Abstract
Despite the high abundance of Archaea in the global ocean, their metabolism and biogeochemical roles remain largely unresolved. We investigated the population dynamics and metabolic activity of Thaumarchaeota in polar environments, where these microorganisms are particularly abundant and exhibit seasonal growth. Thaumarchaeota were more abundant in deep Arctic and Antarctic waters and grew throughout the winter at surface and deeper Arctic halocline waters. However, in situ single-cell activity measurements revealed a low activity of this group in the uptake of both leucine and bicarbonate (<5% Thaumarchaeota cells active), which is inconsistent with known heterotrophic and autotrophic thaumarchaeal lifestyles. These results suggested the existence of alternative sources of carbon and energy. Our analysis of an environmental metagenome from the Arctic winter revealed that Thaumarchaeota had pathways for ammonia oxidation and, unexpectedly, an abundance of genes involved in urea transport and degradation. Quantitative PCR analysis confirmed that most polar Thaumarchaeota had the potential to oxidize ammonia, and a large fraction of them had urease genes, enabling the use of urea to fuel nitrification. Thaumarchaeota from Arctic deep waters had a higher abundance of urease genes than those near the surface suggesting genetic differences between closely related archaeal populations. In situ measurements of urea uptake and concentration in Arctic waters showed that small-sized prokaryotes incorporated the carbon from urea, and the availability of urea was often higher than that of ammonium. Therefore, the degradation of urea may be a relevant pathway for Thaumarchaeota and other microorganisms exposed to the low-energy conditions of dark polar waters.
Keywords: amoA, ureC, Beaufort Sea, Ross Sea, Amundsen Sea
The realization that Archaea were not strict extremophiles but widespread in the environment has become one of the most exciting findings in the recent history of microbial ecology. Since the discovery of a new group of mesophilic archaea, classified as the phylum Thaumarchaeota (1), that prevail in soils, oceans, and freshwater systems (2–4), the unveiling of their biogeochemical role in the environment has remained a challenge (5–7). In the oceans, Thaumarchaeota are very abundant (globally approximately 20% of prokaryotic cells) (8), likely influencing the oceanic biogeochemistry through contributions to the carbon and nitrogen cycles. However, the extreme difficulty in culturing representatives of this phylum has hampered elucidation of their metabolic traits.
The fact that the single planktonic marine Thaumarchaeota cultured to date (Nitrosopumilus maritimus SCM1) is a strict autotrophic ammonia oxidizer (9), and the reports on the abundance of genes encoding archaeal ammonia monooxygenases (amoA) in oceanic waters (4, 10), have led to the belief that marine Thaumarchaeota are predominantly nitrifiers. Indeed, the genetic potential for ammonia oxidation is a common feature of the other two marine Thaumarchaeota with sequenced genomes: Candidatus “Cenarchaeum symbiosum” (11) and Candidatus “Nitrosoarchaeum limnia SFB1” (12). However, experimental data from oceanic samples suggests that marine Thaumarchaeota are metabolically diverse, hinting at heterotrophic or possibly mixotrophic lifestyles (13, 14).
Consistent with the potential for heterotrophy, early single-cell activity measurements showed that the Marine Group I (MGI) Archaea cluster, which is the dominant thaumarchaeal group in marine waters, can incorporate organic compounds such as amino acids (15). Those initial results were confirmed in large-scale samplings across the Atlantic Ocean (13, 16, 17). However, other studies have shown that some MGI Archaea fix carbon autotrophically (18, 19), presumably linked to ammonia oxidation (9, 10, 20), or have provided evidence for mixed autotrophic and heterotrophic metabolisms (14, 21). Although the contribution of MGI Archaea to prokaryotic production and dark CO2 fixation appears to be significant in the global ocean (13), their actual contribution to nitrification has not been resolved yet (22, 23).
Here we focused on the metabolism of marine Thaumarchaeota in polar environments, where these microorganisms are very abundant and exhibit seasonal growth (24–26). Although knowledge on the diversity of polar archaea is rapidly increasing (27–29), their in situ metabolic activities remain virtually unexplored. Two previous studies in Arctic waters obtained contradictory results, reporting high archaeal uptake of organic compounds during summer in the Chukchi Sea (30) while year-round heterotrophic activity was low in the Beaufort Sea (26). Archaeal amoA genes have been detected in Arctic and Antarctic waters (28), but in the Southern Ocean there are no reports of archaeal in situ activities. Hence, the main sources of energy for growth of polar Thaumarchaeota remain unknown. Here, we combined in situ single-cell activity measurements, quantitative PCR (qPCR), and metagenomic analyses to shed light on the metabolism of these enigmatic, uncultivated polar microorganisms.
Results and Discussion
Dynamics of Polar Thaumarchaeota.
We analyzed the dynamics of archaeal abundance along the winter-to-summer transition in surface and halocline waters of the Southeast Beaufort Sea, Western Arctic. Additionally, archaeal abundance was estimated from depth profiles taken in summer in the Ross and Amundsen Seas, Antarctica (SI Appendix, Fig. S1). The water column in the Southeast Beaufort Sea is perennially stratified, with low-salinity Pacific waters forming a halocline between the surface Polar Mixed Layer and deeper warmer Atlantic waters. This oceanic area is oligotrophic (31), and during the sampling period, there were seasonal fluctuations in surface light availability (from 0.01 to 3.29 μmol quanta m−2 s−1), ice cover, and temperature (from −1.7 to −0.4 °C; SI Appendix, Table S1). Chl a values were generally low (<0.05 μg/L during the winter; SI Appendix, Fig. S2). The Southern Ocean sampling included a regional trophic gradient with stations located in open waters or under total ice cover in summer (SI Appendix, Table S1). Surface Chl a ranged from 0.2 to 10.4 μg/L in the Eastern Amundsen Sea and from 0.3 to 8.4 μg/L in the Ross Sea. Surface water temperature ranged from −0.21 to −1.70 °C. Different oceanographic water masses within the depth profiles were analyzed including Antarctic Surface Waters, Thermocline, deep Shelf Waters, and Circumpolar Deep Waters (CDW) to have a wide representation of Antarctic Thaumarchaeota from different habitats.
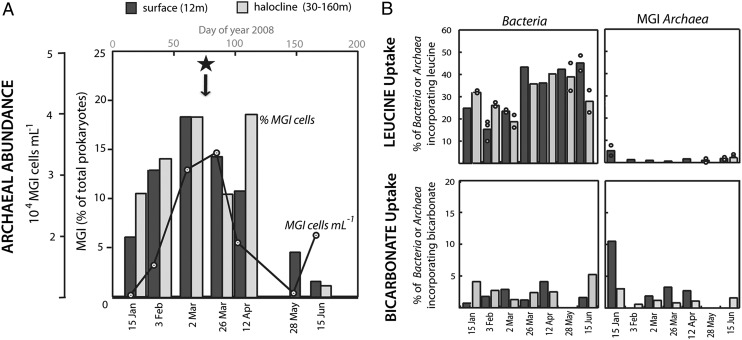
Our results in the Arctic confirmed previous reports of increases in the proportion of Thaumarchaeota in winter polar surface waters (25, 26, 32). The abundance of MGI Archaea in the Southeast Beaufort Sea increased from 6% of prokaryotic cells in January 2008 to 18% in March 2008, as analyzed by catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH; Fig. 1A). Because the winter water column remained stratified (33), our results indicate that the increase of MGI Archaea was due to continuous growth of these populations in situ, and not to mixing with deeper water masses, as hypothesized for Antarctic waters (28, 34). Interestingly, we detected a similar temporal trend in deeper samples from the halocline (Fig. 1A), where MGI Archaea contributed higher abundances (up to 24% of prokaryotes; SI Appendix, Fig. S3). In contrast to surface waters, the abundance of MGI Archaea in this deeper layer remained high until April (18% of prokaryotes; Fig. 1A).
Fig. 1.
Marine Group I (MGI) archaeal in situ abundance and metabolic activity in Arctic waters as analyzed by CARD-FISH and MAR-FISH. (A) Abundance of MGI Archaea expressed as percentage of total prokaryotes (bars) and number of cells in surface waters (line). The star symbol indicates the date at which the Arctic metagenome was retrieved. (B) Single-cell activity of Bacteria and MGI Archaea in the uptake of leucine (Upper) and bicarbonate (Lower). Individual dots represent replicate measurements carried out in some samples. Dark and light gray bars represent samples collected in Arctic surface and halocline waters, respectively.
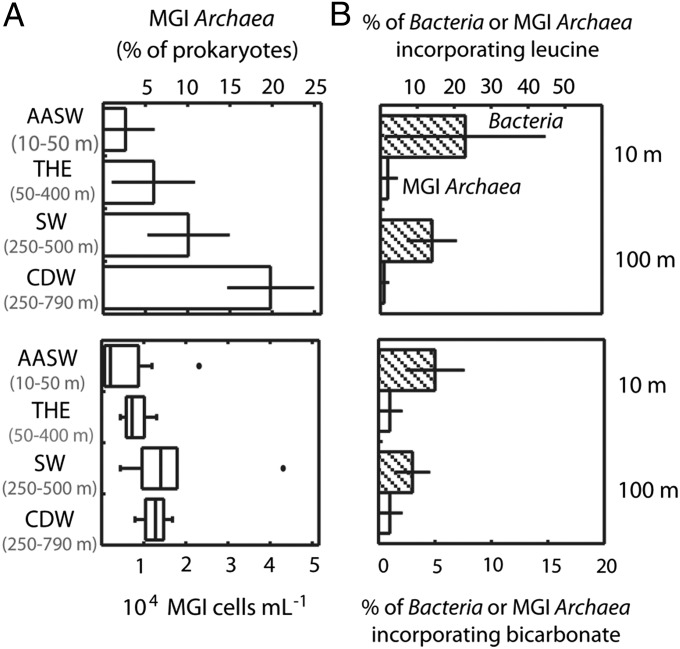
In Arctic surface waters, the abundance of MGI Archaea rapidly declined once light became detectable beneath the sea ice (SI Appendix, Fig. S2), consistent with the idea that Thaumarchaeota experience photoinhibition (35). By May and June, MGI Archaea contributed less than 5% of cells in Arctic surface waters. Similarly, the archaeal abundance in summer Antarctic surface waters was low (< 4% of prokaryotes; Fig. 2A). However, Thaumarchaeota were abundant in deep Shelf Waters and CDW, contributing on average 10% and 19% of total cells, respectively. Thus, our data confirm that Thaumarchaeota are an abundant and dynamic component of polar marine microbial communities by showing growth at both surface and halocline Arctic waters during the winter.
Fig. 2.
MGI archaeal in situ abundance and metabolic activity in Antarctic waters as analyzed by CARD-FISH and MAR-FISH. (A) Abundance of MGI Archaea in waters of the Ross and Amundsen seas expressed as percentage of total prokaryotes (bars in Upper) and number of cells (box plots in Lower). Data in Upper are shown as averages ± SD, and n ranges from 4 to 18. (B) Single-cell activity of Bacteria (hatched bars) and MGI Archaea (open bars) in the uptake of leucine (Upper) and bicarbonate (Lower). n ranges from 4 to 5. AASW, Antarctic Surface Waters; CDW, Circumpolar Deep Waters; SW, Shelf Waters; THE, thermocline.
Single-Cell Metabolic Activity of Polar MGI Thaumarchaeota.
Microautoradiography combined with FISH (MAR-FISH) was used to analyze the metabolism and activity levels of polar MGI Archaea. Bicarbonate and leucine were used as proxies for autotrophic and heterotrophic metabolisms, respectively (13). Throughout the seasons, less than 2% of Arctic MGI archaeal cells took up leucine (Fig. 1B). In contrast, Bacteria increased their heterotrophic activity from winter to spring and summer (i.e., from 25 to 45% of Bacteria actively took up leucine). A similar pattern was found in Antarctic summer samples of the Ross Sea at 10- and 100-m depths, with less than 3% of MGI Archaea and more than 15% of bacterial cells incorporating leucine (Fig. 2B).
The low archaeal heterotrophic activity is consistent with results from a previous seasonal study of surface waters at a close location (Franklin Bay, Beaufort Sea), where other labile organic substrates were also tested (26). However, in summer surface waters of the Arctic Chukchi Sea, MGI Archaea were highly active in the uptake of a variety of organic compounds (30), suggesting regional differences in the metabolism of Arctic Thaumarchaeota, possibly related to differences in the composition of the archaeal communities inhabiting these two Arctic regions (27). It should be noted that the Chukchi Sea is an exceptionally nutrient-rich area and one of the most productive oceanic regions in the world (36). Therefore, we believe that our finding that polar Thaumarchaeota do not contribute significantly to the uptake of labile monomers such as leucine is more representative for the low to moderate productivity conditions found in most polar marine waters.
Reports of archaeal amoA genes in polar environments (28), and recent evidence of active nitrification in Arctic winter waters (31, 37), suggest that polar Thaumarchaeota may be nitrifiers. To test whether polar Thaumarchaeota were growing autotrophically, we measured their activity in bicarbonate uptake. With the exception of a sample collected in surface waters in January, a low proportion of Arctic Bacteria or MGI Archaea incorporated bicarbonate (<5% of active cells) even during winter (Fig. 1B). Similarly, low archaeal bicarbonate uptake was found in Antarctic waters down to 100 m depth (Fig. 2B). Interestingly, low percentages of MGI Archaea active in bicarbonate uptake were also generally found in summer waters of the Chukchi Sea (30). Overall, despite detecting an active growth of MGI Archaea in winter Arctic waters, our in situ measurements revealed paradoxically low autotrophic activities and low incorporation of leucine for this group, pointing to alternative sources for obtaining carbon and energy to sustain growth. To identify those sources and to obtain an unbiased overview of the functional potential of Thaumarchaeotai, we analyzed a winter Arctic metagenome.
Metabolic Capacity of Polar Thaumarchaeota Analyzed via Metagenomics.
The sample for metagenomic analysis was collected under the ice during the period of highest archaeal abundance (March), when Thaumarchaeota made up 18% of total prokaryotic cells near the surface and in the halocline (Fig. 1A). Accordingly, reference genome mapping of the reads revealed that Thaumarchaeota comprised 14% of the total Arctic microbial community (SI Appendix, Fig. S4). The metagenome was collected at 65-m depth in halocline waters, where a peak in nitrite was found, and it was hypothesized that the potential for nitrification was highest (SI Appendix, Fig. S5).
Analysis of thaumarchaeal 16S rRNA genes in the metagenome revealed that the assemblage was represented by closely related genotypes affiliated with the cluster MGI-α, which also comprises the nitrifiers N. maritimus and Ca. N. limnia (SI Appendix, Fig. S4). Most (70%) of the thaumarchaeal phylotypes shared >98% similarity among their 16S rRNA genes and belonged to a cluster that included metagenomic samples retrieved from Antarctic winter waters (38, 39), and several dominant phylotypes from Arctic and Antarctic water masses (SI Appendix, Fig. S4). These results indicate that a single closely related clade of MGI Archaea dominates Thaumarchaeota throughout the seasons in both Arctic and Antarctic waters. At the genomic level, this clade has only 84% and 86% average nucleotide similarity with N. maritimus and Ca. N. limnia, respectively, indicating that it affiliates with hitherto unknown archaeal species (40).
From a total of 338,714 protein coding genes found in the Arctic metagenome, 5,091 were identified in contigs or reads affiliated with Thaumarchaeota (i.e., the Arctic thaumarchaeal metagenome). Thirty-two percent of these thaumarchaeal genes could be functionally annotated and assigned to orthologous groups (OGs) via eggNOG (SI Appendix). Transcriptional regulators and thioredoxins were among the most abundant thaumarchaeal OGs (SI Appendix, Table S2), consistent with the genome analysis of N. maritimus (41). Ammonia permeases (COG0004) and the enzyme glutamine synthetase (COG0174) were also well represented, suggesting the importance of nitrogen metabolism for Arctic Thaumarchaeota.
In the context of carbon and energy sources, we found a 6.5-kb contig containing the complete operon encoding the ammonia monooxygenase with 96% amino acid identities with N. maritimus (SI Appendix, Fig. S6). In total, the abundance of amo genes in the thaumarchaeal metagenome was high (i.e., 17, 19, and 12 copies of amoA, amoB, and amoC genes, respectively). As a reference, the thaumarchaeal metagenome had an average (±SD) of 18 (±7) copies of single-copy prokaryotic essential genes (42). Therefore, we can conclude that a majority of the metagenomic thaumarchaeal population had the ability to oxidize ammonia.
Comparative Thaumarchaeal Metagenomic Analysis.
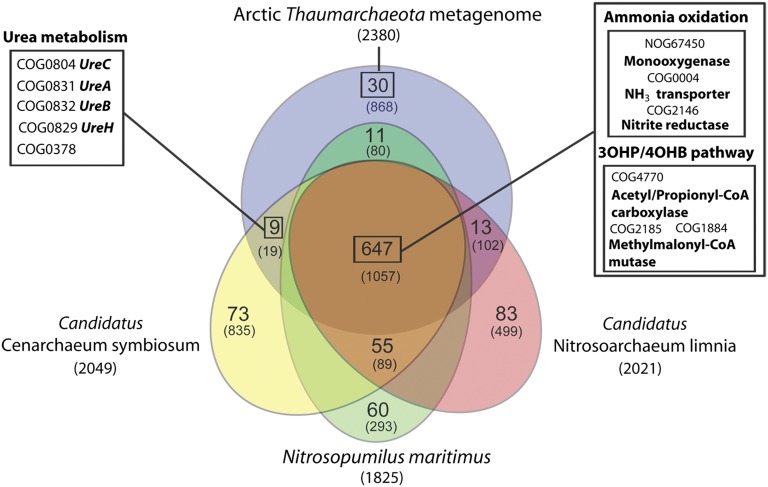
A genomic comparison between the Arctic thaumarchaeal metagenome and the three published MGI archaeal genomes (N. maritimus, Ca. C. symbiosum, and Ca. N. limnia) was carried out to identify common metabolic pathways and potentially unique metabolic features of polar Thaumarchaeota (Fig. 3). We found a functional core of 647 OGs shared by all four thaumarchaeal phylotypes that included genes involved in several central metabolic pathways (SI Appendix, Fig. S7), ammonia oxidation, and the main carbon fixation pathway of Thaumarchaeaota, i.e., the 3-Hydroxypropionate-4-hydroxybutyrate pathway (Fig. 3) (43). Among the 868 nonredundant gene families found exclusively in the Arctic thaumarchaeal metagenome, only a few could be functionally annotated (i.e., 30 OGs). Although some of these genes were involved in widespread processes such as translation or transcription, a key enzyme in fatty acids biosynthesis, which is an unusual pathway in the Archaea domain, was also detected in low abundance (the 3-oxoacyl-acyl-carrier-protein synthase; SI Appendix, Table S3).
Fig. 3.
Comparative genomic analysis between the Arctic thaumarchaeal metagenome and the three sequenced MGI Archaea to date. Nitrosopumilus maritimus SCM1, Candidatus N. limnia SFB1, and Candidatus C. symbiosum A were used to build OGs specific to Thaumarchaeota and, thereafter, the genes identified in Arctic Thaumarchaeota metagenome were assigned to these OGs. The numbers in parentheses represent the total number of OGs for the genomes or regions in the Venn. The large numbers in each region of the Venn represent OGs with functional annotation from eggNOG v2. OGs that did not occur in the Arctic thaumarchaeal metagenome and were shared among only two other genomes are hidden. Some OGs of interest have been highlighted in boxes. 3OHP/4OHB, 3 hydroxyproprionate/4 Hydroxybutyrate.
Arctic Thaumarchaeota also shared some OGs exclusively with each of the MGI archaeal isolates. Ca. C. symbiosum was the genotype that shared the fewest number of OGs with Arctic Thaumarchaeota but, remarkably, most of them were involved in degradation of urea (i.e., ureases, the accessory protein ureH and a GTPase involved in regulation of the expression and maturation of ureases, COG0378; Fig. 3). Thaumarchaeal ureA, ureB, and ureC genes were relatively abundant in the Arctic metagenome (i.e., 8, 7, and 12 gene copies, respectively), and genes encoding urea transporters highly similar (80–90% amino acid identities) to the transporter present in Ca. C. symbiosum (CENSYa_0457) were also found. The presence of these genes implies that urea degradation could be a key pathway used by Thaumarchaeota to obtain carbon and energy in polar environments.
Quantification of amoA and ureC Gene Abundances in Arctic and Antarctic Waters.
The prevalence of amoA and ureC genes in polar Thaumarchaeota was analyzed by qPCR. The copy numbers of archaeal amoA and 16S rRNA genes were highly correlated, and the ratio between these genes was on average 3 and 1 in Arctic and Antarctic water samples, respectively (Fig. 4 and SI Appendix, Tables S4 and S5), in agreement with previous reports from the Chukchi Sea (37) and off the Antarctic Peninsula (28). Our results thus indicate that a genetic potential for ammonia oxidation is widespread in polar MGI Archaea.
Fig. 4.
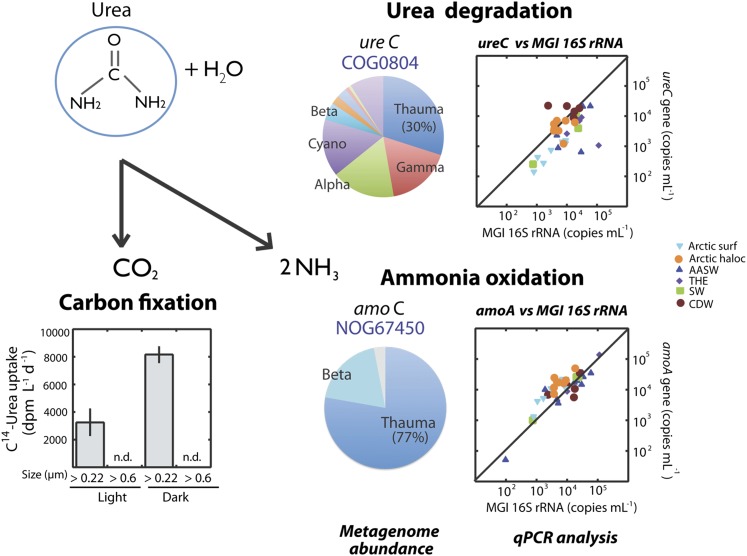
Proposed pathway used by Arctic Thaumarchaeota to obtain carbon and energy from the degradation of urea. The pie charts show the phylogenetic affiliation of reads assigned to the orthologous groups COG0804 (urease) and NOG67450 (ammonia monooxygenase) within the Arctic metagenome. Plots at Right show the abundance of ureC genes (upper plot) and amoA genes (lower plot) versus the abundance of MGI Archaea 16S rRNA genes in Arctic and Antarctic waters, as analyzed by qPCR. Two samples from AASW did not show detectable amplification of ureC genes and were not included in the upper plot. Uptake measurements of 14C-labeled urea in surface Arctic samples of different size fractions (>0.2 and >0.6 μm) under light and dark conditions are shown on the bottom left side. AASW, Antarctic Surface Waters; CDW, Circumpolar Deep Waters; haloc, halocline; n.d, nondetected; surf, surface; SW, Shelf Waters; THE, thermocline.
To analyze the abundance of ureC genes, we optimized a qPCR assay targeting polar Thaumarchaeota ureC genes. Full-length sequences were initially obtained by amplifying several Arctic and Antarctic samples with primers previously designed for marine thaumarchaeal ureC genes (44). Remarkably, most ureC genes retrieved from polar waters formed a tight cluster with more than 90% nucleotide identity. This polar cluster showed only approximately 75% nucleotide similarity with archaeal ureC genes previously found in surface and deep waters from other marine environments (44) (SI Appendix, Fig. S8). The qPCR analysis revealed that ureC genes were abundant in polar Thaumarchaeota, particularly in deep water layers such as the Arctic halocline or Antarctic CDW, where the average ratio of ureC versus MGI 16S rRNA genes was close to or greater than 1 (Fig. 4). This ratio was lower in Arctic surface samples (i.e., average ratio of 0.2), suggesting genetic differences between archaeal populations inhabiting surface and halocline waters. In Antarctic waters, the average ratio between ureC and MGI 16S rRNA genes was 2.7 in deeper CDW, and approximately 0.3 in other water masses. Altogether, these results indicate that the potential for urea degradation is widespread in polar Thaumarchaeota.
Relevance of Urea Degradation for Polar Thaumarchaeota.
Relatively high concentrations of urea have been found in polar seawater and sea ice (45–49), with the principal sources being microbial metabolism of purines (50) or nitrogenous waste excretion by zooplankton and other metazoans (45). Urea is recognized as a significant source of nitrogen for polar phytoplankton (47, 48, 51), but its degradation by polar prokaryotes has received limited attention, although diverse marine microorganisms have the potential to use this compound (52). In an experiment carried out in Arctic winter surface waters, we found detectable urea utilization in the prokaryotic size fraction, but not in the size fraction corresponding to phytoplankton (i.e., >0.6 μm; Fig. 4), supporting the hypothesis that urea is relevant for the metabolism of polar prokaryotes thriving at low energy conditions.
Within Thaumarchaeota, the first report of ureases was in the obligate symbiont Ca. C. symbiosum, which may use the waste-product urea from its host, a marine sponge, to fuel ammonia oxidation (11). However, neither of the nitrifiers N. maritimus (41) or Ca. N. limnia (12) have ureases, showing that the potential to degrade urea is not ubiquitous among marine Thaumarchaeota. Indeed, ureases are rare in the Archaea domain (53). Nevertheless, we show that ureases are widespread in polar Thaumarchaeota and, interestingly, in the Arctic metagenome, Thaumarchaeota was the phylum with the highest abundance of urease genes (Fig. 4).
The potential use of urea to fuel nitrification has been reported for ammonia oxidizing bacteria (54) and, recently, for a soil nitrifying Thaumarchaeota isolate (55). Because synthesizing ureases and urea transporters requires energy, the utilization of urea would be advantageous only when the availability of ammonium were low or intermittent. Although urea is often assumed to be present at a lower concentration than ammonium in marine systems (53), we found the opposite pattern in summer samples collected across the Arctic Beaufort Sea. The availability of urea-N was often higher than ammonium and, remarkably, at many depths where ammonium was below detection levels, urea was still detected (SI Appendix, Fig. S9). Similarly, urea was often found in higher concentration than ammonium in samples collected in summer in the eastern Canadian Arctic (47).
Unfortunately, no winter measurements of ammonium are available from our study; however, this substrate was below the detection limit (<10 nM) in early spring measurements carried out on the second and seventh of April 2008. These results suggest that virtually no ammonium was available for the later part of the winter when Thaumarchaeota were still abundant, at least in deeper halocline waters (Fig. 1). In contrast, urea was present in several Arctic winter profiles, and the average (±SD) concentration (70 ± 80 nM) was similar to that found in summer (July, 60 ± 30 nM; SI Appendix, Fig. S9). Comparable results were obtained in Arctic waters at the end of March (46), and in spring Antarctic samples below the ice (56), where ammonium was below detection and urea concentration and uptake rates were relatively high. In the only station with total ice cover during our Antarctic sampling, the concentration of ammonium was also below detection limit (SI Appendix, Table S1). These observations suggest that urea is a more stable source of energy for ammonia oxidizers in low productive polar systems than ammonium. During polar winter, the input of photosynthetically produced organic matter is halted, but urea can be continuously supplied by other microorganisms and zooplankton (45). The ability to use urea would therefore provide an ecological advantage to nitrifying MGI Archaea.
We made estimates to determine whether the availability of urea would be enough to sustain the observed winter Arctic archaeal growth. The molar ratio of biomass produced per unit of ammonia molecule oxidized (C/N) in Thaumarchaeota was estimated 0.046 based on the growth of N. maritimus (9), using a biomass factor of 12 fg of C per cell (57). The net growth rate of polar MGI Archaea over the winter was similar in surface and halocline waters (≈0.21 d−1) and comparable to the maximal growth rate reported for N. maritimus (0.72 d−1) (9). Using a C/N ratio of 0.046, a consumption rate of 4.8 and 3.9 nM urea/d would be required to sustain the winter growth of MGI Archaea (from January to March) in surface and halocline waters, respectively. Assuming an average winter concentration of urea of 70 nM, the turnover time of urea would be ≈16 d, which is consistent with previous estimates from Arctic waters (47). These calculations indicate that the availability of urea in Arctic waters could support the winter archaeal growth observed, although additional energy would be required to synthesize ureases and urea transporters. We found oligopeptide transporters (COG4608) and different catabolic pathways in the thaumarchaeal metagenome (SI Appendix, Fig. S7). These findings suggest that polar MGI Archaea, in addition to ammonia, may exploit some organic compounds to supplement their growth, as found for the thaumarchaeon Nitrososphaera viennensis (55) or some ammonia oxidizing bacteria (58). Interestingly, urea serves as a source of nitrogen and also carbon for certain ammonia oxidizing bacteria in soil (59, 60). The detectable in situ incorporation of 14C-labeled urea shows that the carbon of urea was assimilated by polar prokaryotes (Fig. 4). The possibility that Thaumarchaeota were actively incorporating the carbon from urea could explain the low activity in bicarbonate uptake detected in situ.
Role for Urea Metabolism in Marine Thaumarchaeota.
Based on the prevalence of ureC and amoA genes in polar Thaumarchaeota, we hypothesize that urea degradation may be a relevant pathway by which these populations meet their carbon and energy demands. Most of the few previous reports of marine thaumarchaeal ureases were carried out in deep Mediterranean and Pacific waters (44, 61) (SI Appendix, Fig. S8), where Thaumarchaeota also thrive under low energy conditions and urea is often present in high concentrations. In Pacific mesopelagic waters, high rates of urea degradation were measured (62). Although at the time of the latter study it was unknown that Thaumarchaeota were dominant members of the mesopelagic (8), it is remarkable that the degradation of urea was paralleled by high nitrification rates (62). Recently, archaeal ureases have been found in both a metagenome and a metatranscriptome of mesopelagic Pacific waters, indicating that ureases are expressed by Thaumarchaeota in deep oceanic waters (63).
A predominance of chemoautotrophic pathways in winter polar waters and the global dark ocean is now accepted (39), but the potential relevance of urea in this scenario has not been recognized. Recent studies suggest that the supply of ammonium is insufficient to fuel marine autotrophic microorganisms and hint at the use of alternative energy sources (22). Given the high abundance of Thaumarchaeota in polar and deep oceanic waters, and their genomic potential to use urea, the contribution of this substrate to the metabolic demands of these and other microorganisms living in low energy environments needs to be revised.
Materials and Methods
Water samples for qPCR, CARD-FISH, and MAR-FISH analyses were collected near the surface and in deeper halocline waters from the Arctic Southeast Beaufort Sea during the overwintering CFL cruise and in the upper 500 m from the Antarctic Amundsen and Ross seas during the summer in the Oden Southern Ocean cruise (SI Appendix, Fig. S1). Samples were collected by using a ship-deployed rosette for open water sampling and through the ship’s internal moon pool for winter ice-covered sampling. All materials and methods are described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank the captains and crew of the icebreakers Oden and CCGS Amundsen and the Swedish Research Polar secretariat for logistic support; Dan Nguyen, Roxane Maranger, and other members of the Circumpolar Flaw Lead (CFL) cruise for collecting samples; A. Niemi, B. Philippe, C. J. Mundy, A. Sallon, C. Michel, and M. Gosselin for chlorophyll data; J. Gagnon for nutrient data; P. Guillot and Y. Gratton for conductivity–temperature–depth (CTD) data; and the European Molecular Biology Laboratory Information Technology core facility and Y. Yuan for managing the high-performance computing resources. 454 pyrosequencing was supported by the K&A Wallemberg foundation. This work is a contribution to the International Polar Year–CFL system study (2007/2008) supported through grants from the Canadian International Polar Year Federal Program Office and the Natural Sciences and Engineering Research Council (NSERC) Canada. Spanish participation was funded by the Spanish Ministry of Science and Innovation Grant Boreal CLG2007-28872-E/ANT (to C.P.-A), and laboratory work was supported by the Swedish Research Council (to S.B.). L.A.-S. was supported by Marie Curie Fellowship PIEFGA-2008-221121 and a “Juan de la Cierva” contract from the Spanish Ministry of Science and Innovation. We acknowledge support by the National Science Foundation Antarctic Organisms and Ecosystems Program Grants ANT-0741409 and ANT-08361440 (to K.B. and P.Y.); Swedish Research Council Environment, Agricultural Sciences, and Spatial Planning Grant 217-2006-342 (to L.R and H.F.); and NSERC of Canada (A.S.W., C.L., and J-É.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 17732.
Data deposition: The metagenomic data have been deposited in the European Bioinformatics Institute (EBI) database (accession no. ERP001178); and the sequences reported in this paper have been deposited in the GenBank database (accession nos. JX512003–JX512021).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201914109/-/DCSupplemental.
References
- 1.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 2.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 4.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 6.DeLong EF. Archaeal mysteries of the deep revealed. Proc Natl Acad Sci USA. 2006;103:6417–6418. doi: 10.1073/pnas.0602079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brochier-Armanet C, Gribaldo S, Forterre P. Spotlight on the Thaumarchaeota. ISME J. 2012;6:227–230. doi: 10.1038/ismej.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 9.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 10.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 11.Hallam SJ, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herndl GJ, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingalls AE, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and Archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teira E, Van Aken H, Veth C, Herndl G. Archaeal uptake of enantiomeric amino acids in the meso-and bathypelagic waters of the North Atlantic. Limnol Oceanogr. 2006;51(1):60–69. [Google Scholar]
- 18.Pearson A, McNichol A, Benitez-Nelson B, Hayes J, Eglinton T. Origins of lipid biomarkers in Santa Monica Basin surface sediment: A case study using compound-specific Δ14C analysis. Geochim Cosmochim Acta. 2001;65:3123–3137. [Google Scholar]
- 19.Wuchter C, Schouten S, Boschker HTS, Sinninghe Damsté JS. Bicarbonate uptake by marine Crenarchaeota. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 20.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansman RL, et al. The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc Natl Acad Sci USA. 2009;106:6513–6518. doi: 10.1073/pnas.0810871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinthaler T, van Aken HM, Herndl GJ. Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic’s interior. Deep Sea Res Part II Top Stud Oceanogr. 2010;57:1572–1580. [Google Scholar]
- 23.Varela MM, van Aken HM, Sintes E, Reinthaler T, Herndl GJ. Contribution of Crenarchaeota and Bacteria to autotrophy in the North Atlantic interior. Environ Microbiol. 2011;13:1524–1533. doi: 10.1111/j.1462-2920.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 24.DeLong EF, Wu KY, Prézelin BB, Jovine RV. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 25.Murray AE, et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso-Sáez L, Sánchez O, Gasol JM, Balagué V, Pedrós-Alio C. Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ Microbiol. 2008;10:2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x. [DOI] [PubMed] [Google Scholar]
- 27.Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- 28.Kalanetra KM, Bano N, Hollibaugh JT. Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ Microbiol. 2009;11:2434–2445. doi: 10.1111/j.1462-2920.2009.01974.x. [DOI] [PubMed] [Google Scholar]
- 29.Alonso-Sáez L, Andersson A, Heinrich F, Bertilsson S. High archaeal diversity in Antarctic circumpolar deep waters. Environ Microbiol Reports. 2011;3:689–697. doi: 10.1111/j.1758-2229.2011.00282.x. [DOI] [PubMed] [Google Scholar]
- 30.Kirchman D, Elifantz H, Dittel A, Malmstrom R, Cottrell M. Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnol Oceanogr. 2007;52:495–507. [Google Scholar]
- 31.Tremblay J-É, et al. Vertical stability and the annual dynamics of nutrients and chlorophyll fluorescence in the coastal, southeast Beaufort Sea. J Geophys Res. 2008;113:C0S790. [Google Scholar]
- 32.Church M, et al. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr. 2003;48:1893–1902. [Google Scholar]
- 33.Forest A, et al. Biogenic carbon flows through the planktonic food web of the Amundsen Gulf (Arctic Ocean): A synthesis of field measurements and inverse modeling analyses. Prog Oceanogr. 2011;91:410–436. [Google Scholar]
- 34.Williams TJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012;6:1883–1900. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merbt SN, et al. Differential photoinhibition of bacterial and archaeal ammonia oxidation. FEMS Microbiol Lett. 2012;327(1):41–46. doi: 10.1111/j.1574-6968.2011.02457.x. [DOI] [PubMed] [Google Scholar]
- 36.Sambrotto RN, Goering JJ, McRoy CP. Large yearly production of phytoplankton in the Western bering strait. Science. 1984;225:1147–1150. doi: 10.1126/science.225.4667.1147. [DOI] [PubMed] [Google Scholar]
- 37.Christman GD, Cottrell MT, Popp BN, Gier E, Kirchman DL. Abundance, diversity, and activity of ammonia-oxidizing prokaryotes in the coastal Arctic ocean in summer and winter. Appl Environ Microbiol. 2011;77:2026–2034. doi: 10.1128/AEM.01907-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Béjà O, et al. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl Environ Microbiol. 2002;68:335–345. doi: 10.1128/AEM.68.1.335-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grzymski JJ, et al. A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters. ISME J. 2012;6:1901–1915. doi: 10.1038/ismej.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker CB, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 43.Berg IA, Kockelkorn D, Buckel W, Fuchs G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science. 2007;318:1782–1786. doi: 10.1126/science.1149976. [DOI] [PubMed] [Google Scholar]
- 44.Yakimov MM, et al. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea) ISME J. 2011;5:945–961. doi: 10.1038/ismej.2010.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conover R, Gustavson K. Sources of urea in arctic seas: Zooplankton metabolism. Mar Ecol Prog Ser. 1999;179:41–54. [Google Scholar]
- 46.Conover R, Mumm N, Bruecker P, MacKenzie S. Sources of urea in arctic seas: Seasonal fast ice? Mar Ecol Prog Ser. 1999;179:55–69. [Google Scholar]
- 47.Harrison W, Head E, Conover R, Longhurst A, Sameoto D. The distribution and metabolism of urea in the eastern Canadian Arctic. Deep Sea Research Part A. 1985;32:23–42. [Google Scholar]
- 48.Waldron HN, Attwood CG, Probyn TA, Lucas MI. Nitrogen dynamics in the Bellingshausen Sea during the Austral spring of 1992. Deep Sea Res Part II Top Stud Oceanogr. 1995;42:1253–1276. [Google Scholar]
- 49.Simpson KG, Tremblay J-É, Gratton Y, Price NM. An annual study of inorganic and organic nitrogen and phosphorus and silicic acid in the southeastern Beaufort Sea. J Geophys Res. 2008;113:C07016. [Google Scholar]
- 50.Vogels GD, Van der Drift C. Degradation of purines and pyrimidines by microorganisms. Bacteriol Rev. 1976;40:403–468. doi: 10.1128/br.40.2.403-468.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansell DA, Goering JJ. Pelagic nitrogen flux in the northern Bering Sea. Cont Shelf Res. 1990;10:501–519. [Google Scholar]
- 52.Collier JL, Baker KM, Bell SL. Diversity of urea-degrading microorganisms in open-ocean and estuarine planktonic communities. Environ Microbiol. 2009;11:3118–3131. doi: 10.1111/j.1462-2920.2009.02016.x. [DOI] [PubMed] [Google Scholar]
- 53.Solomon C, Collier J, Berg G, Glibert P. Role of urea in microbial metabolism in aquatic systems: A biochemical and molecular review. Aquat Microb Ecol. 2010;59:67–88. [Google Scholar]
- 54.Koper TE, El-Sheikh AF, Norton JM, Klotz MG. Urease-encoding genes in ammonia-oxidizing bacteria. Appl Environ Microbiol. 2004;70:2342–2348. doi: 10.1128/AEM.70.4.2342-2348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bury S, Owens N, Preston T. 13C and 15N uptake by phytoplankton in the marginal ice zone of the Bellingshausen Sea. Deep Sea Res Part II Top Stud Oceanogr. 1995;42:1225–1252. [Google Scholar]
- 57.Fukuda R, Ogawa H, Nagata T, Koike I. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl Environ Microbiol. 1998;64:3352–3358. doi: 10.1128/aem.64.9.3352-3358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arp DJ, Chain PSG, Klotz MG. The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu Rev Microbiol. 2007;61:503–528. doi: 10.1146/annurev.micro.61.080706.093449. [DOI] [PubMed] [Google Scholar]
- 59.Burton SA, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marsh KL, Sims GK, Mulvaney RL. Availability of urea to autotrophic ammonia-oxidizing bacteria as related to the fate of 14C- and 15N-labeled urea added to soil. Biol Fertil Soils. 2005;42:137–145. [Google Scholar]
- 61.Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho B, Azam F. Urea decomposition by bacteria in the Southern California Bight and its implications for the mesopelagic nitrogen cycle. Mar Ecol Prog Ser. 1995;122:21–26. [Google Scholar]
- 63.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.