Abstract
Synchronization of the circadian clock in cyanobacteria with the day/night cycle proceeds without an obvious photoreceptor, leaving open the question of its specific mechanism. The circadian oscillator can be reconstituted in vitro, where the activities of two of its proteins, KaiA and KaiC, are affected by metabolites that reflect photosynthetic activity: KaiC phosphorylation is directly influenced by the ATP/ADP ratio, and KaiA stimulation of KaiC phosphorylation is blocked by oxidized, but not reduced, quinones. Manipulation of the ATP/ADP ratio can reset the timing of KaiC phosphorylation peaks in the reconstituted in vitro oscillator. Here, we show that pulses of oxidized quinones reset the cyanobacterial circadian clock both in vitro and in vivo. Onset of darkness causes an abrupt oxidation of the plastoquinone pool in vivo, which is in contrast to a gradual decrease in the ATP/ADP ratio that falls over the course of hours until the onset of light. Thus, these two metabolic measures of photosynthetic activity act in concert to signal both the onset and duration of darkness to the cyanobacterial clock.
Keywords: chronobiology, entrainment, input pathway, pseudoreceiver, jetlag
The circadian clock in cyanobacteria provides an adaptive advantage when the period length of endogenous physiological and genetic cycles resonates with the length of the external day–night cycle (1). In circadian systems from diverse organisms from cyanobacteria to mammals, the physiological processes that oscillate with an endogenous circadian rhythm become synchronized with the environmental variation that is generated by the rotation of Earth, and external cues modulate the relative timing of peaks and troughs of activity (2). However, the mechanisms by which environmental input is conveyed to the central oscillator are diverse. In mammals, plants, fungi, and other eukaryotic species, specific photoreceptors have been identified that directly or indirectly modify the activities of transcription factors, which are components of interlocking feedback loops of the circadian clock (3). The cyanobacterial circadian clock is unique in that the circadian oscillator is a physical entity that can be reconstituted in vitro, and the mechanisms of input can be tested directly (4).
In the circadian model, cyanobacterium Synechococcus elongatus, a kinase called CikA, is required to enable the timing of peaks of circadian rhythms to be reset to an environmental cue that is equivalent to a change of four or more time zones (5). Although CikA shares domain features with phytochromes, which serve as photoreceptors in plants and many bacteria, it does not function as a photoreceptor (6). Moreover, inactivation of any of the photoreceptor genes identifiable in the S. elongatus genome has no effect on circadian rhythms of gene expression (7). These data suggest another route for conveying light information to the clock. Two key metabolic changes accompany the transition from light to darkness in cyanobacteria and have the potential to serve as proxies for light: the ratio of ATP to ADP, which depends on photophosphorylation, and the redox state of the plastoquinone (PQ) pool, which connects photosystem II (PSII) and the cytochrome b6f complex in the photosynthetic electron transport chain.
The cyanobacterial circadian oscillator is composed of three proteins: KaiA, KaiB, and KaiC (4). The phosphorylation state of KaiC oscillates with a 24-h rhythm both in vivo (8) and in a mixture with the other Kai proteins in vitro (hereafter, referred to as the in vitro oscillator mixture) (Fig. 1A). KaiA enhances autophosphorylation of KaiC, whereas KaiB activates KaiC autodephosphorylation by inhibiting KaiA function (9–11). Previous work showed that the ATP/ADP ratio affects KaiC phosphorylation directly and that manipulation of this ratio in the in vitro oscillator mixture can mimic resetting of the circadian phase of rhythms in vivo (12). The C-terminal domain of KaiA is known to enhance KaiC phosphorylation by binding to the A-loops of KaiC (11, 13); the N-terminal pseudoreceiver domain of KaiA is related to signal transduction receiver domains, suggesting a regulatory role (9). We previously showed that the pseudoreceiver domains present in both CikA and KaiA bind quinones (9, 14–16). Importantly, only oxidized quinones bind to the pseudoreceiver domains of KaiA, forming KaiA aggregates that cease to stimulate KaiC phosphorylation (15); this redox selectivity suggests that reversible binding and aggregation is a sensory mechanism for entraining the oscillator. Here, we show that oxidized quinones, markers of the onset of darkness in S. elongatus, reset the clock both in vivo and in vitro. We propose that this sensory mechanism, acting through KaiA, works in concert with [ATP]/[ADP] sensing by KaiC to signal both the onset and duration of darkness to the clock.
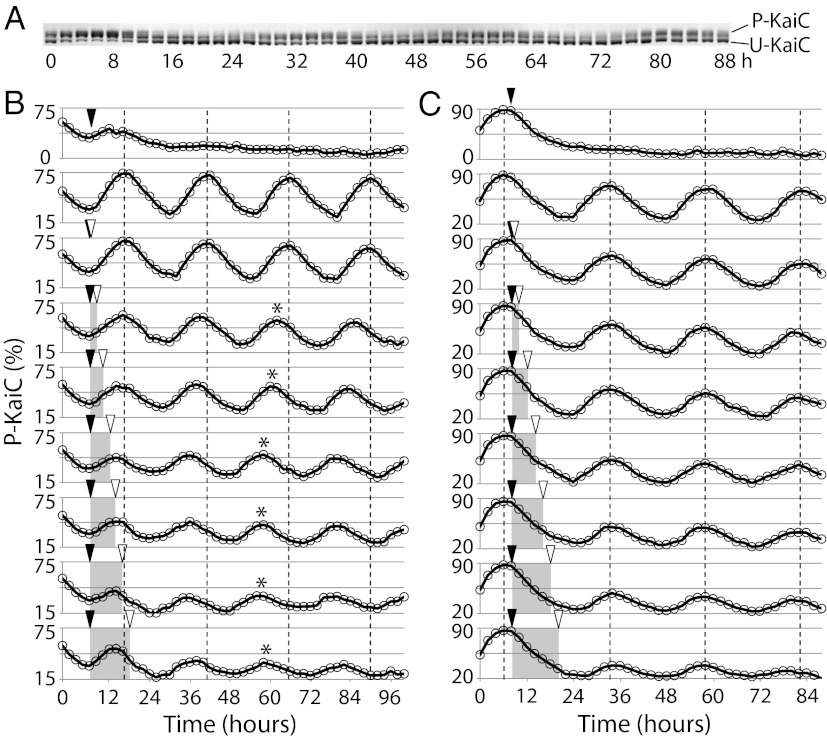
Fig. 1.
KaiC phosphorylation in vitro. (A) SDS/PAGE of KaiC in the in vitro oscillator mixture. The ratio of bands of phosphorylated KaiC (P-KaiC) to unphosphorylated KaiC (U-KaiC) oscillated in an ∼24-h period. This gel corresponds to the second trace in C. (B) Phase-selective effect of addition of oxidized Q0 on the in vitro KaiC phosphorylation cycle. Addition of oxidized Q0 during the phosphorylation phase of KaiC. The dotted lines are aligned on the peak positions of a control in vitro oscillation mixture to which no Q0 was added to aid comparisons of the timing of peaks in each reaction. Addition of oxidized Q0 is marked by a solid triangle, and addition of dithionite to reduce Q0 is marked by an open triangle. The shaded area indicates the duration of oxidized Q0. The observed phase shift is marked by an asterisk. (C) Same as B except that Q0 was added during the dephosphorylation phase.
Results
Oxidized Quinones Applied in the Phosphorylation Phase of KaiC Induce a Phase Shift in the in Vitro Oscillator Mixture.
A circadian rhythm is, by definition, sensitive to environmental cues (2), and treatments given at different times in the clock cycle generate different outcomes for the behavior under study [known as a phase response curve (PRC)] (17). In S. elongates, a 4- to 8-h dark pulse applied during the phosphorylation phase of the KaiC cycle induces striking phase shifts in gene expression rhythms, but it has little effect if applied during the dephosphorylation phase (Fig. 2) (12). We used this property to test the phase-dependent effect of KaiA quinone sensing in the in vitro oscillation mixture by applying quinones (15). We found that 4.8 µM oxidized 2,3-dimethoxy-5-methyl-p-benzoquinone (Q0) was sufficient to trigger complete dephosphorylation of KaiC, and the addition of the reducing agent dithionite recovered KaiC phosphorylation activity (Fig. 1 B and C). Application of oxidized Q0 for different durations during both phosphorylation and dephosphorylation phases, expected to mimic a dark pulse, had phase-dependent effects. The addition of oxidized Q0 for 2–12 h, only during the phosphorylation phase of the KaiC cycle, induced a shift in the phase of subsequent peaks of phosphorylation (Fig. 1B). The addition of oxidized Q0 during the dephosphorylation phase of KaiC (Fig. 1C) or dithionite alone at any time during the cycle (Fig. S1) did not induce a phase shift. The decrease in peak phosphorylation is characteristic of Q0 treatment and likely reflects the inability of dithionite to fully disaggregate KaiA (15).
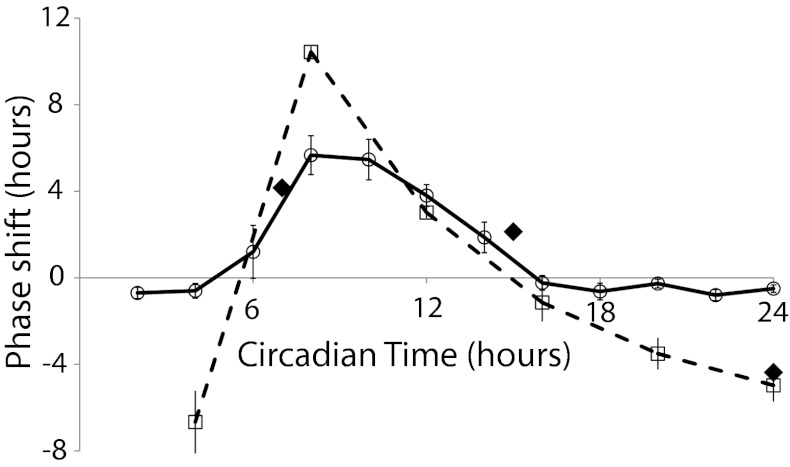
Fig. 2.
Comparisons of PRCs of in vitro and in vivo oscillations to darkness or oxidized quinone. The PRC generated by the addition of oxidized Q0 to the in vitro oscillator mixture (open circles connected by solid line) and the in vivo cultures held under constant light (solid diamond); the PRC is reported in the work by Kiyohara et al. (18), in which 4-h dark pulses were administered to in vivo cultures held otherwise under constant light (open squares connected by dashed line). Error bars placed on the in vitro data points are ±1 SEM and calculated from three different experiments.
Quinone-Induced Phase Advances, but Not Delays, Were Observed in Vitro.
This phase-selective effect corresponds to the previously reported PRC for gene expression rhythms generated by dark-pulse treatment of cyanobacterial cultures (18) and predicted that a 4-h addition of oxidized Q0 should be equivalent to a 4-h dark pulse. To test this hypothesis, 4-h pulses of oxidized Q0 were administered at different points throughout the cycle to generate a PRC (Fig. S2). The data from Q0 pulses in the oscillator mixture matched well with previously reported PRCs from bioluminescence rhythms of reporter genes for phase advances, but phase delays were not observed in vitro (Fig. 2). In contrast, the previously reported method of entrainment by modulating the ratio of ATP/ADP in the oscillator mixture (12), which acts through KaiC, induces both phase advances and delays.
PRC Generated by Administration of Oxidized Quinones in Vivo Corresponds to the PRC Induced by 4-h Dark Pulses.
It is possible that the effect of quinone modulation is limited in vitro, because only KaiA, and not CikA, is affected in the in vitro oscillator mixture. Thus, Q0 administration was used in vivo to determine the PRC of oxidized quinone in the intact system. Q0 was added to cyanobacterial reporter strains at different circadian times, and the timing of subsequent bioluminescence peaks was determined. Phase shifts were observed on addition of a 20-μL drop of Q0 solution at or above 3 mM (Fig. S3). At circadian times (CTs) 9 and 17, phase advances were observed, whereas administration at CT 24 resulted in a phase delay, which is in agreement with the previously reported PRC for 4-h dark pulses (Fig. 2) (18). Thus, oxidized quinones can reset the clock in both directions, but the isolated minimal oscillator is limited in its response to this signal.
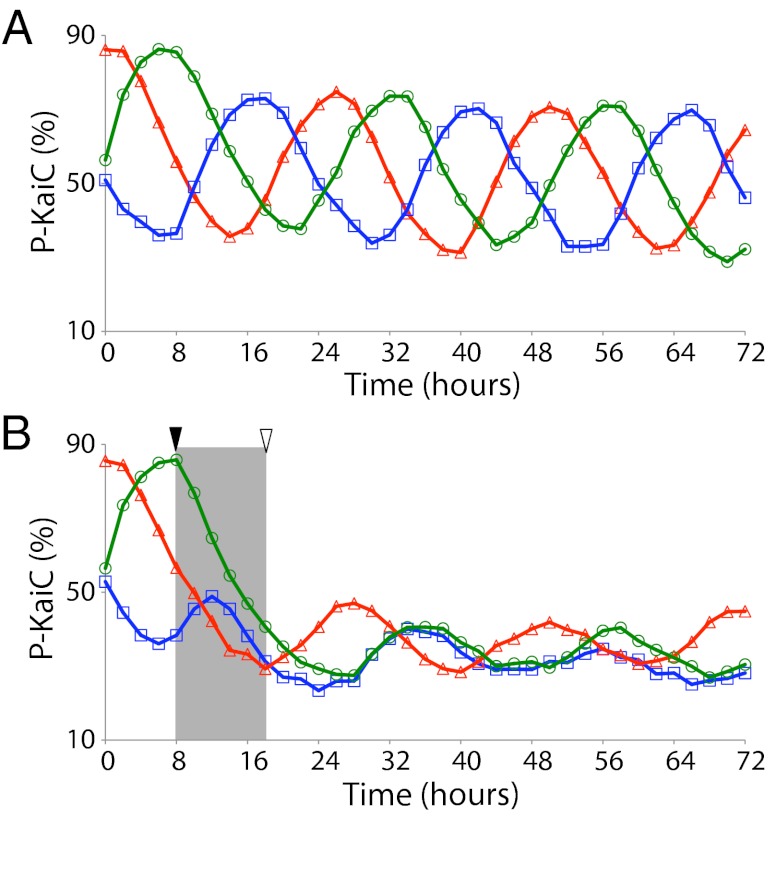
In Vitro Oscillator Mixtures of Different Phases Can Be Synchronized by Applying Oxidized Quinones.
Cyanobacterial cultures that have been entrained to different phases of peak gene expression by offset light–dark (LD) cycles can be synchronized by one 12-h L to 12-h D cycle (19). We tested the ability of oxidized Q0 to mimic the effect of dark synchronization in oscillator mixtures by administering it for 10 h, the duration for which the maximum phase shift was observed in Fig. 1B, to three oscillator mixtures with peaks that were offset by 8 h (Fig. 3A). After the treatment, two, but not all three, of the oscillating rhythms were set to the same phase (Fig. 3B). This result is consistent with the observation that repeated LD cycles are required to change the phase entrainment of a cikA null mutant (14), which is missing one quinone-sensing component.
Fig. 3.
Synchronization of phases in vitro. (A) KaiC phosphorylation in three oscillator mixtures initiated 8 h apart to generate different phases. KaiC phosphorylation was plotted at each time point without addition of oxidized Q0. (B) Same as A except that oxidized Q0 was added (black triangle) simultaneously to all mixtures for 10 h (shaded area). The duration for which the maximum phase shift was observed in Fig. 1B before reduction by dithionite is shown by the open triangle.
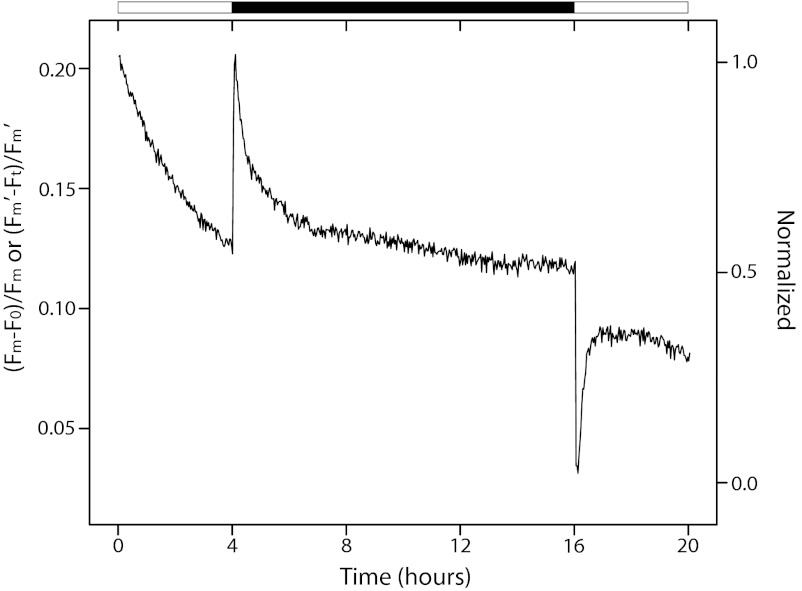
PQ Pool in S. elongatus Is Oxidized on Onset of Darkness.
As previously hypothesized (15), lipid-soluble quinones may interact with KaiA at the periphery of a cellular membrane (20). Thus, the redox state of the PQ pool, which is modulated by light-dependent photosynthetic electron transport, could signal an L/D transition. To test the premise that oxidized quinones signal the onset of darkness, we measured the status of the PQ pool continuously in S. elongatus during LD transitions that simulated diurnal cycling. Redox poise was measured indirectly by monitoring chlorophyll (Chl) fluorescence emission associated with PSII (Fv; variable yield) and total Chl emission intensity using fast repetition rate fluorometry (21) in accordance with previous studies in plants (22). Changes in Chl fluorescence quenching are attributed to the redox state of QA, which is in rapid equilibrium with the PQ pool (23) (SI Text, Fig. S4–S8, Scheme S1–S2). The PQ pool became substantially more oxidized immediately after the LD transition, changing 40.9 ± 3.6% (n = 3) of the total observed range in Fv/Fm (Fig. 4). The redox state of the PQ pool partially recovered quickly and then, slowly became more reduced throughout the 12-h dark period, consistent with the expected transition from photosynthetic to respiratory electron flow through the pool.
Fig. 4.
FRR fluorescence of S. elongatus measured during a simulated diurnal cycle. Illumination was provided by a blue LED at 60 μE m−2 s−1 during hours 0–4 and 16–20 (white boxes). Darkness was given during hours 4–16 (black box). The cell sample was subjected to five 60-μs STFs of saturating intensity every 2 min throughout the entire 20-h experiment. Each data point represents the average of five STFs. The variable fluorescence yield of PSII = (Fm − Fo)/Fm = Fv/Fm was calculated from the raw data (Fig. S5) (21). Data are representative of three biological replicates.
Discussion
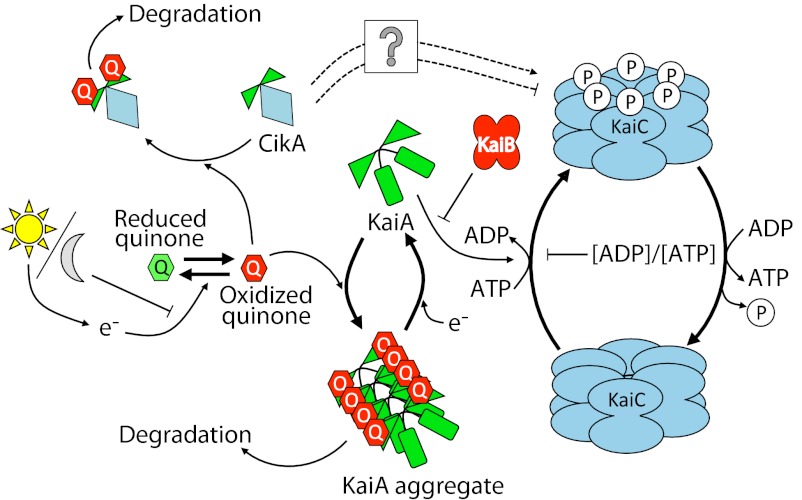
Our data are consistent with a model in which the diurnal L/D solar cycle is sensed by cyanobacteria through the change in electron flux through shared electron carriers of photosynthesis and respiration, which affect both the ATP/ADP ratio and the oxidation state of the PQ pool. The former affects the status of KaiC directly, and the latter affects the ability of KaiA to stimulate KaiC autophosphorylation, providing two sources of metabolic input to the circadian oscillator (Fig. 5).
Fig. 5.
Suggested molecular mechanism of oscillator entrainment by the redox state of quinone. Oxidation and reduction of plastoquinone are controlled by photosynthetic electron transport as a function of light availability and by respiratory electrons in the dark. Oxidized quinone is bound by the pseudoreceiver domain of KaiA and CikA; KaiA aggregates and ceases stimulation of KaiC phosphorylation. The mechanism by which CikA influences KaiC is not known, but the protein is likely degraded on quinone binding (14). The major quinone sensing pathway is emphasized by bold arrows.
As previously reported, manipulation of the ATP/ADP ratio can also reset the phase of KaiC phosphorylation in the in vitro oscillator mixture (12). When cells enter darkness, the ATP/ADP ratio drops gradually rather than acutely and reaches a threshold ratio, which induces a phase shift, after ∼2 h of darkness (12). In contrast, the PQ pool becomes oxidized abruptly on onset of darkness (Fig. 4), and a 2-h administration of oxidized quinones was sufficient to induce a phase shift (Fig. 1B). Together, the quinone oxidation state could provide an acute stimulus marking an L/D transition, whereas the ATP/ADP ratio, changing slowly and steadily, would measure the duration of darkness.
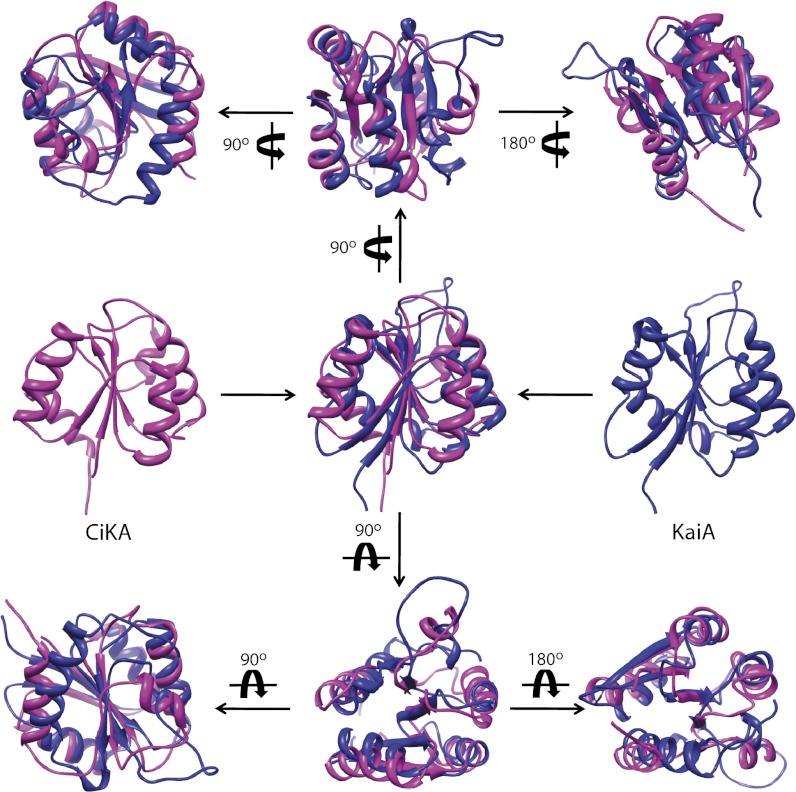
The in vitro oscillator mixture has only the three Kai proteins, and it is not connected to additional components with which the oscillator is likely to interact in vivo, such as CikA, which also is quinone- and L/D-sensitive (14, 24) and necessary for normal resetting of the clock by dark pulses (5). The quinone-induced PRC for the in vitro oscillator mixture is limited relative to the PRC in vivo that is generated either by pulses of darkness or oxidized quinones (Fig. 2). These data may reflect the role of CikA in vivo, which is absent from the isolated oscillator. Consistent with this idea, a cikA mutant requires several L/D cycles to reset and synchronize circadian phase in a cell population compared with a single cycle to reset the WT. Similarly, synchronization of three out-of-phase in vitro oscillator mixtures would require multiple cycles of quinone oxidation reduction (Fig. 3). Overall, the data are consistent with environmental sensing and jetlag in the cikA mutant (5), which occurs at the single-cell level. KaiA and CikA share structural similarity in the pseudoreceiver domain (Fig. 6) (9, 16), and abundance of the CikA protein is affected by the oxidation state of the PQ pool (14). Thus, a possible function of CikA in phase resetting is to work cooperatively with KaiA to generate phase delays in the PRC, which are quinone-induced in vivo but not in the in vitro oscillator mixture (Fig. 2).
Fig. 6.
The superimposed structures of the pseudoreceiver domains of CikA and KaiA. The pseudoreceiver domains of CikA (magenta; Protein Data Bank ID code 2J48) and KaiA (blue; Protein Data Bank ID code 1M2E) were superimposed using the Matchmaker function of UCSF Chamera (27).
The onset of darkness initiates both acute and gradual responses in various metabolites of photosynthesis, and components of the cyanobacterial oscillator have evolved to sense an L/D transition using at least two, the ATP/ADP ratio and the redox state of the quinone pool, through KaiC and KaiA, respectively. Therefore, we propose that the photoreceptive signal transduction input pathway of the cyanobacterial circadian clock is in the well-characterized photosynthesis machinery and the metabolic consequences of its activity.
Materials and Methods
Purification of KaiA and KaiB.
The genes encoding KaiA and KaiB from S. elongatus were amplified using PCR and cloned in-frame with Small ubiquitin-related modifier (SUMO) into the pET-28b expression vector using NdeI and HindIII cloning sites. The resulting plasmids were used to transform Escherichia coli BL21(DE3). Transformed E. coli cultures in log phase in LB at 37 °C were induced to overexpress recombinant KaiA or KaiB with 1 mM isopropyl β-d-thiogalactopyranoside (Calbiochem). Cells were harvested after 6 h, and pellets were resuspended in 50 mM NaCl and 20 mM Tris⋅HCl (pH 7.0). Cell suspensions were passed two times through a chilled French press cell, and lysates were clarified by centrifugation at 20,000 × g for 60 min at 4 °C. Tagged proteins were isolated on an Ni-charged chelating column. Proteases and ATPases were removed by anion-exchange chromatography (buffer A: 20 mM NaCl, 20 mM Tris⋅HCl, pH 7; buffer B: 1 M NaCl, 20 mM Tris⋅HCl, pH 7; gradient: 0–80% buffer B over 80 mL). The SUMO fusion KaiA and KaiB was cleaved after incubation at 4 °C overnight with the Ulp1 protease. KaiA and KaiB were separated from SUMO-His6 tags and uncut proteins by a second passage through an Ni-charged chelating column. All proteins were analyzed for purity by SDS/PAGE and dialyzed against phosphorylation assay buffer (20 mM Tris⋅HCl, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM ATP, pH 8.0). Protein solutions were concentrated, passed through a sterile 0.2-µm filter, and stored at −80 °C. Protein concentrations were determined by using Coomassie Plus-The Better Method Assay Reagent (Pierce).
Purification of KaiC.
The gene encoding KaiC from S. elongatus was amplified using PCR and cloned in-frame with the PreScission Protease (GE Healthcare) cutting site into the pET41a(+) vector (Novagen) between NcoI and XhoI sites; the resulting plasmid was used to transform E. coli BL21(DE3). Transformed E. coli cultures in log phase in LB at 37 °C were cooled to room temperature for 1 h and induced to overexpress recombinant KaiC with 0.1 mM isopropyl β-d-thiogalactopyranoside (Calbiochem). Cells were harvested after 16 h, and pellets were resuspended in 50 mM Tris⋅HCl (pH 7.3) with 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 5 mM ATP. Tagged KaiC proteins were separated from the supernatant fraction on a GSTrap HP column (GE Healthcare) by washing the column with 90 mL buffer. PreScission Protease (GE Healthcare) was used (1 unit/mL in 12 mL) to cut the GST tag. KaiC was separated from the cleaved GST tag, tagged KaiC, and protease by passage a second time through a GSTrap HP column. Purity was analyzed by SDS/PAGE and dialyzed against phosphorylation assay buffer (20 mM Tris⋅HCl, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM ATP, pH 8.0). Protein solution was concentrated, passed through a sterile 0.2-µm filter, and stored at −80 °C. Protein concentrations were determined by using Coomassie Plus-The Better Method Assay Reagent (Pierce).
KaiC Phosphorylation Assay.
The in vitro KaiC phosphorylation assays were performed in sterile 2-mL glass vials in a 30 °C water bath and included KaiA and KaiB at 1.2 µM and 3.5 µM final concentrations in the phosphorylation assay buffer. The in vitro oscillator mixture was divided from the same stock mixture immediately after KaiC (3.5 µM) was added and frozen at −80 °C. Here, 10 mM ATP was used to remove previously reported [ADP]/[ATP] effect on the phase (12). To generate different phases, each reaction mixture was melted at 30 °C at the desired time points. Q0 solution stock (4.8 mM) was prepared by dissolving Q0 into 200-proof ethanol. Dithionite was dissolved into water immediately before use. All of the in vitro reactions were performed under the nitrogen environment to avoid oxygen. Periodically, 20-µL aliquots were removed and denatured at 65 °C for 10 min with 2 µL SDS/PAGE gel loading dye (100 mM Tris⋅HCl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, 400 mM β-mercaptoethanol). A sample (5 µL) of each was loaded onto SDS polyacrylamide gels (4% stacking, 6.5% running) with 15 wells. Densitometry was performed to record KaiC phosphorylation using ImageJ (National Institutes of Health).
Bioluminescence Assay.
S. elongatus AMC 462 is a derivative of a WT isolated PCC 7942 carrying a bacterial luciferase reporter that consists of two neutral site chromosomal insertions: PkaiBC::luxAB at NS1 and PpsbAI::luxCDE at NS2. WT S. elongatus AMC 462 was propagated in BG-11 medium with appropriate antibiotics at 30 °C as described previously (25). A 20-μL drop of Q0 solution (1, 3, 5, or 7 mM) was added to cyanobacteria atop a 300 μL agar pad in the microwell plate used for circadian monitoring at three different time points (CTs 9, 17, and 24). There was no need to add dithionite to reduce Q0 in vivo, because the midpoint potential of Q0 indicates that it will accept electrons from the PQ pool (26). Bioluminescence assays were performed on a Packard TopCount scintillation counter (PerkinElmer Life Sciences) according to a previous protocol (25).
Fast Repetition Rate Fluorometry.
S. elongatus PCC 7942 was grown in BG-11 medium on a 12-h/12-h L/D cycle (100 μE m−2 s−1) at 30 °C in a Photon Systems Instruments FMT 150 photobioreactor (Brno) in turbidostat mode. The culture was mixed by bubbling with humidified air and maintained at OD735 = 0.40 ± 0.01 by automated dilution with fresh medium. Instantaneous fluorescence, Ft, was monitored within the turbidostat every 2 min after a 627-nm measuring pulse (<1 μE m−2 s−1). Chl fluorescence was detected by a built-in positive-intrinsic-negative (PIN) photodiode equipped with 665- to 750-nm bandpass filters.
Fast repetition rate (FRR) fluorometry studies were performed as previously described (21) with the following specific alterations. Aliquots were taken from the turbidostat 4 h before the L to D transition and concentrated to 20 μg Chl mL−1; 50-μL samples (1 μg Chl total) were loaded into the humidified FRR fluorometer sample chamber. During the first 4 h of the experiment, the sample was continuously illuminated at 60 μE m−2 s−1 by a blue light emitting diode (LED). This actinic light source was switched off at t = 4 h and turned on again for 4 h at t = 16 h. This lighting schedule was coordinated to conditions under which the cells were acclimated in the turbidostat. During the entire 20-h experiment, variable Chl fluorescence was monitored by subjecting the sample to five 60-μs single turnover flashes (STFs) at 10 Hz every 2 min. The average light intensity of five STFs over the 2-min period is 0.08 μE m−2 s−1. These conditions are illustrated in Scheme S1. During the periods when the actinic light source was on, fluorescence parameters are defined as light-adapted origin or instantaneous fluorescence (Ft), light-adapted maximum fluorescence (Fm′), and light-adapted variable fluorescence yield [Fv′/Fm′; (Fm′ − Ft)/Fm′]. For the dark period, fluorescence parameters are defined as origin fluorescence (Fo), maximum fluorescence (Fm), and maximal variable fluorescence yield [Fv/Fm; (Fm − Fo)/Fm].
Supplementary Material
Acknowledgments
We thank Drs. J. Bordowitz, S. Cohen, R. Greenspan, N. Keren, and A. LiWang for helpful discussions and S. Kang for technical assistance. D.J.V. is supported by the Department of Defense, Army Research Office through National Defense Science and Engineering Graduate (NDSEG) Fellowship 32CFR168a. Chlorophyll fluorescence assays were supported by Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy Grant DE-FG02-10ER16195 (to G.C.D.). This work was supported by National Institute of General Medical Sciences of the National Institutes of Health Grant R01GM062419 (to S.S.G.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216401109/-/DCSupplemental.
References
- 1.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95(15):8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunlap JC, Loros JJ, DeCoursey PJ, editors. Chronobiology: Biological Timekeeping. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- 3.Devlin PF, Batschauer A. Photoreceptors Resetting the Circadian Clock. Photoreceptors and Light Signalling. Vol 3. Cambridge, UK: Royal Society of Chemistry; 2003. pp. 343–368. [Google Scholar]
- 4.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308(5720):414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289(5480):765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 6.Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278(21):19102–19110. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- 7.Mackey SR, Ditty JL, Zeidner G, Chen Y, Golden SS. Mechanisms for Synchronizing the Cyanobacterial Circadian Clock System with the Environment. Berlin: Springer; 2009. pp. 141–156. [Google Scholar]
- 8.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99(24):15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99(24):15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22(9):2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim YI, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105(35):12825–12830. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rust MJ, Golden SS, O’Shea EK. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science. 2011;331(6014):220–223. doi: 10.1126/science.1197243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pattanayek R, et al. Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J. 2006;25(9):2017–2028. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103(46):17468–17473. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood TL, et al. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci USA. 2010;107(13):5804–5809. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16(3):465–475. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson CH. Phase Response Curves: What Can They Tell Us About Circadian Click? Sapporo, Japan: Hokkaido Univ Press; 1992. pp. 209–249. [Google Scholar]
- 18.Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187(8):2559–2564. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo T, et al. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90(12):5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan J, Kurisu G, Cramer WA. Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex. Proc Natl Acad Sci USA. 2006;103(1):69–74. doi: 10.1073/pnas.0504909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ananyev G, Dismukes GC. How fast can photosystem II split water? Kinetic performance at high and low frequencies. Photosynth Res. 2005;84(1–3):355–365. doi: 10.1007/s11120-004-7081-1. [DOI] [PubMed] [Google Scholar]
- 22.Groom Q, Kramer DM, Crofts AR, Ort DR. The non-photochemical reduction of plastoquinone in leaves. Photosynth Res. 1993;36:205–215. doi: 10.1007/BF00033039. [DOI] [PubMed] [Google Scholar]
- 23.Krause G, Jahns P. In: Chlorophyll a Fluorescence. Papageorgiou GC, Govindjee , editors. Berlin: Springer; 2004. [Google Scholar]
- 24.Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: A component of the circadian clock senses redox state of the cell. EMBO J. 2005;24(6):1202–1210. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackey SR, Ditty JL, Clerico EM, Golden SS. Detection of rhythmic bioluminescence from luciferase reporters in cyanobacteria. Methods Mol Biol. 2007;362:115–129. doi: 10.1007/978-1-59745-257-1_8. [DOI] [PubMed] [Google Scholar]
- 26.Cooley JW, Vermaas WF. Succinate dehydrogenase and other respiratory pathways in thylakoid membranes of Synechocystis sp. strain PCC 6803: Capacity comparisons and physiological function. J Bacteriol. 2001;183(14):4251–4258. doi: 10.1128/JB.183.14.4251-4258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.