sarcolemma membrane on one side and to MLP84B anchored at the myofibril Z-line. In this way, Thin/TRIM32 would function to link costameric proteins, thus stabilizing the structure at a crucial point of intersection between the membrane and myofiber. TRIM32 has been shown to homodimerize (19). It may be that complexes of homodimers interact to form Thin/TRIM32 multimers to fulfill this role. Second, Thin/TRIM32 may also function to ubiquitinate sarcomeric proteins, such as Actin, Dysbindin, and possibly MHC (17, 33). The costamere may be a critical junction for this process, allowing for Thin/TRIM32 to process sarcomeric components as they are either damaged (polyubiquitination) or possibly reintegrated during reloading (monoubiquitination). Future experiments will be needed to elucidate which (or whether both) of these mechanisms predominate in muscle tissue.
Materials and Methods
Standard fly genetics, molecular biology, and immunostaining techniques were used. Antibodies specific for Tn were generated for this project. Details of all constructs, reagents, and protocols are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank John Sparrow for providing the original l(2)tn allele. This work was supported by funding from National Institutes of Health Grants AR059311 and AR060788 (to E.R.G.); Muscular Dystrophy Association Grant 176329 (to R.M.C.); and American Heart Association Grant 080206Z (to E.M.L.-D). E.M.L.-D. was also supported by National Institute of General Medical Sciences/Initiative for Maximizing Student Development (IMSD) Grant 5R25GM060201.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1208408109/-/DCSupplemental.
References
- 1.Emery AE. The muscular dystrophies. Lancet. 2002;359(9307):687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 2.Amato AA, Griggs RC. Overview of the muscular dystrophies. Handb Clin Neurol. 2011;101:1–9. doi: 10.1016/B978-0-08-045031-5.00001-3. [DOI] [PubMed] [Google Scholar]
- 3.Mathews KD, Moore SA. Limb-girdle muscular dystrophy. Curr Neurol Neurosci Rep. 2003;3(1):78–85. doi: 10.1007/s11910-003-0042-9. [DOI] [PubMed] [Google Scholar]
- 4.Sandonà D, Betto R. Sarcoglycanopathies: Molecular pathogenesis and therapeutic prospects. Expert Rev Mol Med. 2009;11:e28. doi: 10.1017/S1462399409001203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrié A, et al. Mutational diversity and hot spots in the alpha-sarcoglycan gene in autosomal recessive muscular dystrophy (LGMD2D) J Med Genet. 1997;34(6):470–475. doi: 10.1136/jmg.34.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bönnemann CG, et al. β-Sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat Genet. 1995;11(3):266–273. doi: 10.1038/ng1195-266. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi S, et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270(5237):819–822. doi: 10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 8.Nigro V, et al. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat Genet. 1996;14(2):195–198. doi: 10.1038/ng1096-195. [DOI] [PubMed] [Google Scholar]
- 9.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet. 1999;21(3):285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 10.Nagano A, et al. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat Genet. 1996;12(3):254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman EP, et al. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N Engl J Med. 1988;318(21):1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 13.Watkins SC, Hoffman EP, Slayter HS, Kunkel LM. Immunoelectron microscopic localization of dystrophin in myofibres. Nature. 1988;333(6176):863–866. doi: 10.1038/333863a0. [DOI] [PubMed] [Google Scholar]
- 14.Frosk P, et al. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am J Hum Genet. 2002;70(3):663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2H and the role of TRIM32. Handb Clin Neurol. 2011;101:125–133. doi: 10.1016/B978-0-08-045031-5.00009-8. [DOI] [PubMed] [Google Scholar]
- 16.Chiang AP, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc Natl Acad Sci USA. 2006;103(16):6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J Mol Biol. 2005;354(2):413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 18.Kudryashova E, Wu J, Havton LA, Spencer MJ. Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component. Hum Mol Genet. 2009;18(7):1353–1367. doi: 10.1093/hmg/ddp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudryashova E, Struyk A, Mokhonova E, Cannon SC, Spencer MJ. The common missense mutation D489N in TRIM32 causing limb girdle muscular dystrophy 2H leads to loss of the mutated protein in knock-in mice resulting in a Trim32-null phenotype. Hum Mol Genet. 2011;20(20):3925–3932. doi: 10.1093/hmg/ddr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chartier A, Benoit B, Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 2006;25(10):2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feany MB, Bender WW. A Drosophila model of Parkinson’s disease. Nature. 2000;404(6776):394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 22.Crowther DC, et al. Intraneuronal Abeta, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience. 2005;132(1):123–135. doi: 10.1016/j.neuroscience.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Ball E, Ball S, Sparrow J. A mutation affecting larval muscle development in Drosophila melanogaster. Dev Genet. 1985;6:77–92. [Google Scholar]
- 24.Metaxakis A, Oehler S, Klinakis A, Savakis C. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171(2):571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark KA, Bland JM, Beckerle MC. The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J Cell Sci. 2007;120(Pt 12):2066–2077. doi: 10.1242/jcs.000695. [DOI] [PubMed] [Google Scholar]
- 26.Ribeiro I, Yuan L, Tanentzapf G, Dowling JJ, Kiger A. Phosphoinositide regulation of integrin trafficking required for muscle attachment and maintenance. PLoS Genet. 2011;7(2):e1001295. doi: 10.1371/journal.pgen.1001295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparrow JC, Schöck F. The initial steps of myofibril assembly: Integrins pave the way. Nat Rev Mol Cell Biol. 2009;10(4):293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- 28.Ervasti JM. Costameres: The Achilles’ heel of Herculean muscle. J Biol Chem. 2003;278(16):13591–13594. doi: 10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 29.Reymond A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudryashova E, Kramerova I, Spencer MJ. Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J Clin Invest. 2012;122(5):1764–1776. doi: 10.1172/JCI59581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicklas S, et al. TRIM32 regulates skeletal muscle stem cell differentiation and is necessary for normal adult muscle regeneration. PLoS ONE. 2012;7(1):e30445. doi: 10.1371/journal.pone.0030445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;11:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 33.Locke M, Tinsley CL, Benson MA, Blake DJ. TRIM32 is an E3 ubiquitin ligase for dysbindin. Hum Mol Genet. 2009;18(13):2344–2358. doi: 10.1093/hmg/ddp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saccone V, et al. Mutations that impair interaction properties of TRIM32 associated with limb-girdle muscular dystrophy 2H. Hum Mutat. 2008;29(2):240–247. doi: 10.1002/humu.20633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.