Abstract
Sodium-coupled substrate transport plays a central role in many biological processes. However, despite knowledge of the structures of several sodium-coupled transporters, the location of the sodium-binding site(s) often remains unclear. Several of these structures have the five transmembrane-helix inverted-topology repeat, LeuT-like (FIRL) fold, whose pseudosymmetry has been proposed to facilitate the alternating-access mechanism required for transport. Here, we provide biophysical, biochemical, and computational evidence for the location of the two cation-binding sites in the sodium-coupled betaine symporter BetP. A recent X-ray structure of BetP in a sodium-bound closed state revealed that one of these sites, equivalent to the Na2 site in related transporters, is located between transmembrane helices 1 and 8 of the FIRL-fold; here, we confirm the location of this site by other means. Based on the pseudosymmetry of this fold, we hypothesized that the second site is located between the equivalent helices 6 and 3. Molecular dynamics simulations of the closed-state structure suggest this second sodium site involves two threonine sidechains and a backbone carbonyl from helix 3, a phenylalanine from helix 6, and a water molecule. Mutating the residues proposed to form the two binding sites increased the apparent Km and Kd for sodium, as measured by betaine uptake, tryptophan fluorescence, and 22Na+ binding, and also diminished the transient currents measured in proteoliposomes using solid supported membrane-based electrophysiology. Taken together, these results provide strong evidence for the identity of the residues forming the sodium-binding sites in BetP.
Keywords: secondary transport, symmetry, membrane protein, alkali metal ion, osmoregulation
Secondary transporters constitute an important class of membrane proteins that use the free energy stored in ion-concentration gradients to drive the transport of various molecules across membranes (1, 2). In recent years a number of X-ray crystallographic structures of secondary transporters have been reported, providing compelling support for the long-standing alternating-access hypothesis (reviewed in ref. 3). According to this hypothesis, transporters must undergo conformational changes that alternately expose a substrate-binding site from one side and then from the other side of the membrane but never both at the same time.
The structural data have revealed that several transporters previously assigned to different families based on sequence homology do, in fact, share common folds. One such group of protein structures includes, to date, seven different transporters originating from five different sequence families, namely, BetP and CaiT from the betaine/carnitine/choline (BCC) transporter family (4–7); AdiC and ApcT from the amino acid/polyamine/organocation (APC) transporter family (8, 9); Mhp1 from the nucleobase/cation symporter-1 (NCS1) family (10, 11); LeuT from the neurotransmitter/sodium symporter (NSS) family (12); and vSGLT from the solute/sodium symporter (SSS) family (13). All these proteins possess similar 3D structures—the five transmembrane-helix inverted-topology repeat, LeuT-like (FIRL) fold (also known as the “LeuT fold”)—and recently have been classified together in the APC superfamily (www.tcdb.org/superfamily.php).
The FIRL-fold is characterized by an internal twofold pseudosymmetry, with an axis running parallel to the membrane plane through the center of the transporter relating the first five transmembrane (TM) helices to the second set of five TM helices, so that their TM topologies are inverted with respect to one another (12, 14). Because some of these transporters also contain additional TM helices either preceding or following the 5-TM repeat (N- or C-terminal helices, respectively), for simplicity of comparison we introduce a numbering scheme for the repeated helices, namely A1–5 for TM helices of the first repeat and B1–5 for helices belonging to the second repeat (Fig. S1).
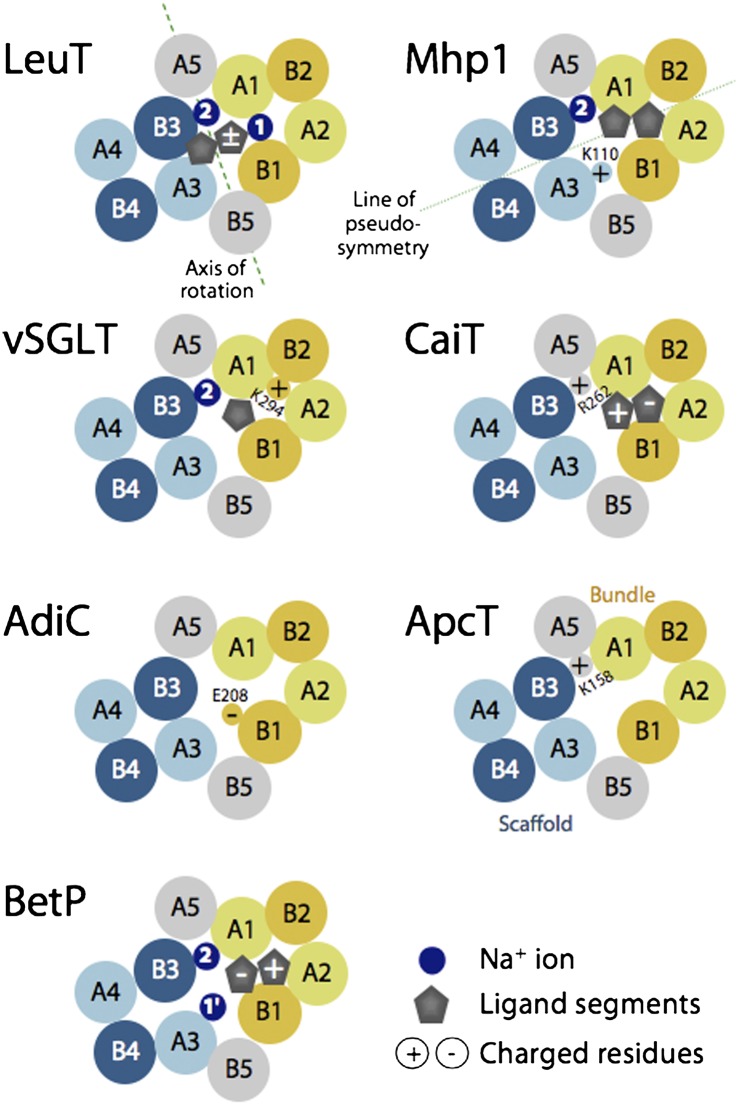
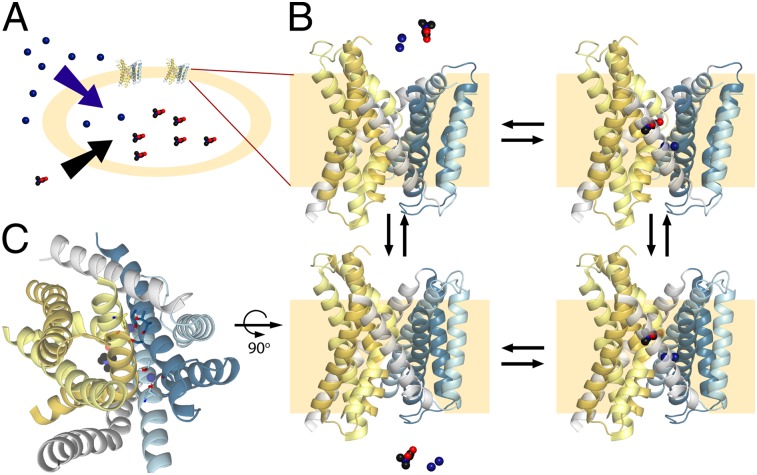
Discrete structural elements within the FIRL-fold have been identified, including a four-helix bundle consisting of the first two helices of each repeat (A1–2 and B1–2) (Fig. 1) and the so-called “scaffold,” consisting of the remaining three helices of each repeat (A3–5 and B3–5) (Fig. 1), which contains within it a smaller element called the “hash domain” (A3–4 and B3–4) (Fig. 1). It has been proposed that the conformational changes required for the alternating-access mechanism involve relative movements of the bundle with respect to elements of the scaffold by the rocking-bundle mechanism (11, 15–17). Other gating-like mechanisms involving local conformational changes (i.e., at the level of individual TM helices) also have been put forward (18, 19), and recent studies suggest mechanisms that combine elements of both types (7, 20).
Fig. 1.
Schematics of helix packing in FIRL-fold transporters of known structure, viewed from the periplasm at a slice through the membrane plane roughly midway across the membrane. The approximate locations of ligands (gray symbols), known or proposed sodium-binding sites (dark blue circles), and selected positively or negatively charged amino acids (circles containing a + or − sign, respectively) are shown. A putative axis of rotation (dashed green line) and the axis of pseudosymmetry (dotted green line) are shown. Bundle helices (A1–2 and B1–2) are colored yellow; scaffold helices (A3–5, and B3–5) are colored blue (for the hash domain) or gray (for the arms). Helices in the first repeat are colored lighter shades. See Fig. S1 for the helix numbering in each family.
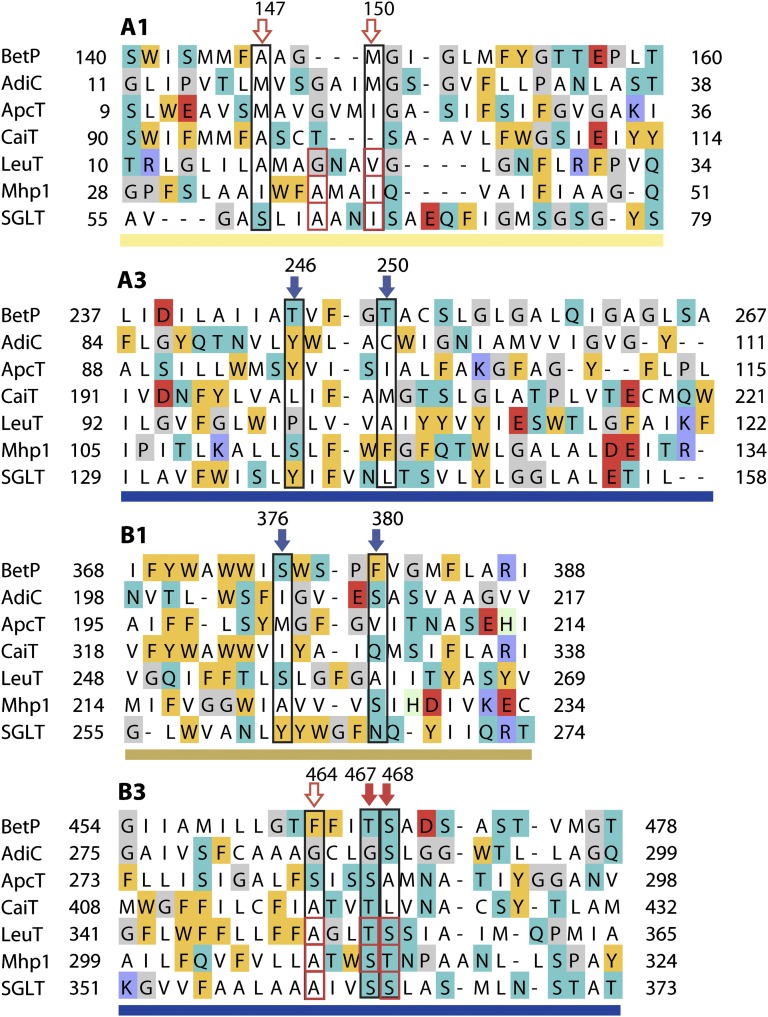
Many FIRL-fold secondary transporters couple substrate transport to sodium flux. Binding sites for those sodium ions have been identified in a few of the FIRL-fold secondary transporters of known structure, namely LeuT, Mhp1, and vSGLT (Fig. 1). In LeuT, the structural and biochemical data indicate that one of the two sodium ion sites, designated “Na1,” is located within the four-helix bundle and is coordinated directly by the carboxylate moiety of the bound substrate molecule and that the second site, designated “Na2,” is formed by two hydroxylic side chains from helix B3, namely T354 and S355, and three carbonyl oxygen atoms from residues G20, V23, and A351 in helices A1 and B3 (Fig. 2) (12, 21). These two sites are occupied in two states of LeuT open to the outside (12, 20), consistent with the notion that sodium binds preferentially to, and presumably stabilizes, outward-facing conformations of the FIRL-fold, as shown by spectroscopic measurements of the effect of sodium on LeuT (18, 19, 22) and by molecular dynamics (MD) simulations of LeuT in the absence of the ion at Na2, during which the cytoplasmic pathway begins to open (23, 24).
Fig. 2.
Structure alignment of regions involved in sodium binding in FIRL-fold transporters, i.e., helices A1, A3, B1, and B3, highlighting positions potentially forming the Na1′ site in BetP (blue arrows, black boxes) and the Na2 site in BetP (red arrows, black boxes). Filled arrows indicate side-chain interactions; open arrows indicate backbone interactions in BetP. Residues forming the Na2 binding site in other transporters are outlined with red boxes. Background colors indicate amino-acid types as follows: negatively charged (red), positively charged (blue), other polar (cyan), aromatic (gold), helix-breaker (gray), and histidines (pale green). Bars under each segment are colored according to Fig. 1.
In a structure of Mhp1 in an outward-facing occluded state, a single positive peak in the electron density difference map was observed at a position structurally equivalent to Na2 of LeuT, where it is surrounded by carbonyl oxygen atoms from A38 and I41 in helix A1 and A309 in helix B3 plus side-chain hydroxyl atoms from S312 and T313 in helix B3 (Fig. 2) (10). A sodium ion was assigned to this density and remained bound in 100-ns MD simulations of this outward-occluded state (11), similar to the behavior of Na2 in simulations of LeuT (25–27). In a structure of the inward-facing state, in contrast, helix B3 is ∼4 Å further away from A1, so that the sodium-binding site appears to be no longer intact (11); indeed, in multiple MD simulations of the inward-facing state, sodium did not remain bound to this site longer than 2 ns (11), as is consistent with the observations discussed above for LeuT.
In structures of vSGLT, which also are inward facing, no suitable densities for sodium ions have been observed (13, 28), although a site was proposed by analogy to Na2 in LeuT (Fig. 1). The proposed coordination involves a hydroxylic side chain from helix B3 (residue S365) and backbone oxygen atoms from helices B3 and A1 (residues A361 from B3 and A62 and I65 from A1) (Fig. 2). Binding to this site is affected by mutagenesis of S365 in vSGLT to alanine and of analogous residues in other SSS proteins (29–31). Consistent with the observations for Mhp1, MD simulations of these inward-facing conformations of vSGLT show that ions are released quickly from the proposed binding site (28, 32–35).
Several of the known FIRL-fold structures are of sodium-independent transporters. For example, the arginine-agmatine antiporter AdiC is independent of sodium, and in its outward-facing structure the side chain of S289 in helix B3 hydrogen-bonds directly to the backbone carbonyl of G21 in helix A1 (8). Interestingly, in other cases the sodium ions have been replaced by basic side chains. In the proton-coupled amino acid transporter ApcT there is a lysine residue (K158) whose amino group is located at the position structurally equivalent to Na2 in LeuT; its amine interacts with the backbone carbonyl of G19 and the side-chain hydroxyl oxygen of S283, from helices A1 and B3, respectively (Figs. 1 and 2) (9). Protonation and deprotonation of K158 were proposed to play the same role as binding of Na2 in LeuT (9), indicating a common mechanistic principle, which was supported by subsequent MD simulations (23). Similarly, in the carnitine:γ-butyrobetaine antiporter CaiT, which also is neither sodium- nor proton-dependent, an arginine residue (R262) located at a position equivalent to Na2 bridges T100 in A1 and T421 in B3 (Figs. 1 and 2) (5).
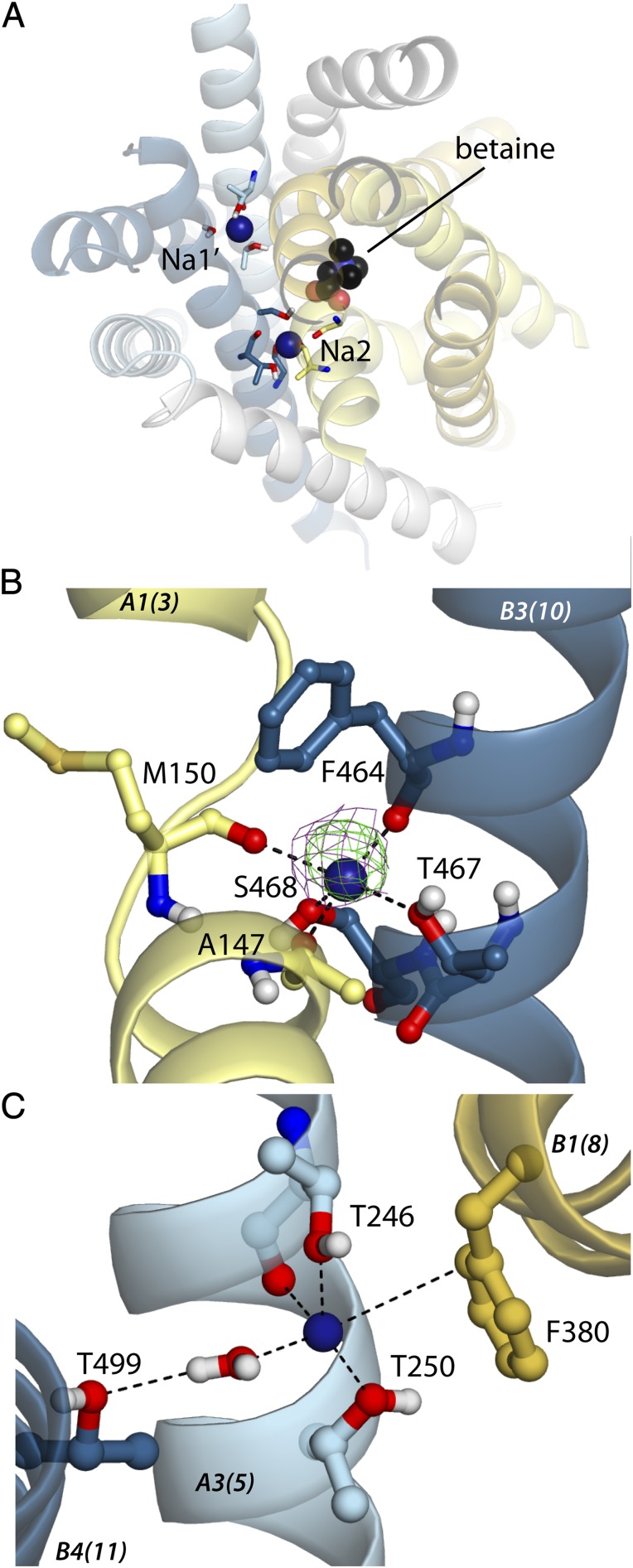
Structural data for the trimeric sodium-dependent betaine symporter BetP have been reported at 3.35-Å resolution in inward-occluded (4) and inward-open conformations (36) and very recently at 3.25- and 3.1-Å resolution in a number of other states, including a substrate-free outward-facing state and a substrate-bound closed state (7). The functional transporting unit of BetP is the protomer, whereas the trimer is a prerequisite for osmoregulation of transport activity (37). Each protomer contains a separate substrate pathway and is expected to bind one betaine molecule along with two sodium ions, reflecting the stoichiometry of substrate transport (4, 38, 39). No density for sodium ions was identified in the earlier structures of BetP (4, 36), presumably because of the moderate resolution or their conformations, or both. However, putative sodium ion-binding sites, which we here denote “pNa1” and “pNa2” (Fig. S2 and Table S1), were tentatively proposed (4) for BetP by comparison with the two sites Na1 and Na2 identified in LeuT; the primary assumption was that one cation would be coordinated directly by the betaine carboxylate group and by a nearby carbonyl group from the unwound segment of helix A1, as found in Na1 of LeuT. However, because BetP has fewer potential coordinating groups in that region, the pNa1 location would require two carbonyl groups from helix A1 (A148 and M150), compared with the one (A22) required for Na1 in LeuT. Therefore these two carbonyl groups were excluded as possible ligands of the second ion, which then was placed at a distance from pNa1 similar to the distance of Na2 from Na1. Thus, pNa2 would involve the side chains of S306 and M310 from helix A5 and the backbone carbonyl of A147 in helix A1 and, unlike Na2 in LeuT, would not directly involve residues from helix B3. In effect, the hypothetical pNa1 and pNa2 sites in BetP (Fig. S2) would be shifted by ∼5–10 Å from their equivalents in LeuT (Fig. 1). However, structural data for the more recent substrate-bound closed state of BetP (7) provide no evidence for the pNa2 and pNa1 sites. Instead, a positive peak in the difference density map is observed at the site equivalent to Na2 of LeuT (see Fig. 3B), and although it is difficult to distinguish between a sodium ion and a water molecule at this modest (3.1-Å) resolution, the conserved pentacoordinate oxygen coordination indicates that this site also is a sodium site in BetP. Unfortunately, however, no other equivalent density was observed that might be assigned to a second ion.
Fig. 3.
Sodium-binding sites in BetP predicted from structural analysis and MD simulations. (A) Simulation snapshot of BetP in a closed state, viewed from the cytoplasm and colored according to Fig. 1. For clarity, only the 10 helices forming the core fold are shown. Bound substrate and sodium ions at Na1′ and Na2 are shown as spheres; coordinating residues and a bound water molecule are shown as sticks. (B and C) Close-up views of simulation snapshots. (B) Simulation snapshot of the Na2 site overlaid with the Fo-Fc difference density map before placing the ion (contoured at 3.0σ, green) and the 2Fo-Fc map (contoured at 1.0σ, purple) for PDB ID 4AIN (7). (C) Simulation snapshot of the Na1′ site. Helices are labeled in italics, according to Fig. S1, with BetP numbering in parentheses.
In this work we first assess the two sodium-binding sites initially proposed for BetP by Ressl et al. (4) using MD simulations and biochemical measurements, which together help rule out ion binding to those locations. Next, we provide supporting evidence that the location of a positive peak in the difference density map for the closed structure of BetP reported by Perez et al. (7) is indeed a sodium-binding site, namely Na2. We then identify a second site inspired by the inverted-topology repeats of the FIRL-fold, at the symmetry-equivalent position of Na2, which we call “Na1′.” We examine and cross-validate these sites using MD simulations of the closed conformation (7). Strong experimental support for these sites is provided by biochemical, biophysical, and electrophysiological measurements. The results show the value of considering the pseudosymmetry in transporter structures and provide important insights into the molecular mechanism of transport by BetP and likely also by other sodium-coupled FIRL-fold transporters.
Results
MD Simulations Suggest Previously Proposed Sites Do Not Bind Sodium.
To assess whether the proposed sites pNa1 and pNa2 from Ressl et al. (4) (Fig. S2 and Materials and Methods) are suitable for binding sodium, we first performed MD simulations of structures of BetP trimers from Ressl et al. (4), embedded into hydrated palmitoyl oleoyl phosphatidylglycerol (POPG) bilayers. The structures of the protomers within the trimers were stable over the course of the simulations, as measured by the rmsd of the Cα atoms in the TM helices from the crystal structure, which were in the range 1.2–1.5 Å (± 0.1 Å), averaged over each simulation (37). It should be noted that although the structure used for these simulations appears to be sterically occluded with respect to the substrate betaine, it nevertheless contains a narrow pathway that is accessible to water molecules from the cytoplasm (4). Consistent with this observation, our MD simulations revealed water molecules solvating the substrates in the binding pocket from the intracellular side of the protein.
In all three simulations and in all protomers, the sodium ions placed at pNa2 diffused away from the protein and into the bulk solution essentially immediately after the constraints imposed on the position of the ions during equilibration were removed (Fig. S3). The sodium ion placed adjacent to the substrate at pNa1 also was poorly coordinated in the simulations (Fig. S4) and in some cases diffused away to the cytoplasmic side.
Mutagenesis at pNa2 Has No Effect on the Sodium Dependence of Uptake.
The MD simulations suggested that neither pNa1 nor pNa2 is a bona fide sodium site in the inward-occluded crystal structure, consistent with their low coordination number and long side chain-to-sodium distances (Fig. S2B) and with the absence of densities at these sites in structures of other states (7). Nevertheless, the simulation results could be attributed to the fact that the structure used has an inward-facing conformation to which ion binding appears to be weak for other FIRL transporters (11, 17, 28, 32–34). Therefore we also measured the apparent sodium affinity of BetP after site-directed mutagenesis of the side chains proposed to coordinate the ions directly, i.e., S306 and M310 from pNa2 (Table S1). Note that no protein side chains would coordinate the ion at pNa1 (Table S1), and therefore this site could not be tested by the same approach. Alanine substitutions at positions in pNa2 did not affect the apparent Km values for sodium (Table 1 and Fig. S5), consistent with the MD simulations. Thus, together with the recent structural data (7), these results suggest that these residues are not involved in the transport of sodium ions and that the sodium-binding sites differ from those proposed in Ressl et al. (4).
Table 1.
Kinetic parameters for apparent sodium affinity measured from betaine uptake of WT BetP and mutants
| Site* | Apparent Km (mM) | Vmax (nmol⋅min−1 ⋅ mg−1 cdw) | |
| WT | 3.8 ± 0.9 | 82.9 ± 4.8 | |
| S306A | pNa2 | 4.3 ± 0.8 | 86.0 ± 3.9 |
| M310A | pNa2 | 11.2 ± 2.0 | 106.4 ± 4.5 |
| T467A | Na2 | 76.4 ± 11.3 | 115.3 ± 8.3 |
| S468A | Na2 | 111.2 ± 14.7 | 91.5 ± 9.2 |
| T467A/S468A | Na2 | BD | BD |
| S409A | – | 3.0 ± 0.8 | 40.6 ± 2.7 |
| S376A | – | 30.6 ± 7.1 | 87.1 ± 7.6 |
| T246L | Na1′ | 40.2 ± 5.4 | 40.8 ± 2.5 |
| T250A | Na1′ | 180.7 ± 21.0 | 114.2 ± 20.8 |
| T246L/T250A | Na1′ | BD | BD |
| F380A | Na1′ | 94.8 ± 18.8 | 52.2 ± 4.1 |
BD, below detection; cdw, cell dry weight.
*The binding site to which this residue is believed to contribute. pNa2 is a site proposed in Ressl et al. (4); Na2 is the site common to other FIRL-fold transporters; Na1′ is a site proposed here for BetP. –, residues that were tested when searching for the Na1′ site (see Table S1 for more details).
MD Simulations Support the Presence of a Sodium Ion at Na2.
Having ruled out the pNa1 and pNa2 sites, we then looked for alternative possible binding sites for the ions in BetP. As mentioned above, in crystals of the BetP trimer that diffracted to 3.1-Å resolution (7), one protomer (chain B) was found to be in a closed, betaine-bound conformation to which we also expect sodium to be bound (see Fig. S6A). Analysis of the Fo-Fc difference density map of this protomer revealed a distinct positive peak between helices B3 and A1 (ref. 7, shown also in Fig. 3B). A sodium ion at this location is coordinated by two carbonyl oxygen atoms from residues in helix A1 (A147 and M150, i.e., one residue each from pNa1 and pNa2) and three oxygen atoms from helix B3, namely the backbone carbonyl of F464 and the side-chain hydroxyl oxygen atoms of T467 and S468 (7) (Figs. 2 and 3B and Table S1). This site is equivalent to the Na2 site, which binds sodium ions in three other sodium-coupled transporters (LeuT, Mhp1, and vSGLT) using two hydroxylic residues from helix B3 and backbone carbonyl oxygens in helix A1 (Figs. 1 and 2).
To cross-validate the coordination of a sodium ion at the Na2-binding site in the closed structure, we also carried out MD simulations of this protomer structure embedded in a hydrated POPG lipid bilayer (Materials and Methods). The protein backbone in these simulations again was stable, with the rmsd for the Cα atoms of the TM domains in the range of 0.9–1.5 Å (± 0.2 Å) averaged over each of six replica simulations. Importantly, the sodium-coordination network in the Na2 site described above was stable on the simulation timescale, both in terms of the coordination number and oxygen-ion distances (Fig. S7A), consistent with the notion that sodium is bound to this site in BetP as it is in LeuT, Mhp1, and vSGLT.
Mutations in the Conserved Na2-Binding Site Affect Sodium Binding.
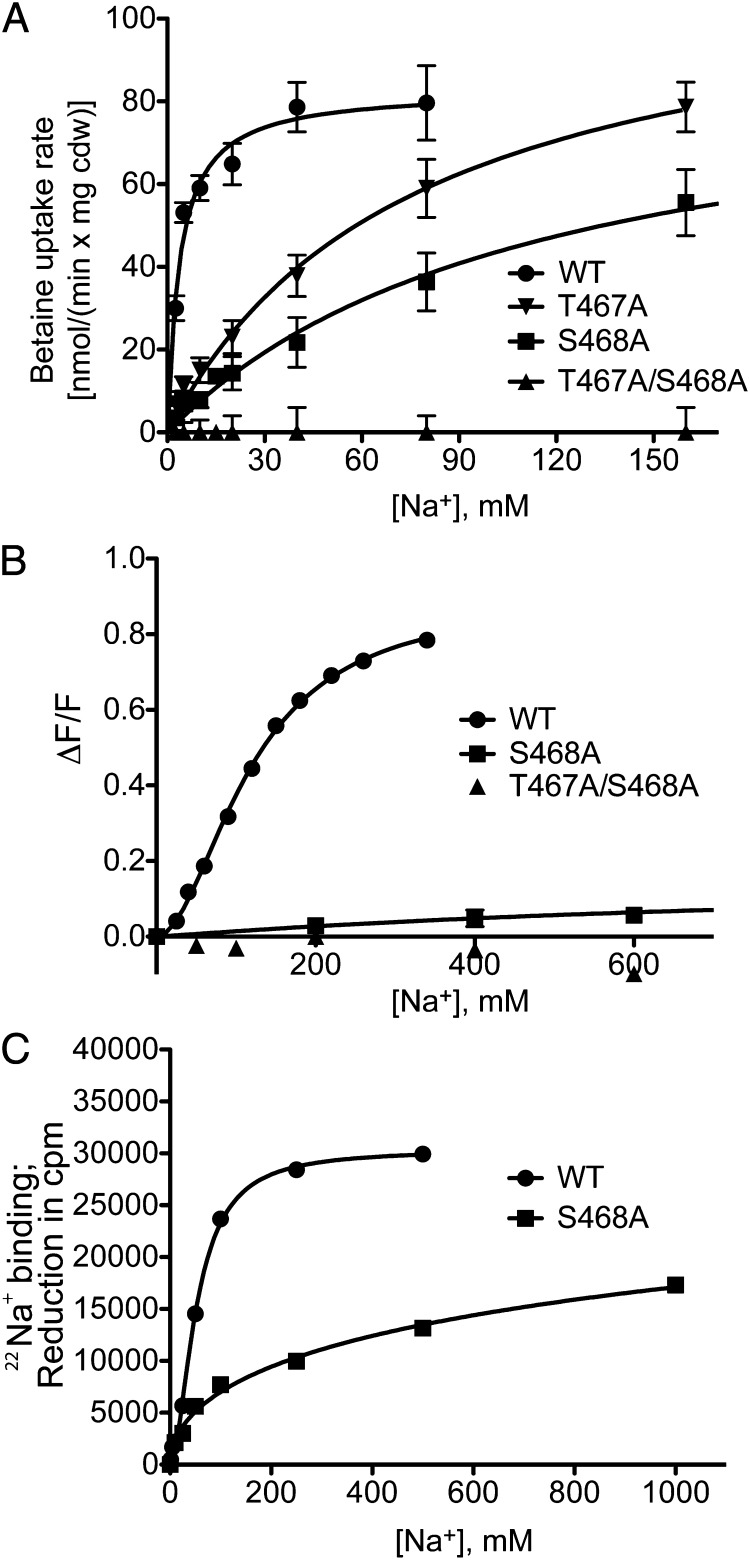
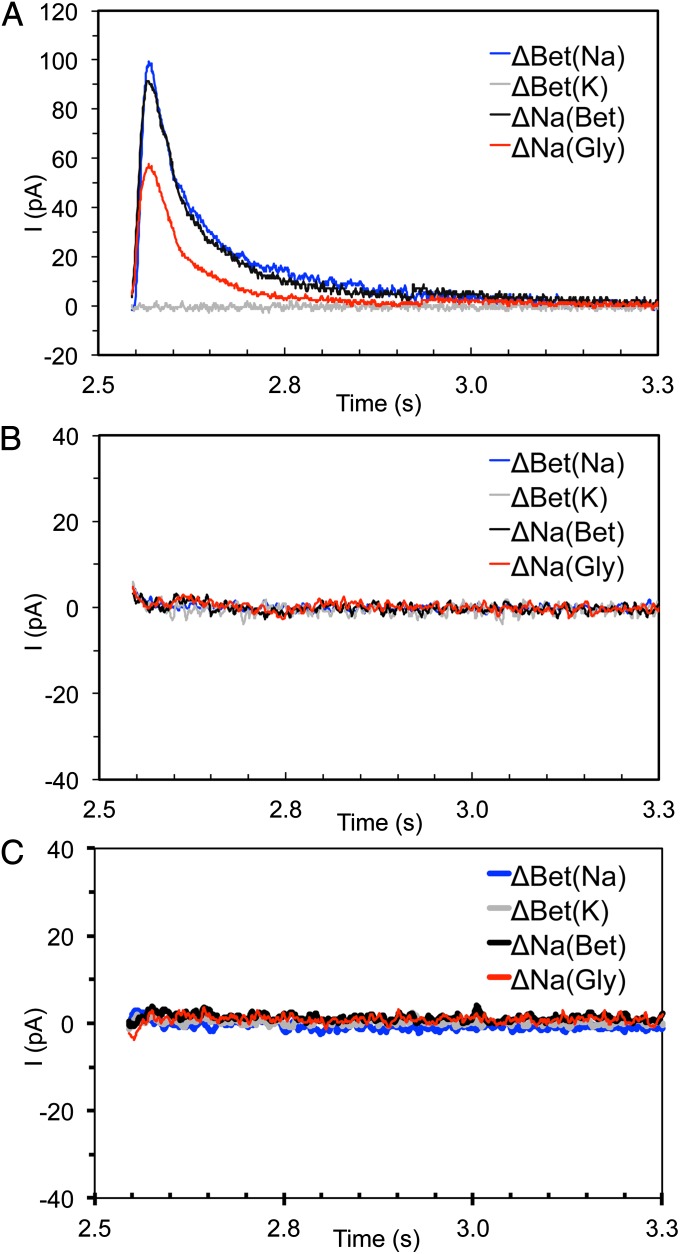
To test the Na2 site further, we next performed single and double substitutions in BetP in which T467 and S468 were replaced by alanine. Uptake assays in Escherichia coli MKH13 cells, tryptophan (Trp) fluorescence in BetP-containing proteoliposomes, and binding of 22Na+ reveal that single-alanine substitutions of T467 and S468 significantly increase the apparent Km and Kd of BetP for sodium (Tables 1 and 2 and Fig. 4). In addition, the double-alanine mutant T467A/S468A shows no uptake into cells or any binding activity as measured by Trp fluorescence and by the scintillation proximity assay (SPA). Solid supported membrane (SSM)-based electrophysiology using proteoliposomes could not detect transient currents corresponding to charge displacement with this double mutant, no matter whether it was activated by betaine [ΔBet(Na)] or by sodium [ΔNa(Bet)] (Fig. 5B). This finding is in stark contrast to that of WT BetP, for which we observed a marked charge displacement triggered by sodium, even in the absence of the substrate betaine [ΔNa(Gly)] (Fig. 5A), with a peak maximum of 57.8 ± 12.5 pA. No charge displacement was triggered by betaine in the absence of sodium [ΔBet(K)]. Together, the conservation analysis (Fig. 2) and the crystallographic (Fig. 3B) (7), simulation (Fig. S7A), and biochemical (Fig. 4) data provide very strong support that a sodium-binding site at Na2 also is present in BetP (Fig. 3B).
Table 2.
Kinetic parameters of Trp fluorescence and 22Na+ binding on WT BetP and mutants
| Trp Fluorescence |
22Na binding |
|||||
| Site | Apparent Kd (mM) | Bmax (ΔF/F) | n (Hill coefficient) | Apparent Kd (mM) | n (Hill coefficient) | |
| WT | 118 ± 3.9 | 0.89 ± 0.02 | 1.9 ± 0.1 | 53.3 ± 2.1 | 2.0 ± 0.2 | |
| S468A | Na2 | 871 ± 113† | 0.16 ± 0.05 | 1.1 ± 0.3 | 147.5 ± 19† | 1.0 ± 0.1 |
| T467A/S468A | Na2 | BD | BD | BD | BD | BD |
| T246L | Na1′ | 305 ± 28 | 0.36 ± 0.02 | 1.9 ± 0.2 | 167.7 ± 15.5 | 1.9 ± 0.2 |
| T246L/T250A | Na1′ | 986 ± 335† | 0.07 ± 0.03 | 2.4 ± 0.8 | 554.8 ± 35.8† | 1.4 ± 0.1 |
| S376A | — | 191 ± 17 | 0.72 ± 0.05 | 1.9 ± 0.2 | 97.7 ± 5.8 | 1.8 ± 0.2 |
| W101A/T351A | ‡ | 153.4 ± 23 | 0.12 ± 0.01 | 2.0 ± 0.4 | 59.2 ± 1.5 | 1.9 ± 0.1 |
BD, below detection.
†Saturation not reached, precluding reliable fitting of the data.
‡Mutations in the trimer interface.
Fig. 4.
Sodium dependence of BetP for mutations introduced at the Na2 site. (A) Betaine uptake rates in nmol⋅min−1⋅mg−1 cell dry weight (cdw) were measured as a function of the external sodium concentration in E. coli MKH13 cells expressing WT BetP (●) or the mutants T467A (▼), S468A (■), and T467A/S468A (▲). Each point shows the average of at least three independent experiments. Error bars represent SD. (B) The maximum of Trp fluorescence at 340 nm as a function of sodium concentration was measured in proteoliposomes. Each point shows the average of at least three independent experiments. Error bars represent SD. (C) 22Na+ binding by given BetP variants measured with the SPA: 1 μM [22Na]Cl binding was measured in the presence of increasing concentrations of nonlabeled NaCl. Error bars indicate SD of triplicate determinations.
Fig. 5.
Transient currents generated by BetP. Using SSM-based electrophysiology, positive transient currents corresponding to the displacement of positive charge into the proteoliposomes were measured. The currents were recorded for WT and mutants in proteoliposomes after concentration jumps of 50 μM betaine or 100 mM sodium at t = 2.5 s. Before these concentration jumps an osmolar gradient of 500–1,000 mM was established to activate BetP. The protein was (A) WT, (B) mutated at T467A/S468A in Na2, and (C) mutated at T246L/T250A in Na1′. Traces in blue and gray correspond to concentration jumps of betaine in the presence of NaCl [ΔBet(Na)] or KCl [ΔBet(K)], respectively. Traces in black and red correspond to concentration jumps of sodium in the presence of betaine [ΔNa(Bet)] or glycine [ΔNa(Gly)], respectively. All traces shown were recorded from one sensor. In A the peak currents are −99.3 ± 10.2 for ΔBet(Na), −91.6 ± 1.7 for ΔNa(Bet), and −57.8 ± 12.5 for ΔNa(Gly). For the complete solution protocol and buffer composition, see SI Materials and Methods.
Identifying the Second Na1 Cation-Binding Site in BetP.
Betaine uptake by BetP is coupled to cotransport of two sodium ions to provide a driving force sufficient to accumulate betaine up to molar concentrations under hyperosmotic conditions (38). Unfortunately, no electron density that might correspond to the second sodium ion was observed in the Fo-Fc difference map for the structure of the closed state [Protein Data Bank (PDB) ID: 4AIN (7)], which at 3.1-Å resolution simply may reflect a more flexible region or lower occupancy than the Na2 site.
In addition, in the closed-state structure (7), the binding configuration of the betaine differs from that in the earlier structures [PDB ID: 2WIT (4)], so that the pNa1 site cannot exist. Specifically, as shown in Fig. S6B, the betaine carboxyl group is hydrogen-bonded by three backbone amine groups in the unwound segment of helix A1 and by the side-chain hydroxyl of S253 in helix A3, leaving no possibility for a sodium ion to coordinate the carboxyl group. This binding arrangement was maintained throughout MD simulations of the closed state, as shown by the distributions of the distances between the betaine oxygen and protein backbone nitrogen atoms (Fig. S7B).
Therefore we searched for a second site that might bind sodium in addition to Na2. Based on an analysis of the water distribution in the MD simulations of the inward-occluded structure, we initially considered a region of BetP, roughly equivalent to Na1 in LeuT, located within the four-helix bundle, which includes the carbonyl oxygen atoms of G149 (in helix A1) and P405, the side chain of S409 (in helix B2), and, perhaps, the carboxylate group of the substrate (Fig. S8A and Table S1). Specifically, S409 is coordinated by a water molecule that entered through the cytoplasmic pathway during these simulations (Fig. S8 A and B). However, in the recent structure of the closed state, the oxygen atoms listed above are far (9–12 Å) from the ligand carboxylate oxygen, and the region around S409 is surrounded by the side chains of F146, M150, and W189 (7). Moreover, when S409 was mutated to alanine, there was essentially no effect on the apparent Km for sodium (Table 1 and Fig. S8C), in marked contrast to the large increases observed upon modification of the Na2-site residues (Fig. 4 and Tables 1 and 2). Therefore, we ruled out S409 as a possible sodium-binding residue.
The next idea for the location of the second cation-binding site was inspired by the observation that the FIRL-fold has a pseudosymmetry axis running through the bundle and the scaffold (Fig. 1, dotted line). This symmetry suggests that, in principle, the role of helices A1 and B3 in forming the Na2-binding site could be recapitulated by helices B1 and A3. Accordingly, we analyzed the transporters of known structure and found that one protein, Mhp1, which requires only one sodium ion to transport its substrate benzyl-hydantoin, contains a positively charged residue (K110) approximately at the symmetry-equivalent position of Na2 (Fig. 1). Specifically, the side chain of K110 bridges the side chains of S114 in helix A3 and of S226 in helix B1. Interestingly, S114 of Mhp1 corresponds to a threonine residue in BetP (T246; Fig. 2). Other structures of FIRL-fold proteins contain either nonpolar or bulky residues at that position of helix A3, consistent with the fact that no cations have been identified there in previous studies. Two additional hydroxylic side chains can be found nearby in helix A3 of BetP, namely T250 and S253 (Fig. 2). As mentioned above, S253 coordinates the betaine carboxylate moiety in the structure and in simulations of closed BetP (Figs. S6B and S7B). In helix B1, BetP also contains a hydroxyl side chain at position S376. Overall, therefore, the region around helices B1 and A3 appears to be a good candidate for binding a second sodium ion.
MD Simulations of BetP Suggest the Coordination in the Proposed Na1′- Binding Site.
To help identify which of the aforementioned residues might coordinate sodium, we carried out MD simulations of the occluded structure of BetP with a sodium ion placed near the symmetry-equivalent position of Na2, i.e., equidistant from residues T246, T250, and S376 (Materials and Methods), and in the presence and absence of a water molecule. The most plausible coordination according to these simulations involved the side chains of T246 and T250, the backbone carbonyl of T246, and a single water molecule (Fig. 3C and Fig. S7 C and D). We name this site “Na1′” to distinguish it from the Na1 site in LeuT (Table S1). The side chain of S376 from helix B1, in contrast, did not coordinate the ion consistently in the simulations (oxygen distance >5 Å; Fig. S7) and instead hydrogen-bonded to the backbone carbonyl of A245 in helix A3 or to the backbone carbonyl of either A372 or W373 from helix B1. In this configuration the sodium ion is also close to the side chain of F380, with the closest Cδ atom at an average distance of 3.9 ± 0.4 Å from the ion (in the presence of a coordinating water), suggestive of a cation–π interaction. Such Na+–π interactions involving aromatic side chains have been reported for several protein structures (40–42) as well as for small molecules (43, 44). However, in these simulations the F380- Na1′ arrangement is not strictly en face, so high-resolution crystallographic data will be required to confirm whether such an interaction is present.
In the absence of explicit waters in the Na1′-binding site, the ion remained coordinated by T246 and T250 in all simulations, but there was a lack of order in other residues in the vicinity (Fig. S7C), and there also was space to accommodate a water molecule adjacent to the ion, similar to observations for the sodium-binding site in F-ATP synthase c-subunits from Ilyobacter tartaricus (45). Instead, when a water molecule was included in that position in the simulations of BetP (Fig. 3C), the side chains around the ion adopted significantly more ordered configurations (Fig. S7D), and the water provided a fourth oxygen ligand (Fig. 3C and Fig. S7E).
Mutations in the Predicted Na1′-Binding Site Affect Sodium Binding.
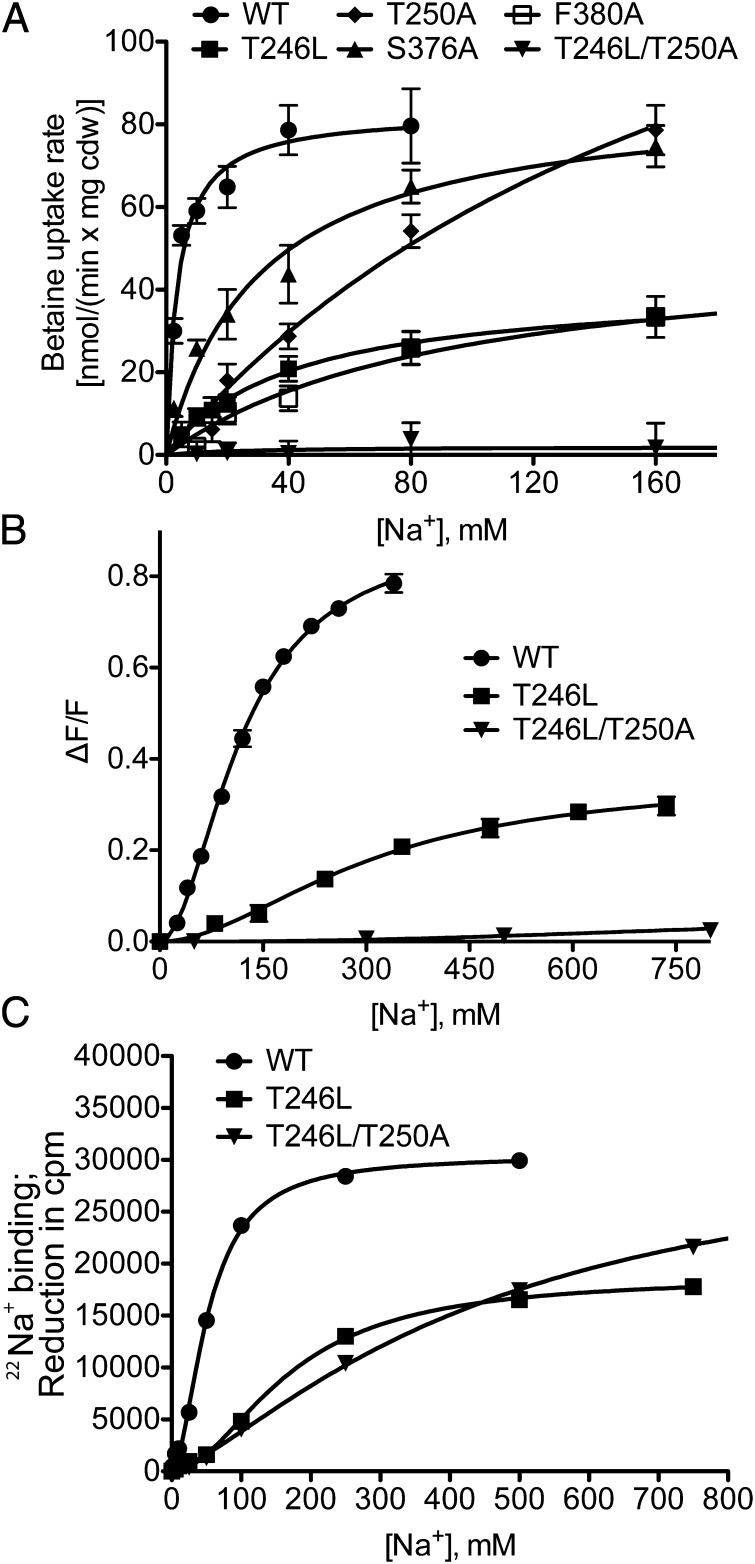
To test the predicted Na1′ site, we performed single and double substitutions at T246, T250, and F380 in BetP as well as at S376, replacing them with either alanine or leucine. Single substitutions of T246, T250, and F380 significantly increased the apparent Km of BetP for sodium by between 10- and 48-fold, whereas an eightfold increase was observed for S376 (Table 1 and Fig. 6A). Trp fluorescence and 22Na-binding assays revealed a threefold increase in the apparent Kd for the T246L single mutant, whereas the S376A mutation had a less significant effect (Table 2). Strikingly, in the double mutant T246L/T250A, transport was completely abolished (Table 1 and Fig. 6A), and only very weak binding could be detected (Table 2 and Fig. 6 B and C).
Fig. 6.
Sodium dependence of BetP for mutations introduced at the Na1′ site. Error bars indicate the SD of at least three independent experiments. (A) Uptake rates in nmol⋅min−1⋅ g−1 cdw were measured as a function of the external sodium concentration in E. coli MKH13 cells expressing WT BetP (●) or the mutants T246L (■), T250A (♦), S376A (▲), F380A (□), and T246L/T250A (▼). (B) The maximum of Trp fluorescence at 340 nm as a function of sodium concentration was measured in proteoliposomes. (C) 22Na+ binding by given BetP variants measured with the SPA: 1 μM [22Na]Cl binding was measured in the presence of increasing concentrations of nonlabeled NaCl.
The double mutant T246L/T250A also was studied by SSM-based electrophysiology. No transient current could be observed for this double mutant (Fig. 5C), in contrast to the currents observed for WT BetP (Fig. 5A) in the presence of sodium [ΔNa(Bet)] and substrate [ΔBet(Na)] betaine.
These data provide strong support for the hypothesis derived from the simulation results, namely that T246, T250, and F380 play a role in sodium binding and that S376 likely does not.
Sodium Binding to BetP Is Cooperative.
The binding isotherm for sodium to WT BetP displays a distinct sigmoidicity (Fig. 4 and Fig. S9) corresponding to a Hill coefficient of ∼2.0 (Table 2), reflecting positive cooperativity in sodium ion binding.
This cooperativity is maintained in T246L, the single mutant of the Na1′ site (Fig. 6 and Fig. S9B), whose binding isotherm also is sigmoidal and shows positive cooperativity with a Hill coefficient of 1.9 (Table 2), indicating that two interacting sodium-binding sites are still present. In contrast, fluorescence measurements on the single mutant S468A at the Na2 site show no sigmoidicity and have a Hill coefficient of 1.1 ± 0.3 (Fig. 4, Fig. S9A, and Table 2). Note, however, that it was not possible to reach saturation of sodium binding for several of the measurements involving T246L/T250A and S468A (Fig. S9 and Table 2), precluding reliable fitting of the data. Nevertheless, it is clear from the titration curves that binding is strongly affected by these mutations (Figs. 4C and 6C).
To rule out the possibility that the sodium cooperativity occurs between protomers in the trimeric complex of WT BetP, we also measured sodium binding in a monomeric mutant of BetP, W101A/T351A (Fig. S9C and Table 2) (37). The monomeric protein retains the sigmoidal binding isotherm of the WT, with a Hill coefficient of ∼2.0 (Table 2), demonstrating that the observed cooperativity occurs between ion-binding sites within a protomer.
Discussion
The binding of sodium ions is a critical property of a large proportion of secondary transporters and of other membrane proteins (2). However, the details of their binding sites are poorly characterized in most cases, in part because of the challenges of membrane protein crystallography and the relatively low resolution of the data currently attainable and also because of the similarity of the scattering factors of sodium ions and water oxygen atoms. In the 3.1-Å resolution X-ray structure of BetP used here, for example, Perez et al. (7) were able to identify only one single positive peak in the difference density map corresponding to one of the two sodium ions, but no second ion could be observed. This situation may reflect increased disorder in the region of the Na1′-binding site, for which the average B-factor is slightly higher (126 ± 15 Å2) than for residues forming the Na2-binding site (115 ± 11 Å2), or may reflect a decreased occupancy and affinity at the Na1′ site in these crystals, e.g., because of dehydration reducing the likelihood of a coordinating water.
In some cases it has been possible to identify cation sites by analogy to sites known from structural data, as for the Na2 site of LeuT (12), which also has been identified in vSGLT (13), Mhp1 (10), and now in BetP (ref. 7 and Figs. 3 and 4). Taken together, these analyses demonstrate that helices A1 and B3 of the FIRL-fold create a well-conserved binding site for a positively charged ion (Fig. 1). Indeed, the same region is occupied by the positively charged moiety of the substrate in an inward-facing structure of the choline-transporting G153D mutant of BetP (36) and in an inward-facing substrate-bound structure of WT BetP (7), as well as by basic side chains in sodium-independent transporters (5, 9). Moreover, hydroxylic side chains are highly conserved in helix B3 in FIRL-fold families, particularly those known to contain many sodium-coupled transporters (Fig. S10). In hindsight, the presence of the charged group K110 in Mhp1 at the homologous location also might have provided a useful starting point (Fig. 1).
The clue that led us to the location of the second Na1′-binding site in BetP was the structural homology between repeated elements of the same protein. A similar concept was used previously to study the electrostatics of ion permeation in ClC channels (46) and to model alternate conformations of secondary transporters (3, 47, 48). In this way, we predicted that the helices corresponding to A1 and B3 in the opposite repeat, i.e., helices B1 and A3 (Fig. 1), also can form a sodium-binding site.
To assess further whether, and in what way, the two proposed sites are likely to coordinate sodium ions in BetP, we first carried out MD simulations on the crystal structure of the closed state at 3.1-Å resolution. In these simulations, the coordination of Na2 observed in the crystal structure is preserved (Fig. S7A). Moreover, the simulations suggest that the Na1′ site includes one aromatic and two hydroxylic side chains, a backbone carbonyl, and potentially a single water molecule (Fig. 3 and Fig. S7 C–E). We then measured experimentally the effect on the apparent sodium affinity of BetP of site-directed mutagenesis at the coordinating residues. Significant effects could be observed not only on the sodium dependence of radiolabeled betaine uptake into cells (Figs. 4A and 6A and Table 1) but also on sodium-dependent Trp fluorescence of BetP in proteoliposomes (Figs. 4B and 6B, Fig. S9, and Table 2), on sodium transient currents also in proteoliposomes (Fig. 5), and on 22Na+ binding to BetP in an SPA (Figs. 4C and 6C and Table 2). It is notable that the mutations with the most dramatic effects are clustered around locations that we predict are suitable for sodium binding, whereas modification of other tested positions such as S306, M310, and S409 had no significant effect on the affinity for sodium (Figs. S5 and S8C and Tables 1 and 2).
Another possible explanation for the effects of the Na1′-site mutations is disruption of interactions required for stabilizing the structure and/or conversion between the different states during transport. Indeed, S376 hydrogen bonds to neighboring helices during the simulations, possibly explaining why mutation to alanine has a nonnegligible effect on transport (Tables 1 and 2). However, 22Na+ binding and transient currents measured in the absence of betaine [Fig. 6C and ΔNa(Gly) in Fig. 5] also were affected by mutations to Na1′. Therefore, any indirect effects these mutations may cause must involve the conformational changes in converting the apo state to the sodium-bound state, rather than the substrate-bound outward-to-inward transition. Because T246 and T250 are not involved in any obviously important networks in the other available structures of BetP (4, 7, 36), it seems plausible that the effects we measure reflect reduced sodium affinity rather than some kind of conformational hindrance.
Together, the evidence appears to support our hypothesis that BetP binds two ions at symmetry-related positions within the FIRL-fold. Both binding sites involve two hydroxyl sidechains originating from the same face of a transmembrane helix. However, the other coordinating residues differ, and thus the symmetry relationship breaks down at the level of the detailed chemistry. Specifically, Na2 includes two backbone oxygen atoms from a nonhelical stretch of A1, whereas Na1′ includes a water molecule as well as a phenylalanine side chain from B1, which is helical (Fig. 3). The possibility that aromatic side chains can coordinate alkali cations is not commonly considered in the protein structure community but has been shown for several small-molecule and protein systems (40, 41, 44, 49). Nevertheless, it should be noted that the simulation data so far do not support a classical en face cation–π interaction between F380 and Na1′. Further experimental evidence is required to assess whether this apparently suboptimal coordination reflects limitations of the force-field used or is indeed the native arrangement in the Na1′-binding site, perhaps arising from conflicting requirements of the ligands in that structure.
Assuming that the Na1′ site is a bona fide sodium site, it also may be present in other transporters. In the APC, NCS1, and SSS transporter families, the positions in helix A3 (BetP residues T246 and T250) that are predicted to coordinate Na1′ do not contain a significant population of hydroxylic side chains (Fig. S10), suggesting that this binding site is not common to those families and perhaps reflecting coupling ratios involving a single sodium ion. In the BCC transporter family, ∼20 proteins have been characterized biochemically to date, although the sodium:substrate stoichiometry is known only for BetP; among the other sodium-dependent BCC transporters only LcoP is conserved at T246, T250, and F380 as well as at T467 and S468 (Fig. S11) and therefore is the only characterized sodium-coupled BCC transporter that would be predicted to bind two sodium ions at the same sites as BetP. Nevertheless, serine and threonine are found in those positions in a substantial fraction of all sequenced BCC and NSS transporters (Fig. S10), and therefore the Na1′-binding site may be found in other members of these families not yet characterized.
The relative positions of helices A1 and B3 change during the transport cycle and appear to be modulated by the presence or absence of Na2 in the sodium-dependent transporters (7, 11, 15, 16, 20). Comparison of inward- and outward-facing structures reveals that helices A3 and B1 forming Na1 also move relative to one other during transport by Mhp1 (Fig. S12 A and B). Strikingly, these conformational changes are accompanied by a change in interactions that center around K110 from helix A3 (TM3), whose positively charged amine group is located at a position similar to that of Na1′ (Fig. 1). Changes also are observed in LeuT in the relative positions of A3 and B1, which are involved in contacts with the substrate amino acid side chain. These movements are most dramatic in the periplasmic helix of A3 (Fig. S12 C and D), but at the region at which Na1′ would bind (i.e., near residues F253 of B1, P101, and V104 of A3 and I359 of B3 of LeuT) these changes are more muted (∼1.5Å), perhaps because of contacts between the cytoplasmic end of B1 and a bound antibody (20) or perhaps reflecting the more hydrophobic interactions in this region of LeuT (Fig. S12 C and D). Overall, we conclude that varying degrees of movements in B1 and A3 are characteristic of the transport cycle in FIRL-fold transporters, in a manner that could be sensitive to the presence or absence of an ion when a binding site is present at Na1′.
The fluorescence and SPA affinity measurements provide clear evidence for cooperativity between the two sodium-binding sites in WT BetP (Fig. S9 and Table 2). Although BetP is trimeric (50), the monomeric W101A/T351A mutant (37) also retains cooperativity (Fig. S9C and Table 2), demonstrating that the cooperativity in sodium binding occurs within each protomer. This cooperativity apparently can be abolished by a single mutation in the Na2-binding site (S468A) but not by single (T246L) or double (T246L/T250A) mutations of the Na1′-binding site, although, as noted above, the lack of saturation in the binding of sodium to S468A and to T246L/T250A may preclude reliable determination of the Hill coefficients in those cases, making interpretation of these effects challenging.
A possible origin of the cooperativity is the location of both ions at the interface between the scaffold and bundle helices (Figs. 1 and 3 and Fig. S12). That is, as helices from those two segments move relative to one another during the inward-to-outward and outward-to-inward transitions (7, 11, 15, 16), binding to either site might stabilize a particular state (e.g., the outward-facing conformation) and thereby facilitate binding to the other site.
The current study illustrates the potential of structure analyses in synergy with MD simulations to provide useful hypotheses regarding the location and likely coordination of ion-binding sites, after which the hypotheses are tested experimentally (3, 51). Theoretical studies also have proposed a cation-binding site in the aspartate transporter GltPh, based on a Monte Carlo search of the structure with a water probe, the results of which were tested by electrophysiological measurements (52). In another case, the identification of a negatively charged side chain in a chloride-independent bacterial homolog led to the prediction of the chloride-binding site in mammalian chloride-dependent NSS transporters; this prediction subsequently was confirmed by biochemical measurements (53, 54).
A clearer characterization of binding sites for coupling ions sets the stage for in-depth investigations into the mechanisms of coupling and alternating access by ion-coupled transporters and is a necessary step toward understanding more complex mechanisms such as regulation. At the same time, the fact that the binding sites in BetP are located at sites that are related to one another by the symmetry in the FIRL-fold suggests that considering the inverted-topology repeats may be useful in studies of various aspects of transport (3, 47, 48).
Materials and Methods
MD Simulations.
MD simulations were carried out on (a) a BetP trimer from PDB ID 2WIT prepared and carried out as described previously (37), with two sodium ions added in each protomer at previously proposed locations pNa1 and pNa2 (Fig. S2) (4); and (b) on a BetP monomer from PDB ID 4AIN chain B, which is in a closed conformation (ref. 7 and Fig. S6C). Betaine and the Na2 ion were placed at the location of positive peaks in the Fo-Fc difference density maps (7), and the Na1′ ion was placed close to helices A3 and B1. MD simulations were performed using NAMD (55), with the all-atom Charmm27 force field (56–58). Full details of the simulation setups are provided in SI Materials and Methods.
Figures of structures and simulation snapshots were prepared with PyMOL 1.1r1 (PyMOL Molecular Graphics System; Schrödinger, LLC).
Structure Alignments.
X-ray models of seven FIRL-fold transporters of known-structure [AdiC, ApcT, BetP, CaiT, LeuT, Mhp1, vSGLT (PDB IDs: 3L1L, 3GIA, 2WIT, 3HFX, 3F3A, 2JLN, and 3DH4, respectively)] were superposed with the structure alignment program SKA (59, 60).
Cell Culturing and Protein Purification.
Cell-culture and protein-preparation methods were described previously (61). Uptake of [14C]-betaine was measured in E. coli MKH13 cells (62). E. coli DH5αmcr (63) was used for the heterologous expression of strep-betP. Cells were grown at 37 °C in LB medium supplemented with carbenicillin (50 μg/mL), and induction was initiated with anhydrotetracycline (200 μg/L). Cells were harvested at 4 °C by centrifugation and resuspended in buffer containing 100 mM Tris⋅HCl (pH 8) and the protease inhibitor Pefabloc (0.24 mg/mL; Sigma). Membranes were isolated from disrupted cells and solubilized with 1.3% (wt/vol) n-dodecyl-β-d-maltopyranoside (DDM). Then the protein was loaded on a StrepII-Tactin macroprep column (Sigma), washed with 50 mM Tris⋅HCl (pH 7.5), 500 mM NaCl, 8.6% (vol/vol) glycerol, 0.05% DDM, and eluted with 5 mM desthiobiotin in the same buffer.
Site-Directed Mutagenesis.
The QuikChange kit (Stratagene) and Pfu Turbo DNA polymerase were applied for nucleotide mutagenesis (primer sequences are listed in Table S2) in pASK-IBA5betP (39). All the plasmids were fully sequenced, and the specific mutations were confirmed.
Protein Reconstitution into Liposomes.
Functional reconstitution of the proteins was performed as described (61). Briefly, liposomes (20 mg phospholipid/mL) from E. coli polar lipid extract phospholipids (Avanti Polar Lipids) were prepared by extrusion through polycarbonate filters (400-nm pore size) and diluted 1:4 in buffer (250 mM KPi, pH 7.5). After saturation with Triton X-100, the liposomes were mixed with purified protein at a lipid/protein ratio of 10:1 (wt/wt). BioBeads at ratios (wt/wt) of 5 (BioBeads/Triton X-100) and 10 (BioBeads/DDM) were added to remove the detergent. Finally, the proteoliposomes were centrifuged and washed before being frozen in liquid nitrogen and stored at −80 °C.
Transport Assays.
Uptake of [14C]-betaine in E. coli cells was performed as described (64). E. coli MKH13 cells expressing a particular strep-betP mutant were cultivated at 37 °C in LB medium containing carbenicillin (50 μg/mL) and induced at an OD600 of 0.5 by adding anhydrotetracycline (200 μg/L). After 2 h the cells were harvested and washed in buffer containing 25 mM KPi buffer (pH 7.5) and then were resuspended in the same buffer containing 20 mM glucose. For uptake measurements the external osmolality was adjusted with KCl at a constant value of 800 mOsmol/kg. Sodium titration was performed by adjusting the NaCl concentration in the buffer. Cells were incubated for 3 min at 37 °C before the addition of 250 µM [14C]-betaine. Betaine uptake was measured at various time intervals, after cell samples were passed through glass fiber filters (APFF02500; Millipore) and were washed twice with 2.5 mL of 0.6 M KPi buffer. The radioactivity retained on the filters was quantified by liquid scintillation counting. Immunoblotting against the N-terminal StrepII-tag of the different BetP variants in membranes of E. coli MKH13 using StrepII-tag–specific antibody confirmed that mutant forms of BetP were synthesized to approximately the same level in cells.
Trp Fluorescence Binding Assay.
Binding assays were performed with 100 μg/mL of purified BetP protein in proteoliposomes resuspended in buffer [250 mM KPi (pH 7.5) and 2 mM β-mercaptoethanol]. Trp fluorescence emission scans between 315 and 370 nm were recorded for titration of proteoliposomes with NaCl on an Hitachi F-4500 fluorescence spectrophotometer and averaged over four readings, with the excitation wavelength set to 295 nm and a slit width of 2.5 or 5.0 nm for excitation or emission, respectively, at a constant betaine concentration of 10 mM. The mean value and SD at the 340-nm emission maximum was plotted for each substrate or ion concentration. Data were fitted using GraphPad Prism version 5.0c for Mac OS X (GraphPad Software).
SPA.
Binding of 22Na+ to purified BetP variants was performed by means of the SPA as described (26). Experiments were preformed in 0.05–1.05 M Tris/MES (pH 7.5), 1.05–0.05 M NaCl (equimolar replacement of Tris/MES with NaCl), 20% (vol/vol) glycerol, 1 mM Tris(2-carboxyethyl)phosphine, 0.1% n-dodecyl-β-d-maltopyranoside using 100 ng of purified BetP variants, 2.5 mg/mL streptavidin-coated YSi SPA beads (RPNQ0012; Perkin Elmer), and 1 μM of [22Na]Cl (19.1 Ci/mmol) (Perkin Elmer).
SSM-Based Electrophysiology.
Proteoliposomes were adsorbed to a sensor equipped with a SSM, and the BetP transporter was activated via a rapid substrate concentration jump (65). Transient currents corresponding to the electrogenic activity of BetP were recorded via capacitive coupling. More details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation Collaborative Research Center 807 “Transport and Communication across Biological Membranes” (L.R.F. and C.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 17754 (volume 109, number 44).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209039109/-/DCSupplemental.
References
- 1.Sobczak I, Lolkema JS. Structural and mechanistic diversity of secondary transporters. Curr Opin Microbiol. 2005;8(2):161–167. doi: 10.1016/j.mib.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Saier MH, Jr, Tran CV, Barabote RD. 2006. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Research 34(Suppl 1):D181–186.
- 3.Forrest LR, Kraemer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807:167–188. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458(7234):47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 5.Schulze S, Koester S, Geldmacher U, Terwisscha van Scheltinga AC, Kuehlbrandt W. Structural basis of cooperative substrate binding and Na+-independent transport in the carnitine/butyrobetaine antiporter CaiT. Nature. 2010;467:233–237. doi: 10.1038/nature09310. [DOI] [PubMed] [Google Scholar]
- 6.Tang L, Bai L, Wang WH, Jiang T. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol. 2010;17(4):492–496. doi: 10.1038/nsmb.1788. [DOI] [PubMed] [Google Scholar]
- 7.Perez C, Koshy C, Yildiz Ö, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012 doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460(7258):1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325(5943):1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322(5902):709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimamura T, et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328(5977):470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl—dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 13.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321(5890):810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khafizov K, Staritzbichler R, Stamm M, Forrest LR. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry. 2010;49(50):10702–10713. doi: 10.1021/bi101256x. [DOI] [PubMed] [Google Scholar]
- 15.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA. 2008;105(30):10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrest LR, Rudnick G. The rocking bundle: A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24(6):377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaikh SA, Tajkhorshid E. Modeling and dynamics of the inward-facing state of a Na+/Cl- dependent neurotransmitter transporter homologue. PLOS Comput Biol. 2010;6(8):e1000905. doi: 10.1371/journal.pcbi.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, et al. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465(7295):188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474(7349):109–113. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zomot E, Kanner BI. Identification of a lithium interaction site in the γ-aminobutyric acid (GABA) transporter GAT-1. J Biol Chem. 2006;281(31):22092–22099. doi: 10.1074/jbc.M602319200. [DOI] [PubMed] [Google Scholar]
- 22.Claxton DP, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat Struct Mol Biol. 2010;17(7):822–829. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi L, Weinstein H. Conformational rearrangements to the intracellular open states of the LeuT and ApcT transporters are modulated by common mechanisms. Biophys J. 2010;99(12):L103–L105. doi: 10.1016/j.bpj.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao C, Noskov SY. The role of local hydration and hydrogen-bonding dynamics in ion and solute release from ion-coupled secondary transporters. Biochemistry. 2011;50(11):1848–1856. doi: 10.1021/bi101454f. [DOI] [PubMed] [Google Scholar]
- 25.Celik L, Schiøtt B, Tajkhorshid E. Substrate binding and formation of an occluded state in the leucine transporter. Biophys J. 2008;94(5):1600–1612. doi: 10.1529/biophysj.107.117580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30(6):667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noskov SY, Roux B. Control of ion selectivity in LeuT: Two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377(3):804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468(7326):988–991. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De la Vieja A, Reed MD, Ginter CS, Carrasco N. Amino acid residues in transmembrane segment IX of the Na+/I- symporter play a role in its Na+ dependence and are critical for transport activity. J Biol Chem. 2007;282(35):25290–25298. doi: 10.1074/jbc.M700147200. [DOI] [PubMed] [Google Scholar]
- 30.Hilger D, Böhm M, Hackmann A, Jung H. Role of Ser-340 and Thr-341 in transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli in ligand binding and transport. J Biol Chem. 2008;283(8):4921–4929. doi: 10.1074/jbc.M706741200. [DOI] [PubMed] [Google Scholar]
- 31.Raba M, et al. Function of transmembrane domain IX in the Na+/proline transporter PutP. J Mol Biol. 2008;382(4):884–893. doi: 10.1016/j.jmb.2008.07.070. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Tajkhorshid E. Ion-releasing state of a secondary membrane transporter. Biophys J. 2009;97(11):L29–L31. doi: 10.1016/j.bpj.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choe S, Rosenberg JM, Abramson J, Wright EM, Grabe M. Water permeation through the sodium-dependent galactose cotransporter vSGLT. Biophys J. 2010;99(7):L56–L58. doi: 10.1016/j.bpj.2010.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zomot E, Bahar I. The sodium/galactose symporter crystal structure is a dynamic, not so occluded state. Mol Biosyst. 2010;6(6):1040–1046. doi: 10.1039/b927492h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazier S, Quick M, Shi L. Conserved tyrosine in the first transmembrane segment of solute:sodium symporters is involved in Na+-coupled substrate co-transport. J Biol Chem. 2011;286(33):29347–29355. doi: 10.1074/jbc.M111.263327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez C, et al. Substrate specificity and ion coupling in the Na+/betaine symporter BetP. EMBO J. 2011;30(7):1221–1229. doi: 10.1038/emboj.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez C, Khafizov K, Forrest LR, Krämer R, Ziegler C. The role of trimerization in the osmoregulated betaine transporter BetP. EMBO Rep. 2011;12(8):804–810. doi: 10.1038/embor.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farwick M, Siewe RM, Krämer R. Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J Bacteriol. 1995;177(16):4690–4695. doi: 10.1128/jb.177.16.4690-4695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiller D, Krämer R, Morbach S. Cation specificity of osmosensing by the betaine carrier BetP of Corynebacterium glutamicum. FEBS Lett. 2004;563(1-3):108–112. doi: 10.1016/S0014-5793(04)00279-0. [DOI] [PubMed] [Google Scholar]
- 40.Wouters J. Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme. Protein Sci. 1998;7(11):2472–2475. doi: 10.1002/pro.5560071127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wouters J, Maes D. Identification of a potential metal cation-π binding site in the structure of a thermophilic Bacillus stearothermophilus triosephosphate isomerase mutant. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 9):1201–1203. doi: 10.1107/s0907444900008659. [DOI] [PubMed] [Google Scholar]
- 42.Matsumura H, et al. Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase. Proteins. 2008;70(2):592–598. doi: 10.1002/prot.21799. [DOI] [PubMed] [Google Scholar]
- 43.Ma JC, Dougherty DA. The Cationminus signpi interaction. Chem Rev. 1997;97(5):1303–1324. doi: 10.1021/cr9603744. [DOI] [PubMed] [Google Scholar]
- 44.De Wall SL, Meadows ES, Barbour LJ, Gokel GW. Synthetic receptors as models for alkali metal cation-pi binding sites in proteins. Proc Natl Acad Sci USA. 2000;97(12):6271–6276. doi: 10.1073/pnas.97.12.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meier T, et al. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J Mol Biol. 2009;391(2):498–507. doi: 10.1016/j.jmb.2009.05.082. [DOI] [PubMed] [Google Scholar]
- 46.Faraldo-Gómez JD, Roux B. Electrostatics of ion stabilization in a ClC chloride channel homologue from Escherichia coli. J Mol Biol. 2004;339(4):981–1000. doi: 10.1016/j.jmb.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 47.Crisman TJ, Qu S, Kanner BI, Forrest LR. Inward-facing conformation of glutamate transporters as revealed by their inverted-topology structural repeats. Proc Natl Acad Sci USA. 2009;106(49):20752–20757. doi: 10.1073/pnas.0908570106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radestock S, Forrest LR. Outward-facing conformation of MFS transporters revealed by inverted-topology repeats. J Mol Biol. 2011;407:698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Ruan C, Yang Z, Hallowita N, Rodgers MT. Cation-pi interactions with a model for the side chain of tryptophan: Structures and absolute binding energies of alkali metal cation-indole complexes. J Phys Chem A. 2005;109(50):11539–11550. doi: 10.1021/jp053830d. [DOI] [PubMed] [Google Scholar]
- 50.Tsai C-J, et al. Structural asymmetry in the betaine transporter BetP trimer indicates three different conformational states. J Mol Biol. 2011;407:368–381. [Google Scholar]
- 51.Faraldo-Gómez JD, Forrest LR. Modeling and simulation of ion- and ATP-driven membrane proteins. Curr Opin Struct Biol. 2011;21:1–7. doi: 10.1016/j.sbi.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 52.Larsson HP, et al. Evidence for a third sodium-binding site in glutamate transporters suggests an ion/substrate coupling model. Proc Natl Acad Sci USA. 2010;107(31):13912–13917. doi: 10.1073/pnas.1006289107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forrest LR, Tavoulari S, Zhang Y-W, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl- dependent transporters. Proc Natl Acad Sci USA. 2007;104(31):12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449(7163):726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 55.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feller SE, Gawrisch K, MacKerell AD., Jr Polyunsaturated fatty acids in lipid bilayers: Intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124(2):318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 57.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 58.Nina M, Beglov D, Roux B. Atomic radii for continuum electrostatics calculations based on molecular dynamics free energy simulations. J Phys Chem. 1997;101(26):5239–5248. [Google Scholar]
- 59.Yang AS, Honig B. An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance. J Mol Biol. 2000;301(3):665–678. doi: 10.1006/jmbi.2000.3973. [DOI] [PubMed] [Google Scholar]
- 60.Petrey D, et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(6) Suppl 6:430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 61.Rübenhagen R, Rönsch H, Jung H, Krämer R, Morbach S. Osmosensor and osmoregulator properties of the betaine carrier BetP from Corynebacterium glutamicum in proteoliposomes. J Biol Chem. 2000;275(2):735–741. doi: 10.1074/jbc.275.2.735. [DOI] [PubMed] [Google Scholar]
- 62.Haardt M, Kempf B, Faatz E, Bremer E. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol Gen Genet. 1995;246(6):783–786. doi: 10.1007/BF00290728. [DOI] [PubMed] [Google Scholar]
- 63.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87(12):4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ott V, Koch J, Späte K, Morbach S, Krämer R. Regulatory properties and interaction of the C- and N-terminal domains of BetP, an osmoregulated betaine transporter from Corynebacterium glutamicum. Biochemistry. 2008;47(46):12208–12218. doi: 10.1021/bi801325r. [DOI] [PubMed] [Google Scholar]
- 65.Schulz P, Garcia-Celma JJ, Fendler K. SSM-based electrophysiology. Methods. 2008;46(2):97–103. doi: 10.1016/j.ymeth.2008.07.002. [DOI] [PubMed] [Google Scholar]