Abstract
MicroRNAs (miRNAs) are important for plant development and stress responses. However, factors regulating miRNA metabolism are not completely understood. SICKLE (SIC), a proline-rich protein critical for development and abiotic stress tolerance of Arabidopsis, was identified in this study. Loss-of-function sic-1 mutant plants exhibited a serrated, sickle-like leaf margin, reduced height, delayed flowering, and abnormal inflorescence phyllotaxy, which are common characteristics of mutants involved in miRNA biogenesis. The sic-1 mutant plants accumulated lower levels of a subset of miRNAs and transacting siRNAs but higher levels of corresponding primary miRNAs than the WT. The SIC protein colocalizes with the miRNA biogenesis component HYL1 in distinct subnuclear bodies. sic-1 mutant plants also accumulated higher levels of introns from hundreds of loci. In addition, sic-1 mutant plants are hypersensitive to chilling and salt stresses. These results suggest that SIC is a unique factor required for the biogenesis of some miRNAs and degradation of some spliced introns and important for plant development and abiotic stress responses.
Keywords: cold stress, hydroxyproline-rich glycoprotein, intron decay, mRNA stability
MicroRNAs (miRNAs) are a class of endogenous small RNAs that function in gene regulation by guiding mRNA cleavage and translational repression and are critical for plant development and stress responses (1–9). The core components involved in miRNA biogenesis have been identified in plants. RNA polymerase II transcribes MIR genes; a 5′ 7-methyl guanosine cap and a 3′ poly(A) tail are added to produce primary miRNA (pri-miRNA) transcripts, which form imperfect stem-loop secondary structures by Watson–Crick base pairing between self-complementary foldback regions. In the Arabidopsis nucleus, the stem-loop structure of the pri-miRNA is processed by the RNase III enzyme DICER-LIKE1 (DCL1) to produce a pre-miRNA, which is further processed to generate a 21-nt-long miRNA/miRNA* duplex. For accurate dicing, DCL1 requires the help of HYPONASTIC LEAVES1 (HYL1, a dsRNA-binding protein) and SERRATE (SE, a C2H2zinc-finger protein) (10). The HUA ENHANCER 1 (HEN1) methyltransferase catalyzes 2'-O-methylation of the ribose sugar in the 3′ termini of miRNA/miRNA* duplexes (11). HASTY (HST), a homolog of mammalian EXPORTIN 5, helps export methylated miRNA/miRNA* duplexes from the nucleus to the cytosol (12). The mature miRNA is incorporated into ARGONAUTE1 (AGO1), forming an RNA-induced silencing complex, which scans for miRNA-complementary mRNAs and directs the cleavage or translational repression of the target mRNAs (1).
miR173 and miRNA390 direct the biogenesis of transacting siRNAs (ta-siRNAs). Noncoding transcripts from TRANS-ACTING siRNA genes (TAS) are cleaved by the miRNA-containing AGO1/AGO7 complex (13, 14). The cleaved transcripts are converted into dsRNA by RDR6, and these dsRNAs are processed by DCL4 to yield ∼21-nt ta-siRNAs. Like miRNAs, ta-siRNAs negatively regulate gene expression posttranscriptionally (15–18).
Nuclear mRNA cap-binding complex proteins, abscisic acid (ABA) Hypersensitive 1 (ABH1)/Cap-Binding Protein 80 (CBP80), and CBP20 also play important roles in pre-mRNA and pri-miRNA processing. abh1/cbp80 and cbp20 mutant plants are impaired in the processing of pri-miRNA transcripts into mature miRNAs, resulting in reduced miRNAs. A significant level of overlap in intron splicing and pri-miRNA processing was observed among se, abh1/cbp80, and cbp20 mutants. Among these, se showed the broadest defect in miRNA biogenesis (19). ABH1 may recruit capped pri-miRNAs to the DCL1/HYL1/SE processing complex or protect the capped pri-mRNA from RNA decay (8). In addition, a nuclear RNA-binding protein, DAWDLE, also interacts with DCL1 and is involved in the biogenesis of a subset of miRNAs and siRNAs in Arabidopsis (20). The Arabidopsis ERECTA mRNA UNDER-EXPRESSED (EMU) encodes a protein homologous to the yeast HPR1, which is a component of the suppressor of the Transcriptional defect of Hpr1 by Overexpression (THO) messenger ribonucleoprotein complex. emu showed altered splicing of serine/arginine-rich protein pre-mRNAs, suggesting that EMU is important for pre-mRNA splicing. Furthermore, emu accumulated lower levels of a subset of miRNAs than the WT (21). Impairment in the RNA decay pathway, as occurs in the ein5 (ethylene insensitive 5) mutant, also leads to a defect in small RNA biogenesis. EIN5 encodes XRN4 (5′→3′ EXORIBONUCLEASE 4), which is involved in the removal of uncapped mRNAs, and thus prevents RDR (RNA dependent RNA polymerase)-dependent small RNA biogenesis from uncapped mRNAs (22).
Among the core components of miRNA biogenesis, null alleles of DCL1 and SE are lethal, whereas null alleles of HYL1 are fertile. Hypomorphic mutations in core components of the miRNA biogenesis, export, and action machinery result in pleiotrophic developmental defects, such as a serrated leaf margin, change in flowering time, abnormal inflorescence phyllotaxy, and reduced fertility (22–31). These findings demonstrated that miRNAs have critical roles in plant development.
Here, an Arabidopsis mutant named sickle-1 (sic-1) was identified on the basis of its enhanced transcript stability of a LUC transgene. sic-1 mutant plants are hypersensitive to chilling and salt stresses and display developmental defects that are hallmarks of mutants defective in miRNA biogenesis. The SICKLE (SIC) gene was cloned and functionally characterized, and our results show that SIC is involved in miRNA biogenesis in Arabidopsis.
Results
sic-1 Mutation Enhances Luciferase mRNA Stability.
A stress-inducible, RD29A promoter-driven luciferase (LUC) reporter (PRD29A:LUC)-based genetic screen (32) was used to isolate mutants with enhanced LUC bioluminescence under cold stress. Seeds harboring PRD29A:LUC transgene (herein after referred to as WT) were mutagenized by ethyl methane sulfonate. The M2 seedlings were screened to obtain putative mutants with enhanced LUC bioluminescence after cold stress (33). One such mutant with high LUC bioluminescence after cold stress at 4 °C for 24 h also displayed sickle-like serrated leaf margins, and hence it was named sickle (sic) and selected for this study. sic-1 plants emitted higher LUC bioluminescence than WT after cold (0 °C, 24 h), ABA (100 μM, 3 h), and NaCl (150 mM, 3 h) treatments (Fig. S1 A and B). A time-course analysis of LUC bioluminescence after cold stress showed higher emissions in sic-1 than in WT plants (Fig. S1C). To determine whether the higher bioluminescence in sic-1 mutant was due to altered expression of the PRD29A: LUC reporter, expression level of the highly unstable LUC transcript was determined (33, 34). LUC transcript was undetectable in WT (Fig. S1D), but a high level of LUC transcript was detected in sic-1 after cold stress (Fig. S1D). In contrast to the high levels of LUC bioluminescence (Fig. S1A) and LUC transcript (Fig. S1D) in sic-1, the endogenous RD29A expression was slightly lower in sic-1 than in WT plants (Fig. S1E). COLD-REGULATED 15A (COR15A) is highly induced by cold stress and is used as common marker gene to study cold-responsive gene expression in addition to RD29A. Hence, we examined whether sic-1 mutation impaired the expression of COR15A. COR15A expression was slightly higher in sic-1 than in WT plants (Fig. S1E). This indicates that the cold-stress signaling pathway that mediates COR15A expression is slightly perturbed by the sic-1 mutation.
A nuclear run-on assay was performed to examine whether the differences in LUC transcript levels were due to differences in the rate of transcription. The result suggests that, in contrast to the several-fold differences in LUC bioluminescence between WT and sic-1, their transcription rates of the LUC transgene were similar (Fig. S2 A and B).
Northern analysis showed that a full-length LUC transcript was detectable in sic-1 (Fig. S2C), whereas only degradation products were detected in WT (Fig. S2C). To confirm that the degradation products observed in WT were LUC specific and to determine the cleaved region of the LUC transcript, three different regions of LUC transcript were used as probe to perform Northern analysis. The results show that both full-length and cleaved transcripts accumulated in sic-1, whereas mainly degradation products of LUC transcript accumulated in WT (Fig. S2D). Together, the results suggest that the sic-1 mutation enhances the stability of LUC transcript.
SIC Encodes a Proline-Rich Protein.
A backcrossed F2 generation from the sic-1 mutant displayed a 3:1 segregation between WT and mutant bioluminescence phenotypes, which suggests that sic-1 is a recessive mutation in a single nuclear gene. The sic-1 mutation was genetically mapped to a 23.3-kb region on chromosome 4. By sequencing genes within this region, a G-to-A change at the 432nd base downstream of the predicted translation initiation codon of At4g24500 was identified, which resulted in a change of tryptophan at 143 to a premature stop codon. Therefore, sic-1 is a loss-of-function mutant and possibly a null allele. To confirm that At4g24500 is SIC, a complementation assay was carried out. Transgenic lines expressing the At4g24500 gene with its native promoter were generated in sic-1 mutant background, and their LUC bioluminescence was compared with WT after cold stress. All of the transgenic lines had a WT LUC phenotype (Fig. S3A), confirming that At4g24500 is SIC.
A second allele of sic-1 was obtained from the SALK collection (SALK_054171). Although annotation from The Arabidopsis Information Resource (TAIR) shows that the T-DNA insertion is in the last exon of SIC, our analysis revealed that the actual insertion is in the 3′ UTR region of SIC. Thus, SALK_054171 is a knockdown allele (sic-2) with reduced SIC expression (Fig. S3B).
SIC is annotated as a hydroxyproline-rich glycoprotein with an unknown function (TAIR 10 release; http://www.arabidopsis.org/). SIC is a single-copy gene in the Arabidopsis genome.
SIC Is Necessary for Normal Plant Development.
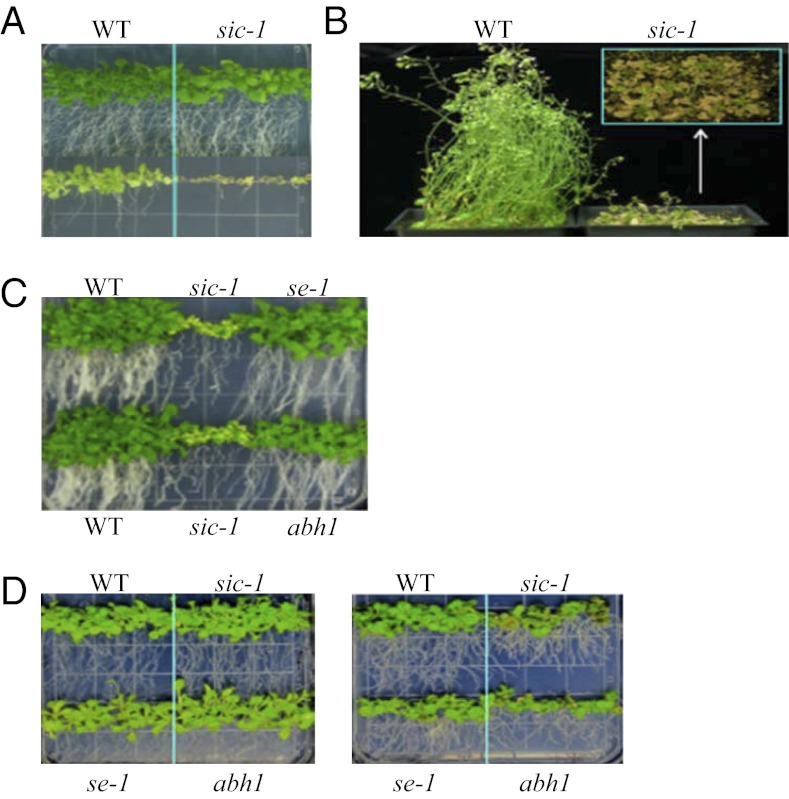
The sic-1 mutant displayed a number of developmental phenotypes (Fig. 1A). The sic-1 mutants showed delayed flowering compared with WT, and this delay was more pronounced under short-day light conditions (Fig. S3 C and D). At maturity, sic-1 exhibited dwarfism compared with WT (Fig. S3E). However, the sic-2 allele did not show strong developmental phenotypes compared with sic-1 (Fig. S3E), likely because expression of the SIC gene was only reduced in the sic-2 mutant. Developmental defects observed in sic-1 are common in mutants impaired in miRNA biogenesis and pre-mRNA processing. For example dcl1, se, hyl1, abh1/cbp80, cpb20, emu, and ein5 all produce leaves with serrated margins (21–25, 27, 29, 31, 35, 36). Abnormal inflorescence phyllotaxy (emergence of flowers from the same internode on the inflorescence) was observed in se-1, hyl1-1, and smu1-2 (suppressors of mec-8 and unc-52 1) plants (24, 31, 37). The leaf-serration phenotype in sic-1 was compared with those in se-1 and abh1-285 mutants. Rosette and cauline leaves of sic-1, se-1, and abh1-285 had deeper serrations compared with WT (Fig. 1A). The sickle leaf margin phenotype of sic-1 mutant was corrected in transgenic lines expressing the WT SIC gene (sic-1:SIC-GFP and sic-1:SIC-FLAG) (Fig. 1B). In addition, sic-1 plants also exhibited an abnormal inflorescence phyllotaxy phenotype similar to se-1 and abh1-285 (Fig. 1C). Although sic-1 plants were shorter than WT, the heights of se-1 and abh1-285 plants were similar to WT (Fig. S3F). The dwarfism and other defects of sic-1 were corrected in the transgenic lines expressing a WT copy of SIC gene (Fig. S3G), supporting that SIC is necessary for normal plant development.
Fig. 1.
Effect of sic-1 on leaf and inflorescence morphology. (A) Leaf morphology. Rosette and cauline leaves of sic-1 have serrated sickle-like margins, in contrast to those of WT. The leaf morphology of sic-1 is similar to that of se-1 and abh1-285. (B) SIC rescues the leaf-shape defect of sic-1. Rosette leaves of WT, sic-1, and sic-1 genetically transformed with native promoter-driven SIC gene (sic-1:SIC-Flag and sic-1:SIC-GFP). (C) Architecture of silique arrangements in WT, sic-1, se-1, and abh1-285. Arrow shows clustering of more than one silique on the same node in sic-1, se-1, and abh1-285.
SIC Is Involved in miRNA Biogenesis.
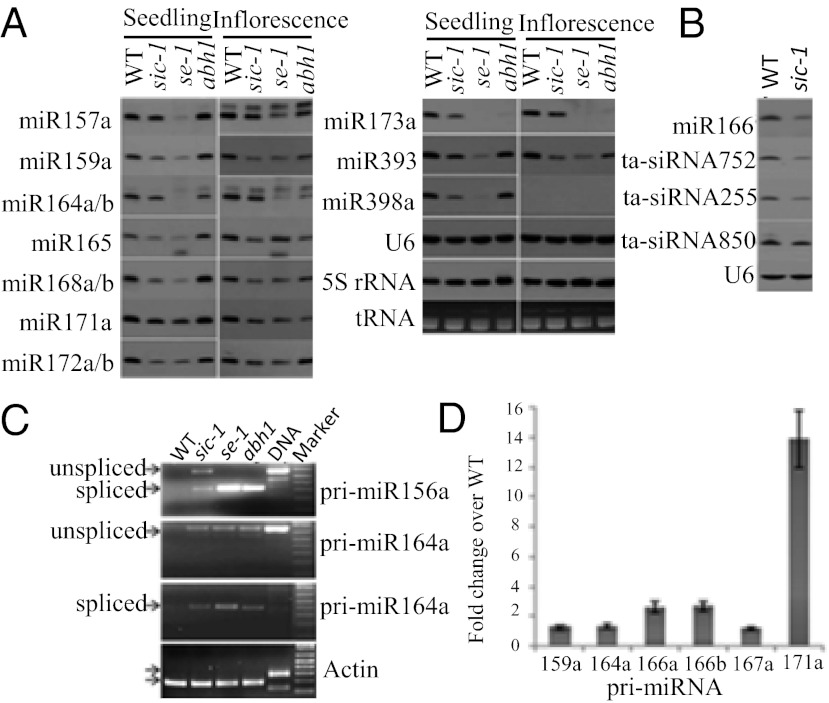
Because the developmental defects of sic-1 were similar to those of mutants impaired in miRNA biogenesis, we hypothesized that SIC may function in miRNA biogenesis. Northern analysis revealed that the accumulation of miR159, miR164, miR165, miR166, miR168, miR171, miR172, miR173, miR393, and miR398 was lower in sic-1 than in WT both in vegetative (seedlings) and reproductive (inflorescence) tissues (Fig. 2A). The level of miRNA accumulation was lowest in se-1 followed by sic-1 in seedling, whereas in inflorescences many miRNAs were similar in sic-1 and se-1 but were lower than in WT. The levels of miRNAs, except for miR164 and miR171, were lower in sic-1 than in abh1-285 plants (Fig. 2A).
Fig. 2.
Effect of sic-1 on the accumulation of miRNAs, ta-siRNAs, and pri-miRNAs. (A) Northern analysis of miRNA accumulation in WT, sic-1, se-1, and abh1-285 of Arabidopsis seedling and inflorescence. (B) Northern analysis of ta-siRNA in WT and sic-1 in Arabidopsis. (C) RT-PCR analysis of pri-miRNA accumulation in WT, sic-1, se-1, and abh1-285 of Arabidopsis (WT, sic-1, se-1, abh1 = RT-PCR, DNA = Columbia genomic DNA was used as template for PCR). (D) Quantitative RT-PCR analysis of pri-miRNA accumulation in WT and sic-1 in seedlings.
In Arabidopsis, TAS1, TAS2, and TAS3 genes can give rise to ta-siRNAs. miR173 and miR390 direct the cleavage of TAS gene transcripts to initiate ta-siRNA biogenesis (15, 17). Because sic-1 plants accumulated lower levels of miR173, the expression levels of TAS1 family of ta-siRNAs, namely ta-siR255, ta-siR752, and ta-siR850, were examined. All three ta-siRNAs accumulated to lower levels in sic-1 than in WT (Fig. 2B). Previous studies have shown that loss-of-function mutations of SE (30), CBP20, and CBP80 (35) lead to reductions in miR173, miR390, and their target ta-siRNAs. Our results provide evidence that ta-siRNA biogenesis also requires SIC. The accumulation level of 24-nt siRNAs was examined to determine whether sic-1 may affect siRNA levels. Accumulation of 24-nt siRNA at the AtSN1 locus did not change in sic-1, se-1, and abh1-285 compared with WT (Fig. S4A). This suggests that SIC is required for biogenesis of DCL1-dependent miRNAs and DCL4-dependent ta-siRNAs, but not for DCL3-dependent repeat-associated siRNAs such as AtSN1.
Previously, se-1, hyl1, dcl1, abh1-285, and cbp20-1 were found to accumulate higher levels of pri-miRNAs but lower levels of mature miRNAs compared with WT (19, 35, 36). Pri-miRNA accumulation in sic-1 was tested, and the results showed that accumulation of unspliced and spliced pri-miR156a and pri-miRNA164a was higher in sic-1 than in WT (Fig. 2C). Real-time RT-PCR analysis showed that pri-miRNA166a, pri-miRNA166b, and pri-miRNA171b were >twofold more abundant in sic-1 than in WT (Fig. 2D). Although pri-miRNA levels were higher in sic-1 than in WT, the levels were much lower in sic-1 than in se-1 and abh1-285 (Fig. S4B).
Expression level of predicted miRNA targets was examined to determine whether the target genes were affected by the sic-1 mutation. Expression levels of the predicted miR159a, miR166, miR171a, and miR390 target genes (At1g53230, At5g37020, At4g00150, and At5g14050, respectively) were higher in sic-1 than in WT (Fig. S4C, Upper), which is consistent with the reduced miRNAs in the mutant. On the other hand, the expression levels of predicted miR165, miR172, miR393, and miR397b target genes (At4g32880, At2g39250, At1g12820, and At2g38080, respectively) did not change despite reduced miRNA levels (Fig. S4C, Lower). This is possibly due to multiple target genes and/or that the miRNAs may cause translational inhibition rather than transcript cleavage of the target genes.
To further confirm that the mutation in SIC is responsible for the reduction in miRNA levels, miRNA accumulation was examined in the sic-1 complementation transgenic lines. miR159a and miR398a were restored to WT levels in the transgenic lines (Fig. S4 D and E). Although miRNA levels in sic-2 were not as low as in sic-1, they were still lower compared with the WT (Fig. S4 D and E), consistent with sic-2 being a weaker allele.
Differentially expressed genes were identified in sic-1 vs. WT by using whole-genome tiling arrays. Among 4,225 differentially expressed genes, 2,135 were up-regulated, and 2,090 were down-regulated in sic-1 relative to WT (Dataset S1). Five up-regulated and five down-regulated genes were validated by real-time RT-PCR, and the results were consistent with the tiling array data for all 10 genes (Fig. S5 A and B). Furthermore, tiling array analysis showed that several miRNA target genes were up-regulated in sic-1 relative to WT (Dataset S2).
sic-1 Mutation Impairs Intron Decay.
The morphological phenotypes of sic-1 plants were largely similar to those of se-1, abh1, cbp20, smu-1, and sta1 plants. These latter mutants are impaired in not only miRNA biogenesis but also in pre-mRNA processing. Hence, pre-mRNA processing (intron splicing) was examined by tiling array analysis. We found that sic-1 mutant plants retained significantly higher levels of introns than the WT (Dataset S3). Thirty-one percent of the accumulated introns were at the first position (Fig. S6). For verification of tiling array data, the top two genes, CRB (At1g09340, CHLOROPLAST RNA BINDING) and CAX1 (At2g38170, a high affinity vacuolar calcium antiporter), which retained higher levels of introns in sic-1 compared with WT (Dataset S3), were selected for Northern blot analysis. In addition to exon-specific probes, for CRB and CAX1 genes first and second intron-specific probes, respectively, were used. CAX1 was up-regulated in both WT and sic-1 in response to cold stress, but sic-1 accumulated more cleaved introns compared with WT (Fig. 3). Similarly, cleaved introns of CRB also accumulated in sic-1 (Fig. 3). The accumulation of excised introns of CAX1 and CRB were evident in sic-1 mutant without stress, but the accumulation was increased in response to cold stress (Fig. 3).
Fig. 3.
Effect of sic-1 on the accumulation of spliced introns. Accumulation of mRNA and spliced intron of CAX1 (CATION EXCHANGER 1/RARE COLD INDUCIBLE 4) and CRB4 (CHLOROPLAST RNA BINDING) genes was determined by Northern analysis.
SIC Colocalizes with HYL1 in Distinct Subnuclear Bodies.
The above results suggest that SIC is localized at the site of RNA processing (i.e., the nucleus). The SIC-GFP fusion exhibited a nuclear localization in root cells (Fig. 4A). HYL1 has previously been shown to interact and colocalize with DCL1 in distinct nuclear bodies referred to as “dicing bodies” (38–40). We found that SIC protein was concentrated in distinct nuclear bodies, in addition to diffuse distribution in the nucleoplasm (Fig. 4B). Simultaneous immunolocalization of SIC and HYL1 revealed that the two proteins were colocalized in the nuclear bodies (Fig. 4B).
Fig. 4.
Colocalization of SIC and HYL1 in distinct nuclear bodies. (A) Nuclear localization of the SIC-sGFP. Confocal microscope images of root cells of SIC-sGFP transgenic Arabidopsis plants. GFP, GFP fluorescence; FM4-64, fluorescence of FM4-64-stained cells; Merged, GFP+FM4-64 superimposed images. (B) Colocalization on SIC and HYL1 in distinct nuclear bodies. DAPI, DAPI-stained nuclei; HYL1, immunolocalization of HYL1 with anti-HYL antibody; SIC-FLAG, immunolocalization of SIC-FLAG with anti-FLAG antibody; Merged, merged field images of DAPI, HYL1, and SIC-FLAG.
SIC Is Important for Chilling and Salt Stress Tolerance.
In addition to having developmental defects, some mutants involved in miRNA biogenesis and pre-mRNA processing show altered stress responses. For example, hyl1, se, abh1, cbp20, sta1, and emu mutants are hypersensitive to ABA during seed germination (21, 23, 33, 35, 41). sic-1 was less sensitive to ABA than se-1 and abh1-285 during germination and was only slightly more sensitive than WT (Fig. S7 A and B). Interestingly, sic-1 was hypersensitive to chilling stress imposed at the germination stage or seedling stage (Fig. 5 A and B). sic-1 was more sensitive to chilling stress than se-1, abh1-285, or WT (Fig. 5C). The basal freezing tolerance (before cold acclimation) was higher in sic-1 than in WT (Fig. S7C), but the sic-1 mutation did not affect acquired freezing tolerance (after acclimation) (Fig. S7D). In addition, sic-1 and abh1-285 were hypersensitive to 150 mM NaCl, whereas se-1 was less sensitive than sic-1 and abh1-285 (Fig. 5D).
Fig. 5.
Abiotic stress responses of the sic-1 mutant plants. (A) Chilling stress response of WT and sic-1. Seeds were germinated and grown on MS medium supplemented with 3% sucrose at 22 °C for 3 wk (Upper) or at 4 °C for 5 wk (Lower). (B) WT and sic-1 seeds were germinated and grown in soil at 22 °C for 3 wk, then transferred to and grown at 4 °C for 2 mo. Inset: Top view of the sic-1 under cold stress. (C) Chilling stress response of sic-1, se-1, and abh1-285. (D) NaCl stress response of sic-1, se-1, and abh1-285. Six-day-old seedlings were transplanted on to MS medium supplemented with 3% sucrose (Left) or with 3% sucrose with 150 mM NaCl (Right) and grown for 3 wk.
Discussion
The sic-1 mutant was isolated on the basis of enhanced stability of LUCIFERASE transgene in Arabidopsis. SIC (At4g24500) encodes a hydroxyproline-rich protein with unknown biochemical function. SIC loss-of-function mutants showed pleiotropic developmental defects including reduction in plant height, delay in flowering, increase in serration at the leaf margin, and abnormal inflorescence phyllotaxy. Mutations in several miRNA biogenesis factors are known to exhibit both serrated leaf margins and abnormal inflorescence phyllotaxy (22–24, 30, 31). Importantly, sic-1 was deficient in miRNA and ta-siRNA accumulation and accumulated higher levels of unprocessed pri-miRNAs as well as spliced introns from protein-coding genes. miRNA accumulation levels in sic-1 were lower than in WT but intermediate between se-1 and abh1-285. Like hyl1 and hen1 (29, 41), sic-1 displayed dwarfism, although plant height was not reduced in se-1 and abh1-285. These results suggest that compared with the other miRNA biogenesis factors, SIC performs shared as well as specific roles in miRNA biogenesis and development. The colocalization of SIC with HYL1 in distinct nuclear bodies further supports that SIC is involved in miRNA biogenesis.
Expression analysis confirmed that sic-1 has lower levels of miRNAs and ta-siRNAs but enhanced accumulation of pri-miRNAs and higher expression of some predicted miRNA target genes. Tiling array analysis showed that sic-1 plants accumulated high levels of introns for 456 loci. se-1, abh1-285, and cbp20 plants showed a significant overlap: 140 loci were commonly affected (19). Like other miRNA biogenesis mutants, sic-1 accumulated unspliced pri-miRNA, albeit to a lower level than se-1 and abh1, which suggests that SIC is another component involved in miRNA biogenesis. However, in contrast to the other mutants, sic-1 mutants accumulated spliced introns rather than the unspliced pre-mRNAs. The result suggests that the degradation of some RNAs is slower in sic-1. This is consistent with the observation of enhanced stability of LUC transcript in sic-1.
Reduced biogenesis of miRNAs in the hyl1 mutant resulted in higher stability of their target transcripts (42). A previous report showed that a mutation in pre-mRNA splicing in STABILIZED1 (STA1) gene led to enhanced stability of the LUC transcript (33). sta1 also has serrated leaf margins and defects in mRNA splicing (33). Hence, the enhanced stability of the LUC transcript in sic-1 seems to be due to its defects in RNA metabolism, which includes miRNA and ta-siRNA biogenesis as well as spliced intron decay. These results suggest a possible common mechanism for the cleavage of pri-miRNA hairpins and decay of spliced introns. Although a link between miRNA biogenesis and spliced intron decay was not known before, a connection between nonsense-mediated decay of mRNAs and small RNA biogenesis was previously found in the ein5-6 mutant of Arabidopsis, which is defective in the decay of uncapped mRNAs and showed a marginal reduction in miRNAs but an increase in the 21-nt small RNAs processed from transcripts of proteins coding genes. Defects in miRNA biogenesis and abnormal inflorescence phyllotaxy were further enhanced in the ein5-6 abh1-1 double mutants (21). Determining the exact mechanisms of action of SIC in miRNA biogenesis, intron splicing, and decay of spliced introns will require further study.
Besides development, miRNAs also regulate abiotic stress responses of plants through posttranscriptional regulation of gene expression (2, 43, 44). The sic-1 mutation enhanced ABA sensitivity only marginally relative to the se-1 and abh1-285 mutations. Interestingly, only sic-1 showed a dramatic hypersensitivity to chilling. Both sic-1 and abh1-285 were more salt sensitive than se-1 or WT. Although the abundance of stress-regulated miRNAs, such as miR159, miR165, miR168, miR171, miR172, miR393, and miR398 (43, 45, 46), were found to be constitutively low in sic-1, se-1, and abh1-285, only sic-1 is compromised in chilling tolerance, whereas se-1 and abh1-285 were more sensitive to ABA than sic-1. This suggests that although SIC, SE, and ABH1 have shared functions in miRNA biogenesis, SE and ABH1 are more important for ABA responses and SIC-dependent RNA metabolism is more important for chilling tolerance in Arabidopsis. Therefore, the developmental defects and stress sensitivity of sic-1 are caused by not only a deficiency in miRNA biogenesis but also other impairments in RNA metabolism.
Postsplicing intron degradation after endonucleolytic cleavage of the excised lariat is not well understood. In yeast, introns in some pre-mRNAs form hairpin structures, which trigger degradation of unspliced pre-mRNAs and lariat introns through RNaseIII (Rnt1p)-mediated cleavage (47). In Arabidopsis, a chloroplast polynucleotide phosphorylase (cpPNPase) with an RNase-PH domain is required for degradation of the excised lariat. This cpPNPase also has poly(A) polymerase activity, which may affect stability of poly(A) mRNA, because poly(A) mRNA stability is low in chloroplasts (48). A feature common to the pri-miRNA hairpin and the lariat in the spliced intron is the loop structure. SIC may assist RNase III in the cleavage of the loop to open the lariat. Excised introns can also form a stem-loop structure, which can be processed by small RNA biogenesis machinery to produce miRNAs or siRNAs. Approximately 2% and 7% of small RNAs are derived from the intron hairpins in Arabidopsis and rice, respectively (49). We speculate that SIC may bind to the loop structure of the pri-miRNA and lariat, thereby enhancing the cleavage of pri-miRNAs, biogenesis of intron-derived siRNAs, and lariat degradation. It is of interest to test the genetic relationship between SIC and known components involved in miRNA biogenesis, such as SE, ABH1, and DCL1, to understand whether SIC and other known components may work together in the same pathway to regulate miRNA biogenesis.
Materials and Methods
Plant Materials and Mutant Isolation.
The Columbia ecotype (with gl-1 mutation) harboring RD29A promoter-driven LUCIFERASE reporter gene (here after referred to as WT) was mutagenized with ethyl methanesulfonate. The M2 seeds were germinated and screened for putative mutants with high LUC bioluminescence under cold stress using a low-light luminescence imaging system with WinView software (Princeton Instruments) (50). One such mutant with high bioluminescence after cold stress at 0 °C for 24 h was designated sickle-1 (sic-1) and was selected for further study.
RNA Analysis.
Seedlings were grown on MS agar plates supplemented with 3% sucrose in a growth chamber for 10 d. Cold treated (4 °C) or untreated (24 °C) seedlings were used for total RNA extraction with the RNeasy plant mini kit (Qiagen). Northern analysis and the probes used for RD29A, COR15A, LUC, and TUBULIN were described previously (33). For CAX1 (At2g38170) and CRB (At1g09340), the probes from exon and intron region were PCR amplified and confirmed by sequencing.
Accumulation of small RNA was determined by Northern analysis. Total RNA was first extracted from 10-d-old seedlings, rosette leaves, and inflorescence by using TRIzol (Invitrogen). Low-molecular-weight RNA was purified from total RNA, and small RNA Northern analysis was carried out as described previously (51).
Construction of GFP- and FLAG-Tagged Transgenic Plants.
A 3,589-bp amplicon consisting of SIC with its native promoter (without termination codon and 3′ UTR) was PCR amplified from WT genomic DNA by using Platinum Taq (Invitrogen). This PCR product was cloned into pENTR-D-TOPO (Invitrogen). The donor vector with SIC was used for cloning into pEG302 (for FLAG fusion) (52) and pGWB504 (for GFP fusion) (53) by using site-specific recombination with Gateway LR Clonase II enzyme mix (Invitrogen). These constructs were verified by sequencing. The binary construct was then introduced into Agrobacterium strain GV3101 and was used for genetic transformation of the sic-1 plants by the floral dip method. T1 transformants from sic-1 plants transformed with pEG302-SIC-FLAG were selected on BASTA-containing MS medium. The pGEB504-SIC-GFP transformants were selected on the basis of hygromycin resistance. These transgenic lines were further confirmed by PCR.
RT-PCR Analysis of Gene Expression.
Total RNA (2 μg) was reverse-transcribed using oligo-dT primers and the SuperScript III First-Strand Synthesis System (Invitrogen). A 1-μL quantity of the cDNA was used as a template for all of the downstream applications, such as standard PCR reactions for analysis of pri-miRNAs, and real-time PCR analysis of pri-miRNA accumulation was carried out using iQ SYBR Green Supermix (Bio-Rad). Gene-specific oligonucleotides used in PCR analysis are given in Table S1.
Immunolocalization of FLAG-SIC and HLY1 Proteins.
The roots of pGEB504-SIC-GFP transgenic plants were imaged with a Zeiss 510 confocal microscope to localize SIC-GFP fusion protein. SIC and HYL1 localizations were examined by immunostaining as described previously (54). After postfixation, the slides were blocked in KPBS [10× KPBS: 1.28 M NaCl, 20 mM KCl, 80 mM Na2HPO4, and 20 mM KH2PO4 (pH 7.2)] with 1% BSA and 0.1% Triton X at 37 °C for 30 min. The slides were then washed three times (5 min each time) with KPBS containing 0.1% Triton X. Slides were incubated with mouse monoclonal anti-FLAG antibody (1:400; Sigma-Aldrich) or anti-HYL1 antibody (1:500) overnight at 4 °C. The slides were washed three times with KPBS and then blocked with blocking solution for 30 min at 37 °C in a humid chamber. The slides were incubated for 2 h at 37 °C in KPBS with 0.1% Triton containing secondary antibody, anti-mouse-FITC (1:100; Sigma) for FLAG-SIC, and goat anti-rabbit-Alexa 594 (1:100; Invitrogen) for HYL1 X. The slides were then washed in KPBS and mounted in antifade reagent with DAPI (Invitrogen). Stained nuclei were observed with a Leica SP2 confocal microscope.
Supplementary Material
Acknowledgments
We thank Y. Adam Yuan, Department of Biological Sciences, National University of Singapore, Singapore for providing HYL1 antibody, and Avnish Kapoor for assistance with the nuclear run-on analysis. This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 and US Department of Agriculture Grant 2008-35100-04518 (to J.-K.Z) and by the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216199109/-/DCSupplemental.
References
- 1.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhu JK. Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):9851–9852. doi: 10.1073/pnas.0805207105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 5.Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. Regulation and functional specialization of small RNA-target nodes during plant development. Curr Opin Plant Biol. 2009;12(5):622–627. doi: 10.1016/j.pbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19(4):374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Curr Opin Plant Biol. 2009;12(1):81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Chen X. A silencing safeguard: Links between RNA silencing and mRNA processing in Arabidopsis. Dev Cell. 2008;14(6):811–812. doi: 10.1016/j.devcel.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105(29):9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307(5711):932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102(10):3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008a;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery TA, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008b;105(51):20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15(16):1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19(18):2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102(36):12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(25):8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furumizu C, Tsukaya H, Komeda Y. Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA. 2010;16(9):1809–1817. doi: 10.1261/rna.2265710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106(4):477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- 24.Prigge MJ, Wagner DR. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13(6):1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12(17):1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutet S, et al. Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr Biol. 2003;13(10):843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol. 2004;55(5):679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- 28.Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18(10):1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14(4):346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 30.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7(10):1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47(6):841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 32.Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9(11):1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee BH, Kapoor A, Zhu J, Zhu JK. STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell. 2006;18(7):1736–1749. doi: 10.1105/tpc.106.042184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK. HOS1, a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell. 1998;10(7):1151–1161. doi: 10.1105/tpc.10.7.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, et al. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 2008;49(11):1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laubinger S, et al. Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome. Proc Natl Acad Sci USA. 2010;107(41):17466–17473. doi: 10.1073/pnas.1012891107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung T, Wang D, Kim CS, Yadegari R, Larkins BA. Plant SMU-1 and SMU-2 homologues regulate pre-mRNA splicing and multiple aspects of development. Plant Physiol. 2009;151(3):1498–1512. doi: 10.1104/pp.109.141705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17(9):818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujioka Y, Utsumi M, Ohba Y, Watanabe Y. Location of a possible miRNA processing site in SmD3/SmB nuclear bodies in Arabidopsis. Plant Cell Physiol. 2007;48(9):1243–1253. doi: 10.1093/pcp/pcm099. [DOI] [PubMed] [Google Scholar]
- 40.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104(13):5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu C, Fedoroff N. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell. 2000;12(12):2351–2366. doi: 10.1105/tpc.12.12.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101(4):1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips JR, Dalmay T, Bartels D. The role of small RNAs in abiotic stress. FEBS Lett. 2007;581(19):3592–3597. doi: 10.1016/j.febslet.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18(8):2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danin-Kreiselman M, Lee CY, Chanfreau G. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol Cell. 2003;11(5):1279–1289. doi: 10.1016/s1097-2765(03)00137-0. [DOI] [PubMed] [Google Scholar]
- 48.Germain A, et al. Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J. 2011;67(3):381–394. doi: 10.1111/j.1365-313X.2011.04601.x. [DOI] [PubMed] [Google Scholar]
- 49.Chen D, et al. Plant siRNAs from introns mediate DNA methylation of host genes. RNA. 2011;17(6):1012–1024. doi: 10.1261/rna.2589011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chinnusamy V, Stevenson B, Lee BH, Zhu JK. Screening for gene regulation mutants by bioluminescence imaging. Sci STKE. 2002;2002(140):pl10. doi: 10.1126/stke.2002.140.pl10. [DOI] [PubMed] [Google Scholar]
- 51.Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26(6):1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45(4):616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 53.Nakagawa T, et al. Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71(8):2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 54.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126(1):79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.