Abstract
Variation in relative brain size is commonly interpreted as the result of selection on neuronal capacity. However, this approach ignores that relative brain size is also linked to another highly adaptive variable: body size. Considering that one-way tradeoff mechanisms are unlikely to provide satisfactory evolutionary explanations, we introduce an analytical framework that describes and quantifies all possible evolutionary scenarios between two traits. To investigate the effects of body mass changes on the interpretation of relative brain size evolution, we analyze three mammalian orders that are expected to be subject to different selective pressures on body size due to differences in locomotor adaptation: bats (powered flight), primates (primarily arboreal), and carnivorans (primarily terrestrial). We quantify rates of brain and body mass changes along individual branches of phylogenetic trees using an adaptive peak model of evolution. We find that the magnitude and variance of the level of integration of brain and body mass rates, and the subsequent relative influence of either brain or body size evolution on the brain–body relationship, differ significantly between orders and subgroups within orders. Importantly, we find that variation in brain–body relationships was driven primarily by variability in body mass. Our approach allows a more detailed interpretation of correlated trait evolution and variation in the underlying evolutionary pathways. Results demonstrate that a principal focus on interpreting relative brain size evolution as selection on neuronal capacity confounds the effects of body mass changes, thereby hiding important aspects that may contribute to explaining animal diversity.
Keywords: phylogenetic, comparative method, energetics, trait coevolution
Large brains and advanced cognitive abilities distinguish modern humans from other species, including our closest primate relatives. Consequently, brain size evolution has attracted the attention of generations of scientists (1). However, the human brain is not the largest in absolute mass or volume, but only under consideration of our rather moderate body mass (2–4). Increased “intelligence” is generally attributed to a deviation from a taxon-specific allometric relationship between brain and body (1, 5–7) (Fig. 1). The main interest of studies in the past has thus been to understand which selective forces led to an increase in brain size relative to body size (8–14).
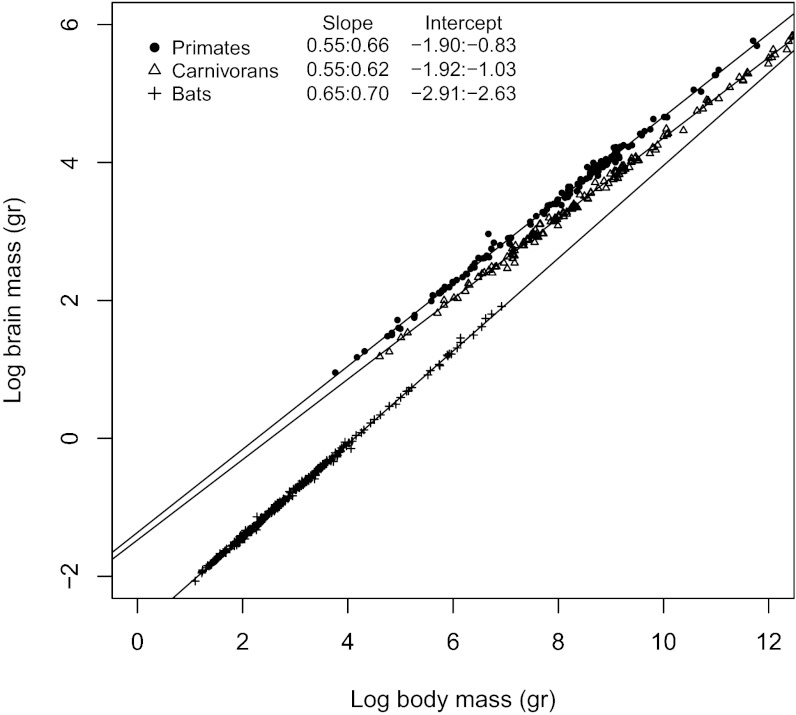
Fig. 1.
Phylogenetic generalized least-squares models of (log) brain to (log) body mass for bats, carnivorans, and primates.
Although the relationship between encephalization and intelligence is intuitive, it is not void of contention (5, 6, 15). Recent research on measures of “general intelligence” in primates has, for example, found more robust correlations with total brain mass than with encephalization (15). The complex relationship between brain mass, body mass, and intelligence has thus been the subject of considerable debate (5, 6, 16), partly because allometric slopes are taxon-specific (17–19). Regardless of these issues, deviations from the general allometric brain–body relationship continue to be commonly interpreted as a result of ecological, behavioral, or social selection pressures directly acting on neuronal capacity (18, 20, 21). Moreover, brain and body size have been suggested to be subject to different (but not mutually exclusive) selective forces (22). Whereas brain size is subject to behavioral pressures that stabilize energy input and/or meet the computational demands to survive in novel environments, body size is subject to lifestyle pressures that increase birth rate (e.g., by exploiting abundant and reliable food resources) and decrease death rate (e.g., by reducing predation risk) (23, 24). How strong selective forces act on brain size relative to body size is central to the interpretation of relative brain size; thus, when interpreting deviations from the brain–body allometry, it should be natural to consider the evolutionary patterns of change for both brain and body mass equally.
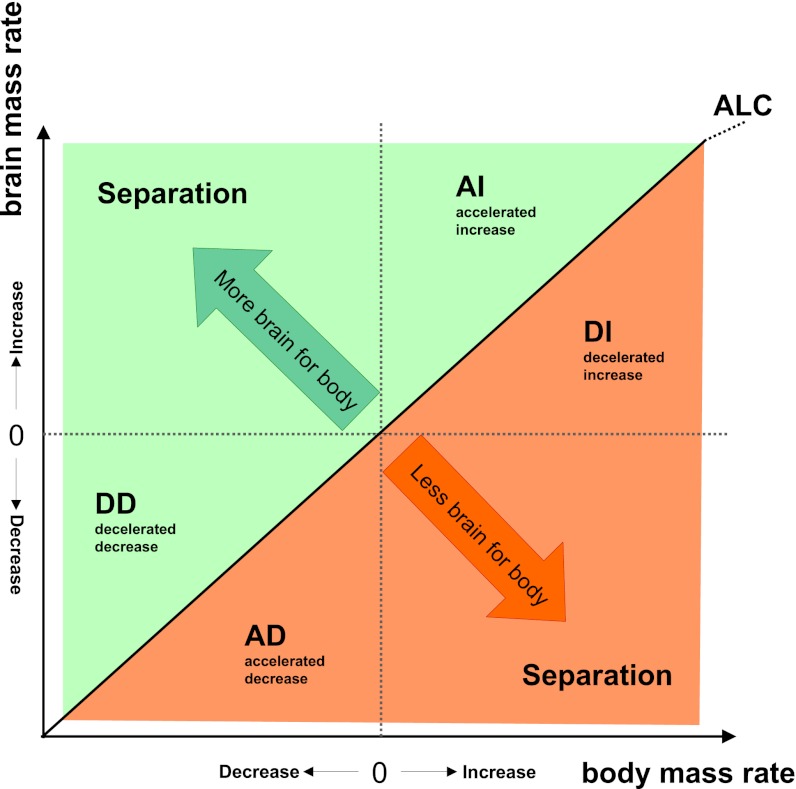
We consider that, as with any combination of two traits, the relative change of one (e.g., brain mass) compared with the other (e.g., body mass) along a single evolutionary branch (from ancestor to descendant) can be attained by several processes related to their combined increase and/or decrease (Fig. 2). For example, brain mass can increase relative to body mass (Fig. 2, green) if brain mass increases relatively more than body mass [accelerated increase (AI)], brain mass decreases relatively less than body mass [decelerated decrease (DD)], or brain mass increases while body mass decreases (separation). Vice versa, brain mass can decrease relative to body mass (Fig. 2, red) when brain mass increases relatively less than body mass [decelerated increase (DI)], brain mass decreases relatively more than body mass [accelerated decrease (AD)], or brain mass decreases while body mass increases (separation). Only a minority of these evolutionary pathways involve pure brain size changes. Allometric relationships reveal taxon-specific patterns of coevolution but fail to reveal which of these processes underlie species’ residual deviation from the general allometric trend. The traditional principal focus on allometry to study trait coevolution may thus hide potentially important adaptive information on the nature of the coevolutionary process.
Fig. 2.
Schematic diagram of the relationship of possible evolutionary changes in two traits (body and brain mass). Body and brain mass are measured as rates of change between all ancestor-descendant pairs. Possible scenarios are plotted with respect to the y axis (rate of change in brain mass). The green area indicates increasing relative brain mass, and the orange area indicates relative brain mass reduction. ALC, allometric change. Within each quadrant, a distinction can be made as to which trait (X or Y) increases or decreases more or less than the other, reflecting all possible evolutionary scenarios of change between traits. An important attribute of this approach is that the isometric line represents the allometric relationship between the traits under investigation, independent of the value of the allometric coefficient. Allometric change (ALC), indicates the line along which the quotient of the rates of change remains equal. Deviations from isometry in this figure therefore represent deviations from the allometric relationship between the traits under investigation (Fig. 3). Possible patterns of relative brain size change include that brain and body mass change in: (i) opposing directions (separation); (ii) the same direction but with brain changing faster than body mass (AI and AD); (iii) as (ii) but with body changing faster than brain mass (DI and DD). For closely related traits (e.g., brain and body mass), increased vs. decreased separation mostly occurs on sister branches: When a trait indicates a burst of change in one branch, the sister branch often indicates a smaller opposite trend.
To determine the processes that underlie changes in relative brain size, we quantify evolutionary rates of both brain and body mass, considering all possible evolutionary scenarios (Fig. 2). We model the evolution of brain and body mass independently and investigate the relative changes of each trait on individual branches of a phylogenetic tree using an adaptive peak (AP) model (25), including fossil values, to increase the accuracy of model predictions (SI Text, Fig. S1, and Table S1). An important attribute of our approach is that the isometric line in the brain-rate against body-rate plot (Fig. 2) represents the allometric relationship between the traits under investigation, irrespective of the value of the allometric coefficient (Fig. 3). Deviations from isometry in Fig. 2 therefore represent deviations from allometry between the traits under investigation (Fig. 3).
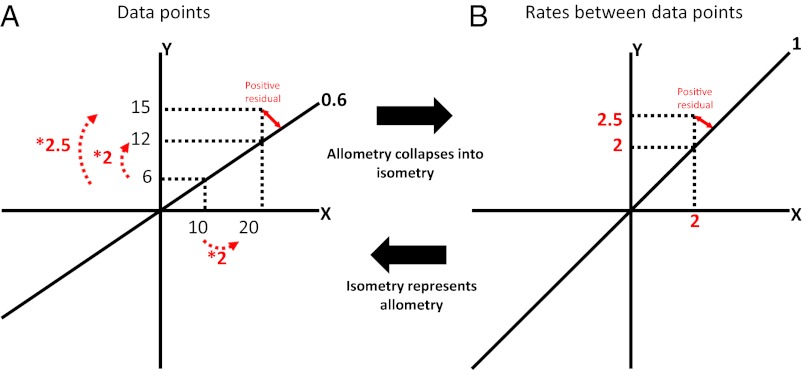
Fig. 3.
Schematic explanation of how the allometric slope of the brain–body relationship collapses into the isometric line when plotting rates of change between data points (B) rather than actual data points (A). Two data points are hypothesized with Y and X values of 6, 10 and 12, 20, respectively, producing an allometric slope of 0.6. Because the allometric slope describes the proportional change from Y to X, plotting rates of change between data points rather than the actual data points results in a collapse of the allometric line into isometry. Equally, positive or negative residuals relative to the allometric line in the “data points plot” (A) are represented as positive or negative (orthogonal) residuals, respectively, relative to the isometric line in the “rates plot” (B). This assumes that the quantification of a rate has the same properties as a proportion (an explanation for how this is true for our model is provided in figure 1 in ref. 25).
Against the background that deviations from the general brain–body allometry are commonly interpreted as selection on neuronal capacity, we investigate three mammalian orders that are expected to be the subject of different selective pressures on body mass due to differences in their locomotor adaptations: bats, which engage in powered flight; primates, which are primarily arboreal but with some terrestrial taxa; and carnivorans, which are primarily terrestrial but with some semifossorial, arboreal, and aquatic taxa. We also contrast echolocating and nonecholocating (fruit) bats; (part-)terrestrial and arboreal primates; and terrestrial, arboreal, and aquatic carnivorans separately due to the differences in selective pressures on body mass evolution expected between those groups. These comparisons will allow us to draw conclusions on the generality, or lack thereof, of the evolutionary processes and adaptive responses involved in changes of relative brain size in mammals. Generally, we expect to see differences between taxa in the relationship between brain and body size evolution, reflecting the varying constraints and demands that different lifestyles, including differences in locomotion, exert on both brain and body size. We further expect some phyletic variation in relative brain size to be driven by variation in body mass rather than brain mass.
Results and Discussion
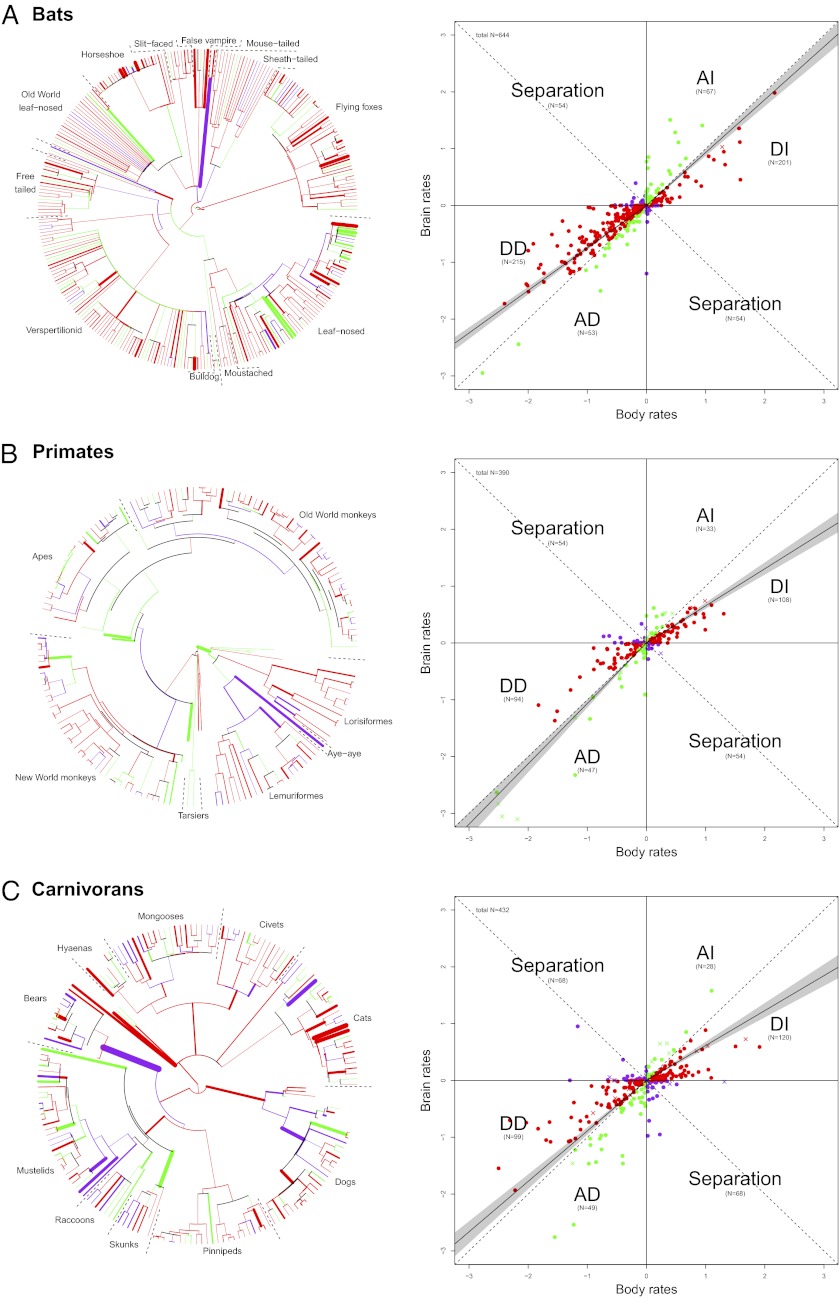
Results show that in all three mammalian orders, brain mass primarily indicates concerted increase or decrease with body mass in the majority of lineages (bats: 536 of 644, primates: 282 of 390, and carnivorans: 296 of 432). The majority of branches indicate a decelerated rate of brain mass change compared with body mass, suggesting that body mass significantly outpaced brain mass changes in most lineages (in line with the allometric relationships indicating significant hyposcaling of brain to body size; Fig. 1). Our analysis, however, also suggests that the relationship between brain and body mass changes is far from being a simple and general principle. First, we find differences between the relationship of brain and body mass changes depending on whether the brain and body mass are increasing or decreasing, resulting in different patterns of symmetry in some taxa (Fig. 4; Fig. S2 presents a high-resolution image of Fig. 4, including species names, and Fig. S3 is based on extant species only). Second, when quantifying differences in the relationship between the rates of brain and body mass changes within and between orders, we find differences in the relative magnitude and variance by which either brain or body mass changes deviate from allometry (Fig. 5 and Tables S2 and S3). These results suggest that the brain–body mass relationship is driven by different mechanisms in different lineages.
Fig. 4.
Independent reconstruction of brain and body mass according to an AP model. Red data points represent branches where body mass change (increase and decrease) was greater than brain mass change (crosses indicate branches leading to fossils, closed circles indicate branches leading to extant species), corresponding to red branches in the phylogenies. Green data points represent branches with brain mass changes greater than body mass changes, corresponding to green branches in the phylogenies. Purple data points represent branches that indicate separation (increase in body size and decrease in brain size, or vice versa), corresponding to purple branches in the phylogenies. The solid line in the scatter plot indicates the regression line of an RMA correlation forced through the origin based on the branches of the upper right and lower left quadrants separately with the 95% confidence interval of the slopes indicated as the shaded gray area (values presented in Table S2). Numbers in brackets are the number of branches in each category. The thickness of branches in the tree represents the deviation from the isometric line (larger residuals have thicker branches). Bats (including fossils) (A), primates (including fossils) (B), and carnivorans (including fossils) (C) are shown.
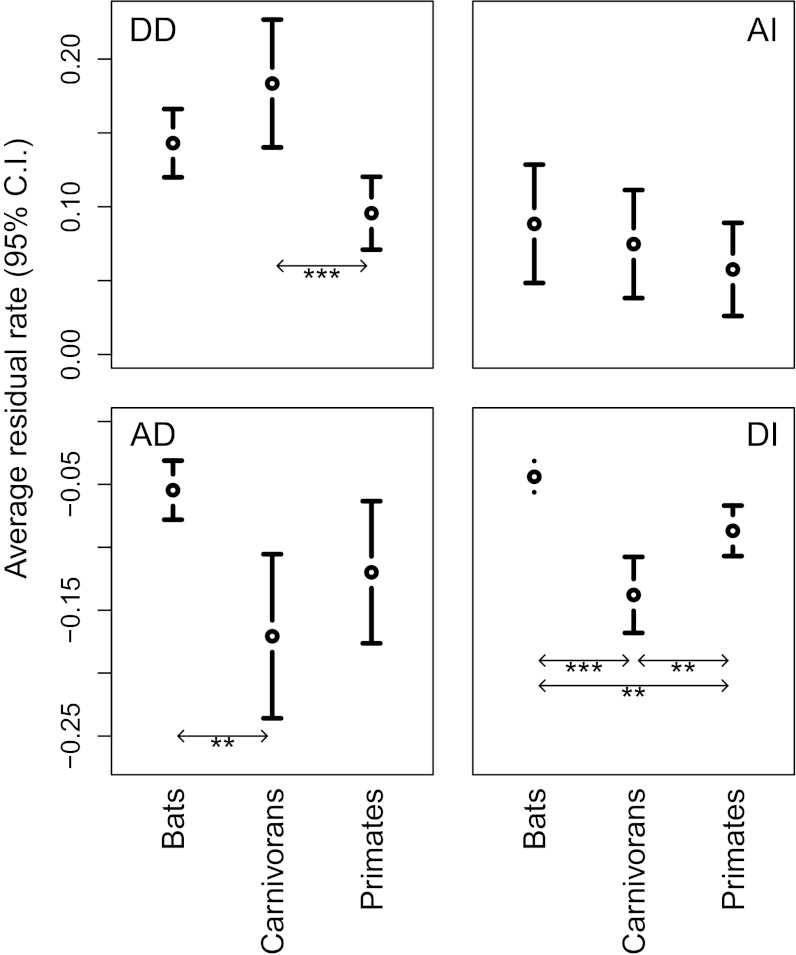
Fig. 5.
Mean and 95% confidence interval (C.I.) of major axis residuals (positive residuals for AI and DD and negative residual values for DI and AD) relative to the isometric line. These values represent the magnitude and variance in the deviation from allometry for each evolutionary scenario (AI, DI, AD, and DD) quantifying “evolvability” (deviations from allometry) in the different mammalian orders. *P < 0.05; **P < 0.01; ***P < 0.001.
Of the three investigated orders, bats are characterized by the strongest deviation from underlying allometric scaling when both brain and body mass decrease and by the weakest deviation when both brain and body mass increase (Fig. 4 and Figs. S2 and S3). Body mass reduction in bats clearly outpaces brain mass reduction, leading to an increased relative brain size (DD). When both traits increase, however, both tend to contribute equally to the brain–body relationship. Overall, this leads to a higher constraint on a decrease in relative brain size compared with primates or carnivorans, which is supported by the fact that bats show significantly lower negative residuals relative to isometry in both size increase and decrease scenarios compared with primates or carnivorans (Fig. 5; detailed results are presented in Table S3). In consequence, compared with primates and carnivorans, when both traits increase, the rate in body mass change is much less pronounced in bats, although marginally more variable than in brain mass (Fig. 4 and Table S2). The independent evolvability (i.e., deviations of brain or body mass changes from allometry) of body mass relative to brain mass in bats is thus higher when both are decreasing, leading to an overall pattern of reductions in body mass driving variability in relative brain size during phyletic dwarfing. For bats, a decrease in body mass, with everything else being equal, can lead to a decrease in physiological costs of powered flight and results in increased maneuverability, allowing bats to forage in cluttered space (26, 27). At the same time, navigation and orientation in cluttered environments are sensorially, and thus neurologically, relatively more demanding, preventing bats from equally high rates of brain mass reduction during dwarfing (14, 28). Overall, bats thus indicate higher evolvability of increased relative brain size (Fig. 4 indicates a more pronounced trend toward DD vs. AD and toward AI vs. DI; detailed results are presented in Table S2) compared with primates and carnivorans, a trend that is reflected in their elevated allometric exponent (Fig. 1). It is interesting to note in this context that in the larger nonecholocating fruit bats, body mass rates do tend to outpace brain mass rates when both increase (Fig. S4), confirming the logical expectation that the energetic constraints imposed by increasing body mass in aerial species will be strongest in larger taxa (29).
Primates stand out compared with bats and carnivorans in that they show the lowest degree of independent evolvability of body mass relative to brain mass when both decrease (Fig. 5, DD scenario). In fact, rates of change in brain mass marginally outpace rates of change in body mass when both decrease (a more pronounced trend toward AD vs. DD; Figs. 4 and 5; detailed results are provided in Tables S2 and S4). In contrast, and as in carnivorans, body mass changes outpace brain mass when both increase (Fig. 4 and Table S2). Variation in relative brain size is thus inferred to be driven primarily by variation in brain mass during phyletic dwarfing, but primarily by variation in body mass in lineages where overall size is increasing. It is important to note that both a separate analysis of species with a terrestrial element of locomotion (Fig. S5) and the exclusion of fossil taxa from the original dataset (Fig. S5) result in a symmetrical pattern, where changes in body mass outpace changes in brain mass when lineages increase in size (DI > AI) as well as when they decrease in size (DD > AD). The relative constraint on the evolvability of body mass in arboreal primates during phyletic dwarfing may be the result of competitive exclusion (e.g., from birds) and of fundamental biological adaptations to fill an arboreal niche at a medium body size (30, 31), resulting in a lower barrier to body mass reduction. There is some support for this suggestion from a plot of lineage-specific rates against ancestral node values, which suggests that lineages derived from ancestral taxa with higher body mass values evolved with a higher variability of negative rates than lineages derived from ancestral taxa with lower body mass values, for which negative rates were largely restricted to lower values (Fig. S6). It should be noted, however, that an earlier analysis of patterns of body mass change restricted to the primate fossil record failed to detect any such trend (32). Alternatively, if phyletic dwarfing is a response to resource limitations, it may be that the generally larger brain mass of primates leads to a faster reduction in brain mass compared with other mammalian orders because of the primate brains’ larger relative contribution to species-specific metabolic rates. This constraint may be less important in species with a terrestrial element of locomotion due to the lower metabolic cost of horizontal compared with vertical locomotion and the reduced need for climbing (33, 34).
As a group, carnivorans display high levels of independent evolvability of body mass relative to brain mass compared with bats and primates (Fig. 4; comparison of homogeneity of variance: carnivorans vs. bats, P < 0.0001; carnivorans vs. primates, P < 0.0001; and bats vs. primates, P = 0.212), corresponding to previous studies demonstrating diverse encephalization allometries across carnivorans (18). When both brain and body mass increase, carnivorans demonstrate significantly higher negative than positive residuals relative to isometry (Fig. 5; detailed results are provided in Table S4), and also demonstrate significantly higher negative residuals than both primates and bats (Fig. 5, DI scenario), which suggests a pronounced influence of body mass rates on the brain–body relationship. In the case of phyletic dwarfing, rates of change in brain and body mass are relatively balanced (although more variable than in primates and bats). Phyletic changes in encephalization thus appear to have evolved mainly through selection for body size in cases of overall size increase. These general carnivoran trends are also expressed in separate analyses of terrestrial and arboreal carnivorans (Fig. S7 A, B, D, and E). In aquatic species (Fig. S7 C and F), the influence of body mass change on the brain–body relationship is more pronounced than in arboreal and terrestrial lineages, with sevenfold more lineages showing body size selection than those demonstrating selection for brain size, and the branch leading to elephant seals displaying a particularly strong trend in this direction. However, brain evolution drives the signal in a few unusual aquatic carnivorans, specifically the walrus and the river otter. These analyses are based on species averages for pinnipeds, but results were similar when males and females of this sexually dimorphic group were analyzed. Pinnipeds are rarely included in analyses of carnivoran brain evolution. This extensive analysis suggests that the brain–body relationship in pinnipeds is driven much more by body size evolution than in their terrestrial and arboreal relatives, which may also reflect the extreme sexual dimorphism in body size observed in some species (35). Our results suggest that for most carnivorans, changes in relative brain size reflect, in large part, body size evolution rather than selection for neuronal capacity. It has been similarly suggested that changes in body size may drive life history evolution in carnivorans (35, 36), and the framework presented here will allow for rigorous testing of this and other hypotheses concerning the mechanisms underlying macroevolutionary trends.
Whether the inclusion of fossils adds valuable information to macroevolutionary studies of brain and body size evolution or not remains a moot point. Fossil specimens can add valuable information to reconstructions of phylogenetic relationships and macroevolutionary patterns by providing primary data on the tempo of evolution (37) and introducing chronological depth into a sample, as well as an element of control of their accuracy; however, including fossils of uncertain phylogenetic affinity and age can also be problematic. In addition, body mass is difficult to infer from fossils (38, 39). For example, three of the fossil specimens included in our primate sample are clear outliers and exert a particularly strong influence on the regression model for decreasing rates (Parapithecus grangeri, brain rate: −3.10; Catopithecus browni, brain rate: −3.05; and Chilecebus caracoensis, brain rate: −2.84). This may reflect a systematic overestimation of body mass in extinct species. Alternatively, there may be a fundamental difference in the relationship between brain and body mass evolution in extinct (or perhaps long extinct) species compared with extant species (Fig. S6), particularly because relative brain size is reported to have increased independently in different primate lineages through time (40). If so, exclusion of fossil taxa may yield misleading reconstructions of the evolutionary patterns and mechanisms leading to the diversity observed in the modern biota. Exploration of these potential issues is critical and deserves further analysis, and particularly more (and more accurate) fossil data.
In conclusion, we find that the magnitude and variance by which either brain or body mass changes deviate from allometry differ between orders and subgroups within orders, suggesting that the relationship between brain and body mass is driven by different mechanisms in different lineages. It is clear that comparative correlations involving relative brain size cannot be interpreted as selection on neuronal capacity alone. Relative brain size is the compromise of two traits taking potentially different evolutionary pathways involving different combinations of brain–body adaptations. Hypotheses based on simple one-way tradeoff mechanisms are unlikely to explain fully the evolution of relative brain size, and the predominant use of relative brain size as a proxy for intelligence thus inherently masks the different possible evolutionary pathways underlying adaptations in body mass. Traditional allometric approaches, however, cannot provide information on the mechanisms underlying species’ deviations from general allometric trends. By comparing rates of evolution along individual branches of a phylogenetic tree, our approach allows a more detailed interpretation of all possible pathways underlying phenotype coevolution, supplementing traditional allometric approaches.
Materials and Methods
Data.
Brain and body mass data were collected for 293 extant bats, 4 fossil bats, 146 extant primates, 23 fossil primates, 162 extant carnivorans, and 25 fossil carnivorans (Table S5). All data are species-level (mixed-sex). The bat phylogeny comes from Bininda-Emonds et al. (41); the primate phylogeny comes from the 10k Trees Project (version 3) (42); and the carnivoran phylogeny is taken from the study by Nyakatura and Bininda-Emonds (43), with fossil carnivorans placed according to references in the study by Finarelli and Flynn (18). Polytomies (the bat phylogeny was 79.7% resolved, the primate phylogeny was 98.6% resolved, and the carnivoran phylogeny was 98.0% resolved) were resolved randomly [using the Analyses of Phylogenetics and Evolution R-package (44)], and a branch length of 0.01 was assigned to the new branches. Primate summary data on arboreality and terrestriality come from Rowe (45).
Inferring Branch-Specific Rates of Evolution.
We used an AP model to estimate rates of evolution of brain and body mass for each branch in a phylogenetic tree. AP models are preferable when modeling traits that are subject to multiple selective pressures because they allow independent rate estimation for individual branches. This overcomes the problem of “inherited maladaptation” (due to the averaging of inferred changes between sister branches) inherent to more traditionally used Brownian motion-based methods (46). Moreover, the AP model collapses into Brownian motion and Ornstein–Uhlenbeck models under relevant conditions (47), and can therefore be considered more flexible with less stringent data assumptions (25). We use the AP-based method of independent evolution (IE) (25), which has been shown to estimate body sizes of extinct primates more accurately than other methods. Because the IE algorithm already includes a proportional distance metric between data points (with equivalent properties to the log-scale), data are not logged before analysis. The IE method infers positive rates for trait increase and negative rates for trait decrease, allowing modeling of trait covariation as suggested in Fig. 2. Rates can be interpreted as proportional changes over time and refer to the (proportional) amount of change that has occurred along the entire length of a branch. More information on AP vs. Brownian motion and Ornstein–Uhlenbeck models of evolution and the use of the IE method can be found in SI Text, Fig. S1, and Table S1.
Analysis.
With rates of evolution of brain mass and body mass for individual branches of a phylogenetic tree as our primary data, we calculated the slope of two reduced major axis (RMA) regressions forced through the origin, once for all the branches falling in the upper right quadrant containing the AI and DI processes and once for the branches falling in the lower left quadrant (DD and AD; Fig. 2). The RMA slopes and 95% confidence intervals indicated whether rates were biased toward acceleration (∞ > slope > 1) or deceleration (0 < slope < 1) of changes in brain mass relative to those in body mass. Because the phylogenies used are not fully resolved for our samples, we investigated whether resolving polytomies randomly affected our results by repeating the RMA analyses 1,000 times, each time resolving polytomies randomly (results are presented in Table S2). This analysis reveals that our procedure to resolve polytomies randomly does not significantly alter our results, due, in part, to the fact that our approach quantifies rates of evolution along an entire branch rather than per unit of time for each branch (the latter would necessarily lead to rate inflation for branches <1).
To quantify the level of independent evolutionary change between brain and body mass rates further, we computed t tests (using Tukey’s honestly significant difference to adjust P values for multiple comparisons) and ANOVA on the major axis residuals relative to the isometric line in Fig. 2. This procedure captures significant differences in the magnitude (t test) and variance (F-ratio test) of the deviation from allometry for each evolutionary scenario. High magnitude and variance in residual values are interpreted as high independent evolvability (high deviations from allometry), and low magnitude and variance in residual values are interpreted as low independent evolvability (low deviations from allometry). Tests were performed within and between orders to provide a complete picture on putative phylogenetic specializations.
Model Accuracy and Incorporation of Fossil Values.
To increase accuracy of model predictions, we incorporated estimates for body and brain mass from fossils (Table S5). Analyses were performed with and without the inclusion of fossils to test the reliability of model predictions. Overall results did not differ significantly between the fossil and extant models, with a few exceptions described in the main text, because the extant model provided an accurate range for fossil values, further demonstrating the accuracy of the AP-based method and the robustness of the results.
Supplementary Material
Acknowledgments
We thank K. Isler, A. Lindholm, and the other organizers of the CooPeerAction meeting in 2011 in Gitschenen, Switzerland. This work was supported by the UK Natural Environment Research Council (Grant NE/H022937/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212181109/-/DCSupplemental.
References
- 1.Roth G, Dicke U, Rotha G. Evolution of the brain and intelligence. Trends Cogn Sci. 2005;9:250–257. doi: 10.1016/j.tics.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Schoenemann PT. Brain size scaling and body composition in mammals. Brain Behav Evol. 2004;63:47–60. doi: 10.1159/000073759. [DOI] [PubMed] [Google Scholar]
- 3.Harvey PH, Krebs JR. Comparing brains. Science. 1990;249:140–146. doi: 10.1126/science.2196673. [DOI] [PubMed] [Google Scholar]
- 4.Martin RD. Relative brain size and basal metabolic rate in terrestrial vertebrates. Nature. 1981;293:57–60. doi: 10.1038/293057a0. [DOI] [PubMed] [Google Scholar]
- 5.Van Valen L. Brain size and intelligence in man. Am J Phys Anthropol. 1974;40:417–423. doi: 10.1002/ajpa.1330400314. [DOI] [PubMed] [Google Scholar]
- 6.Willerman L, Schultz R, Rutledge JN, Bigler ED. In vivo brain size and intelligence. Intelligence. 1991;15:223–228. [Google Scholar]
- 7.Lefebvre L, Reader SM, Sol D. Brains, innovations and evolution in birds and primates. Brain Behav Evol. 2004;63:233–246. doi: 10.1159/000076784. [DOI] [PubMed] [Google Scholar]
- 8.Harvey PH, Clutton-Brock TH, Mace GM. Brain size and ecology in small mammals and primates. Proc Natl Acad Sci USA. 1980;77:4387–4389. doi: 10.1073/pnas.77.7.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar RIM. The social brain hypothesis. Evol Anthropol. 1998;6:178–190. [Google Scholar]
- 10.Oxnard CE. Brain evolution: Mammals, primates, chimpanzees, and humans. Int J Primatol. 2004;25:1127–1158. [Google Scholar]
- 11.Sol D, Lefebvre L, Rodríguez-Teijeiro JD. Brain size, innovative propensity and migratory behaviour in temperate Palaearctic birds. Proc Biol Sci. 2005;272:1433–1441. doi: 10.1098/rspb.2005.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar RIM, Shultz S. Understanding primate brain evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362:649–658. doi: 10.1098/rstb.2006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor AB, van Schaik CP. Variation in brain size and ecology in Pongo. J Hum Evol. 2007;52:59–71. doi: 10.1016/j.jhevol.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Dechmann DKN, Safi K. Comparative studies of brain evolution: A critical insight from the Chiroptera. Biol Rev Camb Philos Soc. 2009;84:161–172. doi: 10.1111/j.1469-185X.2008.00067.x. [DOI] [PubMed] [Google Scholar]
- 15.Deaner RO, Isler K, Burkart J, van Schaik C. Overall brain size, and not encephalization quotient, best predicts cognitive ability across non-human primates. Brain Behav Evol. 2007;70:115–124. doi: 10.1159/000102973. [DOI] [PubMed] [Google Scholar]
- 16.Deaner RO, Nunn CL, van Schaik CP. Comparative tests of primate cognition: Different scaling methods produce different results. Brain Behav Evol. 2000;55:44–52. doi: 10.1159/000006641. [DOI] [PubMed] [Google Scholar]
- 17.Martin RD, Genoud M, Hemelrijk CK. Problems of allometric scaling analysis: Examples from mammalian reproductive biology. J Exp Biol. 2005;208:1731–1747. doi: 10.1242/jeb.01566. [DOI] [PubMed] [Google Scholar]
- 18.Finarelli JA, Flynn JJ. Brain-size evolution and sociality in Carnivora. Proc Natl Acad Sci USA. 2009;106:9345–9349. doi: 10.1073/pnas.0901780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisbecker V, Goswami A. Brain size, life history, and metabolism at the marsupial/placental dichotomy. Proc Natl Acad Sci USA. 2010;107:16216–16221. doi: 10.1073/pnas.0906486107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappelman J. The evolution of body mass and relative brain size in fossil hominids. J Hum Evol. 1996;30:243–276. [Google Scholar]
- 21.Pérez-Barbería FJ, Gordon IJ, Pagel M. The origins of sexual dimorphism in body size in ungulates. Evolution. 2002;56:1276–1285. doi: 10.1111/j.0014-3820.2002.tb01438.x. [DOI] [PubMed] [Google Scholar]
- 22.Navarrete A, van Schaik CP, Isler K. Energetics and the evolution of human brain size. Nature. 2011;480:91–93. doi: 10.1038/nature10629. [DOI] [PubMed] [Google Scholar]
- 23.Sibly RM, Brown JH. Effects of body size and lifestyle on evolution of mammal life histories. Proc Natl Acad Sci USA. 2007;104:17707–17712. doi: 10.1073/pnas.0707725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JH, Sibly RM. Life-history evolution under a production constraint. Proc Natl Acad Sci USA. 2006;103:17595–17599. doi: 10.1073/pnas.0608522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smaers JB, Vinicius L. Inferring macro-evolutionary patterns using an adaptive peak model of evolution. Evol Ecol Res. 2009;11:991–1015. [Google Scholar]
- 26.Norberg RA. Temporary weight decrease in breeding birds may result in more fledged young. Am Nat. 1981;118:838–850. [Google Scholar]
- 27.Norberg UM. In: Ecological Morphology, Integrative Organismal Biology. Wainwright PC, Reilly SM, editors. Chicago: Univ of Chicago Press; 1994. pp. 205–239. [Google Scholar]
- 28.Safi K, Seid MA, Dechmann DKN. Bigger is not always better: When brains get smaller. Biol Lett. 2005;1:283–286. doi: 10.1098/rsbl.2005.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNab BK, Eisenberg JF. Brain size and its relation to the rate of metabolism in mammals. Am Nat. 1989;133:157–167. [Google Scholar]
- 30.Soligo C, Martin RD. Adaptive origins of primates revisited. J Hum Evol. 2006;50:414–430. doi: 10.1016/j.jhevol.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Soligo C, Müller AE. Nails and claws in primate evolution. J Hum Evol. 1999;36:97–114. doi: 10.1006/jhev.1998.0263. [DOI] [PubMed] [Google Scholar]
- 32.Soligo C. Correlates of body mass evolution in primates. Am J Phys Anthropol. 2006;130:283–293. doi: 10.1002/ajpa.20298. [DOI] [PubMed] [Google Scholar]
- 33.Hanna JB, Schmitt D. Locomotor energetics in primates: Gait mechanics and their relationship to the energetics of vertical and horizontal locomotion. Am J Phys Anthropol. 2011;145:43–54. doi: 10.1002/ajpa.21465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor CR, Caldwell SL, Rowntree VJ. Running up and down hills: Some consequences of size. Science. 1972;178:1096–1097. doi: 10.1126/science.178.4065.1096. [DOI] [PubMed] [Google Scholar]
- 35.Fitzpatrick JL, et al. Sexual selection uncouples the evolution of brain and body size in pinnipeds. J Evol Biol. 2012;25:1321–1330. doi: 10.1111/j.1420-9101.2012.02520.x. [DOI] [PubMed] [Google Scholar]
- 36.Webster AJ, Gittleman JL, Purvis A. The life history legacy of evolutionary body size change in carnivores. J Evol Biol. 2004;17:396–407. doi: 10.1046/j.1420-9101.2003.00664.x. [DOI] [PubMed] [Google Scholar]
- 37.Simpson GG. Tempo and Mode in Evolution. New York: Columbia Univ Press; 1944. [DOI] [PubMed] [Google Scholar]
- 38.Strait SG. Dietary reconstruction of small-bodied omomyoid primates. Journal of Vertebrate Paleontology. 2001;21:322–334. [Google Scholar]
- 39.Parr WCH, Chatterjee HJ, Soligo C. Inter- and intra-specific scaling of articular surface areas in the hominoid talus. J Anat. 2011;218:386–401. doi: 10.1111/j.1469-7580.2011.01347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montgomery SH, Capellini I, Barton RA, Mundy NI. Reconstructing the ups and downs of primate brain evolution: Implications for adaptive hypotheses and Homo floresiensis. BMC Biol. 2010;8:9. doi: 10.1186/1741-7007-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bininda-Emonds ORP, et al. The delayed rise of present-day mammals. Nature. 2008;456:274. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- 42.Arnold C, Matthews LJ, Nunn CL. The 10k Trees website: A new online resource for primate phylogeny. Evol Anthropol. 2010;19:114–118. [Google Scholar]
- 43.Nyakatura K, Bininda-Emonds OR. Updating the evolutionary history of Carnivora (Mammalia): A new species-level supertree complete with divergence time estimates. BMC Biol. 2012;10:12. doi: 10.1186/1741-7007-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 45.Rowe N. The Pictorial Guide to the Living Primates. New York: Pogonias Press; 1996. [Google Scholar]
- 46.Hansen TF, Orzack SH. Assessing current adaptation and phylogenetic inertia as explanations of trait evolution: The need for controlled comparisons. Evolution. 2005;59:2063–2072. [PubMed] [Google Scholar]
- 47.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.