Abstract
Ikaros is a zinc finger-containing DNA-binding protein that plays a pivotal role in immune homeostasis through transcriptional regulation of the earliest stages of lymphocyte ontogeny and differentiation. Functional deficiency of Ikaros has been implicated in the pathogenesis of acute lymphoblastic leukemia, the most common form of childhood cancer. Therefore, a stringent regulation of Ikaros activity is considered of paramount importance, but the operative molecular mechanisms responsible for its regulation remain largely unknown. Here we provide multifaceted genetic and biochemical evidence for a previously unknown function of spleen tyrosine kinase (SYK) as a partner and posttranslational regulator of Ikaros. We demonstrate that SYK phoshorylates Ikaros at unique C-terminal serine phosphorylation sites S358 and S361, thereby augmenting its nuclear localization and sequence-specific DNA binding activity. Mechanistically, we establish that SYK-induced Ikaros activation is essential for its nuclear localization and optimal transcription factor function.
Keywords: systems immunobiology, bioinformatics, site-directed mutagenesis, signaling, molecular modeling
Ikaros (IK) is a zinc finger (ZF)-containing sequence-specific DNA-binding protein that plays a pivotal role in immune homeostasis through transcriptional regulation of the earliest stages of lymphocyte ontogeny and differentiation by both (i) gene transcriptional activation via efficient transcription initiation and elongation, and (ii) gene repression (1–6). IK also exhibits a tumor-suppressor function in lymphocyte precursors (1–4, 7, 8). Functional deficiency of IK because of expression of non-DNA binding dominant-negative IK isoforms caused by aberrant splicing (9) or genomic mutations (10) has been detected in leukemic lymphocyte precursors from patients with acute lymphoblastic leukemia (ALL), the most common form of childhood cancer. Currently, our knowledge regarding the upstream regulators of IK function is very limited (1–4, 11–14). IK function, stability, and subcellular localization are generally thought to be regulated by posttranslational modification and heterodimerization with other members of the IK family of DNA binding proteins (1–4). Besides the casein kinase II (CK2)-protein phosphatase 1 (PP1) molecular complex (12–14), other upstream regulators of IK in its function as a transcription factor that activates gene expression have not been deciphered. Spleen tyrosine kinase (SYK) is a physiologically important kinase that serves as a key regulator of multiple biochemical signal-transduction events and biologic responses in B-lineage lymphoid cells throughout B-cell ontogeny (15–21). SYK is generally known as an integral part of effective B-cell antigen receptor (BCR) signaling in mature B-cells (15–21). SYK also has important functions in BCR-independent signaling pathways because of its enhanced tyrosine kinase activity in the context of oxidative stress (22), as well as its nonenzymatic interactions with other regulatory proteins (23). Furthermore, multiple centrosomal substrates for SYK were identified by using sensitive kinase assays linked with phosphoproteomics, suggesting that SYK negatively affects cell division through its centrosomal kinase activity (24). Recently, SYK has been identified as a dual-specificity kinase that not only phosphorylates tyrosine (Y) but also serine (S) residues (21). Our results presented herein provide genetic and biochemical evidence for a previously unknown regulatory function of SYK as an activating partner of IK, that phosphorylates IK at serine phosphorylation sites S358 and S361, thereby augmenting its nuclear localization and sequence-specific DNA binding activity. This evidence is a demonstration of posttranslational phosphorylation as a unique mechanism of IK activation.
Results
SYK Phosphorylates IK at Unique Phosphorylation Sites S358 and S361.
In our search for potential partners of IK, we discovered that 14 transcripts representing 11 IK-regulated lymphoid priming genes were significantly up-regulated in human lymphocyte precursor cells from primary bone marrow specimens of pediatric patients with ALL expressing high levels of the SYK gene, which prompted the hypothesis that SYK may be involved in the regulation of IK function (Fig. S1). High-resolution confocal microscopy demonstrated that native IK and SYK are colocalized in both the nucleus and cytoplasm of human cells (Fig. S2A). In coimmunoprecipitation experiments, SYK immune complexes contained not only SYK (Fig. S2B) but also IK (Fig. S2C), indicating that native IK constitutively exists in a stable physical association with native SYK. This association was further confirmed by demonstrating that IK immune complexes contained SYK (Fig. S2B) as well as IK (Fig. S2C).
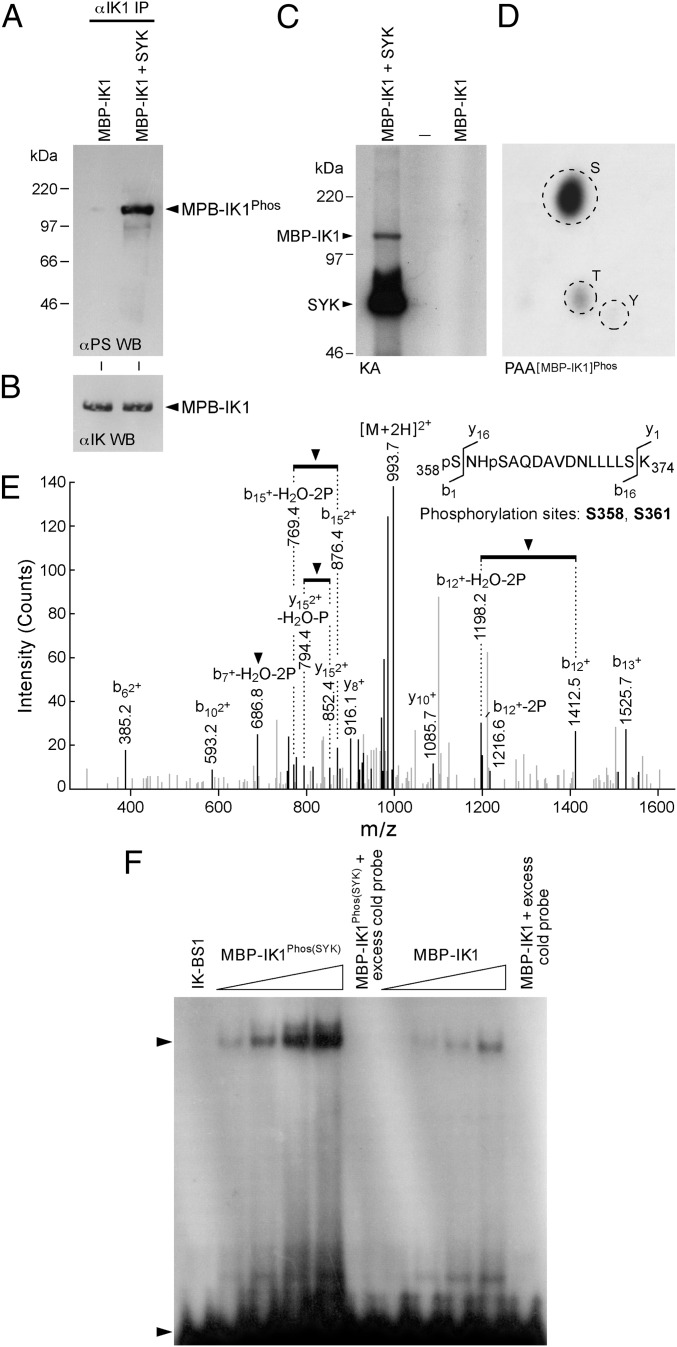
We next performed kinase assays to determine if purified recombinant SYK can phosphorylate purified recombinant IK in vitro. In cold kinase assays, the antiphosphoserine (α-PS) Western blot analysis of IK immunoprecipitated from the SYK plus IK kinase reaction mixtures showed markedly enhanced S-phosphorylation (Fig. 1 A and B). Similarly, phosphoamino acid analysis of the SYK-phosphorylated IK in hot kinase assays confirmed that SYK phosphorylates IK almost exclusively on S-residues (Fig. 1 C and D). A MS analysis was performed on trypsin-digested recombinant IK after an in vitro kinase reaction with purified recombinant SYK. We identified Ser358 (S358) and Ser361 (S361) as two SYK phosphorylation sites within the mouse IK peptide 358SNHSAQDAVDNLLLLSK374, corresponding to a consensus sequence encoded by exon 8 (Fig. 1E and Fig. S3). S358 corresponds to S361 and S361 corresponds to S364 of human IK. According to our structural model of SYK in complex with the human IK peptide (residues 341–375), the α-helix of the target IK peptide would readily bind to SYK catalytic site with a compact conformation because of its narrow and deep shape (Fig. S3), and the terminal phosphate of ATP can easily be transferred by SYK to either S361 or S364 of IK. Notably, S361 and S364 are positioned at the N terminus of an α-helical secondary structure. S364 forms a close contact with the side chain of R360. Therefore, SYK-induced phosphorylation of S364 would enhance the side-chain interaction by a positive-negative salt bridge formation, and thereby stabilize the folded protein conformation of IK. S361 is located at the N-terminal cap of the α-helix and the positively charged environment constituted by the macrodipole of the α-helix would favor the interaction with a SYK-phosphorylated S361, thereby further stabilizing the local protein conformation of IK. Our model posits that SYK-induced phosphorylation of IK could therefore improve its DNA binding function. In agreement with these predictions, SYK-induced phosphorylation of recombinant IK augmented its sequence-specific DNA binding activity in a cell-free EMSA platform (Fig. 1F).
Fig. 1.
Serine phosphorylation and activation of recombinant IK by recombinant SYK. (A) After a cold kinase reaction, maltose-binding protein (MBP)-tagged recombinant IK was immunoprecipitated from the kinase reaction mixture and subjected to α-PS Western blot analysis. (B) Anti-IK Western blots of the samples shown in A. (C) Recombinant SYK showed autophosphorylation and it also phosphorylated MBP-IK1 in hot kinase assays. (D) Phospho amino acid analysis of the SYK-phosphorylated MBP-IK1 band showed S-phosphorylation. (E) MS analysis on trypsin-digested recombinant IK after an in vitro kinase reaction with purified recombinant SYK. (F) EMSAs were performed using increasing amounts (50, 100, 150, 200 ng per sample) of purified MBP-tagged recombinant IK1 protein that has been phosphorylated by recombinant SYK, C. (lane 1) IK-BS1 probe only. (lanes 2–5) Increasing amounts of SYK-phosphorylated MBP-IK1 were used in EMSA. (lane 6) MBP-IK1Phos(SYK) (200 ng) was mixed with 1 ng radiolabeled IK-BS1 in the presence of 100-fold excess unlabeled (cold) IK-BS1 for homologous competition. (lanes 7–10) Increasing amounts of unphosphorylated MBP-IK1 were used in EMSA. (lane 11) MBP-IK1 (200 ng) was mixed with 1 ng radiolabeled IK-BS1 in the presence of 100-fold excess of unlabeled (cold) IK-BS1.
SYK Causes Serine Phosphorylation, Activation, and Nuclear Localization of Native IK.
We next examined the effects of the SYK kinase inhibitor piceattanol (PCT) on S-phosphorylation of native IK in fetal liver-derived human pro–B-cell line FL112. Western blot analysis of IK and SYK immune complexes using a mixture of polyclonal Abs recognizing IK, SYK, and CD19 revealed that CD19 receptor engagement in FL112 cells using a CD19-receptor–specific monoclonal Ab homoconjugate (CD19×CD19) promotes the formation of a multimolecular complex containing CD19, IK, and SYK (Fig. S4). The increased association between IK and SYK in CD19×CD19-stimulated FL112 cells was accompanied by increased S-phosphorylation of IK as documented by detection of larger amounts of IK among S-phosphorylated proteins immunoprecipitated with an α-PS Ab (Fig. S4A), as well as by α-PS Western blot analysis of IK immune complexes (Fig. S4B). SYK-inhibitor PCT blocked this response to CD19 engagement and prevented CD19×CD19-induced S-phosphorylation of IK (Fig. S4). The negative effect of PCT on this activation-induced phosphorylation indicated that SYK was directly responsible for BCR-independent S-phosphorylation of native IK after CD19-receptor engagement.
Confocal fluorescence images of primary leukemic B-cell precursors from newly diagnosed B-lineage ALL patients with wild-type SYK exhibited normal punctate nuclear staining, consistent with the localization of IK to the pericentromeric heterochromatin (PC-HC). In contrast, SYK-deficient primary leukemic cells (25) showed an aberrant, predominantly cytoplasmic localization of IK (Fig. S5A), despite the absence of any IK mutations or exonic deletions that could impair its DNA binding and nuclear localization (Fig. S6). Because the nuclear localization of IK is determined by its DNA binding activity, these results uniquely indicated that the IK–SYK interaction favorably affects the DNA binding activity of native IK in primary human lymphocyte precursors. We next sought direct and unequivocal genetic evidence for a regulatory role of SYK in nuclear localization of native IK in lymphoid cells using SYK-deficient DT40 chicken B-cell lymphoma clones that were established by homologous recombination knockout (22). When analyzed by high-resolution fluorescence microscopy, SYK-deficient DT40 cells exhibited an abnormal IK localization profile, with much of the IK protein found in the cytoplasm, in contrast to the normal speckled staining pattern for IK in wild-type DT40 cells. The abnormal subcellular localization of IK in the SYK-deficient DT40 cells was the direct result of lack of SYK, as evidenced by the fact that SYK− DT40 cells reconstituted with wild-type SYK showed a normal nuclear localization of IK (Fig. S5B). We also examined the regulatory role of SYK in nuclear localization of native IK in human cells using an ecdysone-inducible mammalian expression system (22). Induction of SYK resulted in S-phosphorylation of IK, as documented by detection of larger amounts of IK among S-phosphorylated proteins immunoprecipitated with an α-PS Ab as well as by α-PS Western blot analysis of IK immune complexes (Fig. S7 A–D). SYK induction by Pon-A was sufficient to trigger the nuclear translocation of native IK without any additional treatments (Fig. S7 E and F). These findings provided direct evidence that SYK plays an indispensable role in nuclear localization of native IK.
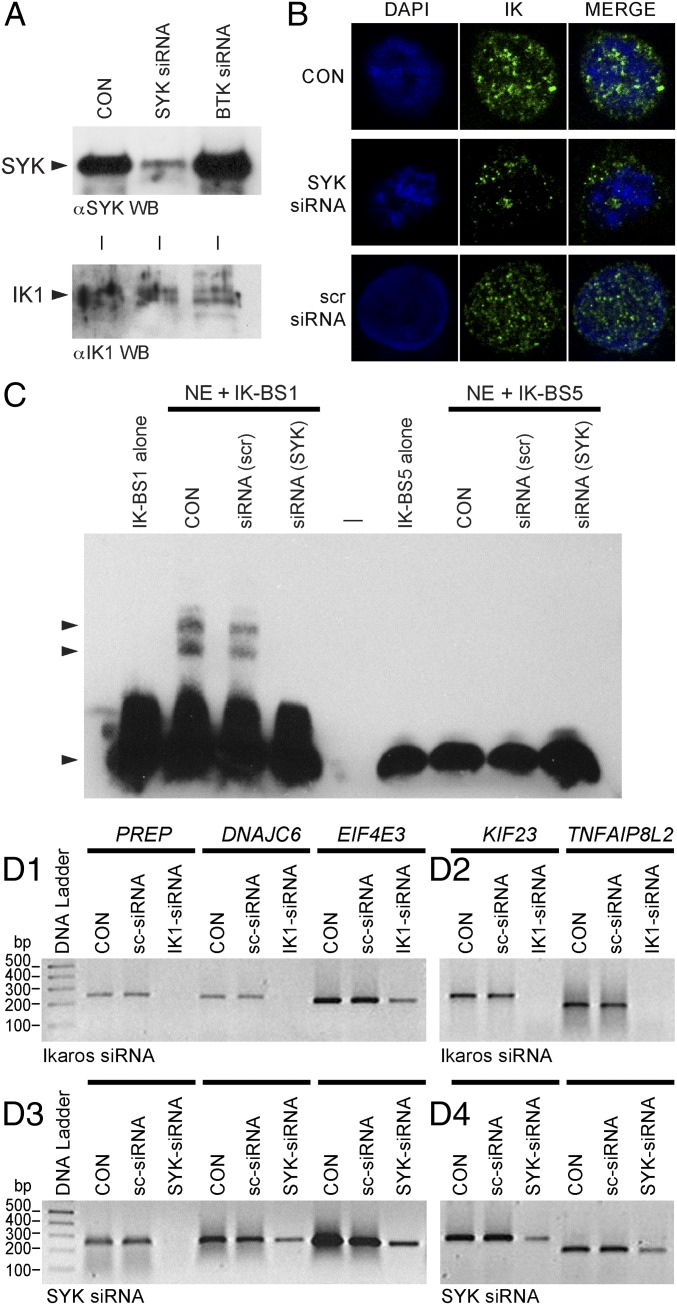
We further explored the role of SYK in regulation of the transcription factor function of native IK in human lymphocyte precursors by comparing the expression levels of 45 recently validated IK target genes harboring IK binding sites (7) (Table S1) in primary samples of lymphocyte precursors from ALL patients with high vs. low SYK expression levels. Of the 60 transcripts representing the 45 IK target genes, 50 were up-regulated in ALL samples with high SYK expression. Notably, the expression levels of 34 transcripts representing 22 IK target genes showed a striking and statistically significant increase in cases with high SYK and IKZF1 expression (Fig. S5C and Table S2). Very similar results were obtained when the B-lineage ALL subset was separately examined for consistent differences between high vs. low SYK expression groups (Fig. S8). These findings indicated that SYK expression levels regulate the transcription factor function of IK. We also found that 23 of 36 transcripts representing 19 validated IK target genes were up-regulated with SYK induction in U373 cells (Fig. S7G). Thus, SYK is capable of causing S-phosphorylation, nuclear translocation, and activation of transcription factor function of IK in human cells. Notably, selective depletion of SYK after treatment with SYK siRNA (but not scrambled control siRNA) markedly diminished the nuclear localization of native IK (Fig. 2 A and B) and abolished its DNA binding activity (Fig. 2C) in human 293T cells. To formally document the importance of SYK in the regulation of IK transcription factor function, we examined the effects of SYK depletion by RNA interference on validated IK target gene expression in human 293T cells using RT-PCR (Fig. 2D). Notably, the expression levels of five randomly selected IK target genes were reduced by siRNA-mediated depletion of SYK. Included as a positive control, IK siRNA (but not scrambled siRNA) also abrogated or reduced the expression of these genes. The striking SYK-dependency of the IK target-gene expression levels demonstrates that SYK plays a critical role in regulation of the native IK function.
Fig. 2.
Effects of siRNA-mediated depletion of native SYK on nuclear localization, sequence-specific DNA binding activity, and transcription factor function of native IK in human cells. (A) SYK vs. IK Western blot analysis of whole-cell lysates from 293T cells treated with medium only (CON), SYK siRNA, or BTK siRNA that was used as a control. (B) Confocal images of 293T cells stained with IK mAb and the blue fluorescent DNA dye DAPI following 72-h treatment with SYK siRNA or scrambled (scr) siRNA (included as a control). CON: No treatment. (Magnification: 630×.) (C) EMSAs measuring IK activity of nuclear extracts (NE) from untreated control (CON) 293T cells as well as 293T cells treated for 72 h with SYK siRNA, or scr-siRNA. (D) RT-PCR results for five randomly selected IK target genes after 72-h treatment with medium alone (CON), scrambled siRNA (sc-siRNA), IK siRNA, vs. SYK siRNA.
Site-Directed Mutagenesis of IK at SYK Phosphorylation Sites Alters Its Subcellular Localization, Sequence-Specific DNA Binding Activity, and Transcription Factor Function.
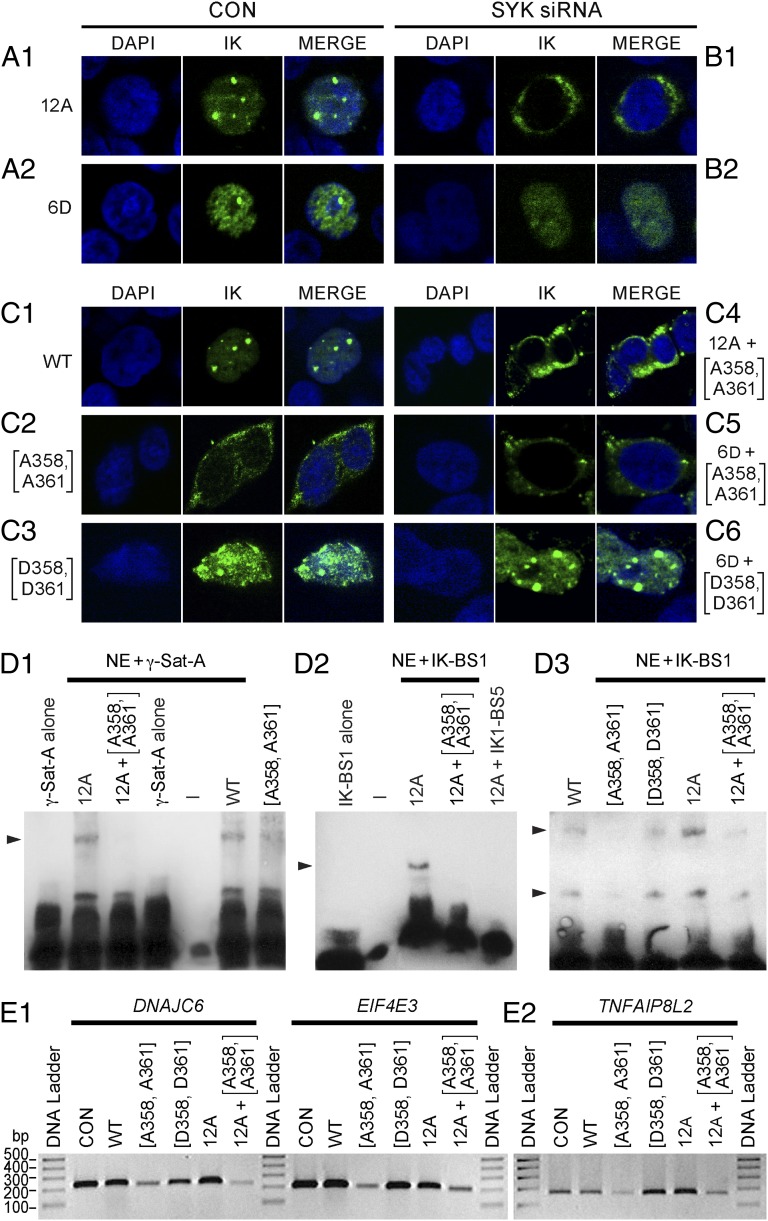
In contrast to the SYK-induced S-phosphorylation of IK, which results in augmented DNA binding activity, CK2-mediated phosphorylation of IK at 11 previously published serine/threonine phosphorylation sites has been shown to reduce the DNA binding activity of IK (12). Protein phosphatase PP1 binds and activates IK by dephosphorylating it on CK2-phosphorylated residues (14). IK mutant 12A containing alanine (A) mutations at the 11 CK2 target sites along with the PP1 recognition motif (A465/467) is able to bind DNA (14) and exhibits a normal PC-HC localization when overexpressed in 293T cells, as reflected by a speckled nuclear staining pattern (Fig. 3A, 1). Notably, siRNA-mediated depletion of native SYK completely blocks the nuclear localization of IK mutant 12A in 293T cells, as evidenced by a strictly cytoplasmic staining pattern (Fig. 3B, 1). Thus, A-substitution of CK2-phosphorylated inhibitory S-residues fails to restore the DNA binding activity of IK in the absence of SYK. The combined aspartate (D) phosphomimetic mutation of six N-terminal CK2 phosphorylation sites causes an abnormal nuclear localization of IK when overexpressed in 293T cells, as characterized by both speckled and diffuse nuclear IK fluorescence staining (Fig. 3A, 2). siRNA-mediated SYK depletion totally abrogated the speckled staining indicative of residual PC-HC localization (Fig. 3B, 2). These results demonstrate that SYK-mediated activation of IK is both indispensable for its normal PC-HC localization in the nucleus and capable of overriding its CK2-mediated inhibition. The contribution of SYK-induced phosphorylation of S358 and S361 residues to IK function was determined by performing site-directed mutagenesis to replace these amino acids with either A for eliminating the effects of SYK-induced phosphorylation or negatively charged D for causing a phosphomimetic effect that mirrors constitutive phosphorylation. Wild-type IK displayed a predominantly nuclear localization with a speckled nuclear immunofluorescence when overexpressed in a subclone of 293T cells lacking native IK expression at detectable levels by confocal fluorescence microscopy (Fig. 3C, 1).
Fig. 3.
Functional studies on mutant IK proteins generated by site-directed mutagenesis. (A–C) Confocal images of 293T cells expressing the mutant IK proteins following treatment with SYK siRNA. (Magnification: 630×.) (D) EMSAs measuring IK activity of nuclear extracts (NE) from 293T cells transfected with expression vectors for wild-type or mutant IK proteins. (E) RT-PCR results for three randomly selected Ikaros target genes in 293T cells expressing wild-type or mutant IK proteins.
In contrast to wild-type IK, SYK-resistant IK protein M1-1 carrying A-mutations at both SYK phosphorylation sites (A358, A361) showed no detectable nuclear localization when overexpressed in 293T cells (Fig. 3C, 2). By comparison, IK protein M2-2 carrying D-mutations at these sites showed markedly enhanced PC-HC localization in the nucleus (Fig. 3C, 3). As with siRNA-mediated SYK depletion, A-substitution of S358 and S361 residues of CK2-resistant mutant IK protein 12A (M5-3) (Fig. 3C, 4) or mutant IK protein 6D with phosphomimetic D-mutations of six N-terminal CK2-phophorylation sites (M6-6) (Fig. 3C, 5) completely abrogated their nuclear localization in transfected 293T cells. Introduction of phosphomimetic D-mutations at SYK phosphorylation sites S358 and S361 restored the normal PC-HC localization of IK with phosphomimetic mutations at CK2 phosphorylation sites to baseline levels (Fig. 3C, 6). These results confirm and extend the results of RNAi experiments and provide direct genetic evidence that SYK-induced phosphorylation of IK at unique phosphorylation sites S358 and S361 controls its normal subcellular localization and binding to the PC-HC.
IK has been shown to bind to repetitive sequences within the PC-HC that contain consensus IK binding sites, and its localization to the PC-HC in the nucleus is directly related to its ability to bind to these sequences (2, 12). Therefore, we next performed EMSAs to directly examine the effect of mutations at SYK phosphorylation sites on the binding of IK to the biotin labeled γ-satellite A probe (2) derived from the centromeric γ-satellite repeat sequences. In agreement with the results of the subcellular localization studies shown in Fig. 3C, A-substitution of the SYK phosphorylation sites S358 and S361 abolished the ability of both wild-type IK and the CK2-resistant mutant IK protein 12A to bind to the γ-satellite A probe (Fig. 3D, 1). Optimal DNA binding is essential for the subcellular localization and transcription factor function of IK because it binds to the regulatory elements of its target genes in a sequence-dependent manner. Unlike wild-type IK or IK with phosphomimetic D-substitutions at SYK phosphorylation sites, mutant IK protein M1-1, which cannot be phosphorylated by native SYK because of A-substitutions at S358 and S361, did not exhibit detectable binding to the IK-specific IK-BS1 probe (Fig. 3D, 2). Similarly, A-substitutions at SYK phosphorylation sites abrogated the sequence-specific DNA binding activity of mutant IK protein 12A that is resistant to inhibitory S-phosphorylation by native CK2 (Fig. 3D, 3). In contrast to the documented effects of the S-to-A mutations at the identified SYK phosphorylation sites of IK, phenylalanine substitutions of Y292, Y409, Y493, and Y499—predicted to be the most likely of the 16 Y-residues in IK to serve as putative Y-phosphorylation sites based on their NetPhos prediction scores (SI Text)—did not affect the binding of IK to the γ-satellite A or IK-BS1 DNA probes (Fig. S9).
We also compared the transcription factor function of wild-type vs. mutant IK proteins expressed in 293T cells. Whereas A-substitutions of IK at inhibitory CK-phosphorylation sites and phosphomimetic D-substitutions of IK at activating SYK-phosphorylation sites were associated with increased expression levels of three of three randomly selected IK target genes (DNAJC6, EIF4E3, TNFAIP8L2) in transfected 293T cells, A-substitutions of SYK phosphorylation sites S358 and S361 caused reduced expression levels of these IK target genes (Fig. 3E). In agreement with the EMSA data, A-substitutions at SYK phosphorylation sites markedly diminished the transcription factor function of the 12A mutant with A-substitutions at inhibitory CK2 phosphorylation sites, as evidenced by the reduced expression levels of IK target genes in 293T cells expressing the M5-3 mutant IK protein (Fig. 3E, 2). Taken together, these functional studies on mutant IK proteins provide unique genetic evidence that SYK-induced phosphorylation of IK at S358 and S361 can control its nuclear localization and transcription factor function by augmenting its sequence-specific DNA binding activity.
Effects of Wild-Type and SYK-Resistant Mutant Ikaros Proteins on B-Cell Precursor Differentiation in Vitro.
We next examined the effects of overexpression of wild-type vs. SYK-resistant mutant IK proteins on the differentiation program of human B-cell precursors by using RT-PCR and multiparameter flow cytometry. Transfection of the pre-pre–B-cell line ALL-1 lacking IK1/IK2 and expressing only truncated non-DNA binding IK isoforms with a plasmid encoding wild-type IK protein induced differentiation, as measured by increased gene and protein expression levels of the mature B-cell surface antigen CD20 at 96 h posttransfection (Fig. S10). In contrast, transfection of ALL-1 cells with a plasmid encoding SYK-resistant IK protein M1-1 carrying A-mutations at both SYK phosphorylation sites (A358, A361) exhibited an opposite effect and interfered with their limited differentiation capacity, as reflected by reduced expression levels of CD20 (Fig. S10). Although CD20 was found on more than 50% of the ALL-1 cells transfected with the wild-type IK plasmid, less than 20% if ALL-1 cells transfected with M1-1 plasmid were surface CD20+. These findings indicate that the SYK-mediated S-phosphorylation of IK is likely important for not only the transcription factor function of IK, but for its function as a key regulator of differentiation in B-cell ontogeny as well.
Discussion
The SYK-phosphorylation sites S358 (S361 in human IK1) and S361 (S364 in human IK1) are outside the main DNA binding domain of IK containing the N-terminal ZFs 1–4. SYK-mediated phosphorylation of the IK protein may induce a conformational change that affects the accessibility and DNA binding affinity of the distant N-terminal ZFs or the more adjacent C-terminal ZFs. Our modeling studies indicate that SYK-induced phosphorylation of S361 and S364 would stabilize the local folded protein conformation of IK in this segment (Fig. S3). Furthermore, phosphorylation of the C-terminal ZF domain may also affect the overall protein conformation of IK and the DNA binding affinity of its N-terminal ZF. Although high-affinity DNA interactions of IK have been generally attributed to its N-terminal ZF 1–3 (26), the C-terminal ZF domain has been shown to bind to the enhancer (δ-A element) of the CD3-δ gene in a sequence-specific manner (27). It has been proposed that the C-terminal ZF domain enables the IK5 isoform lacking three of the four N-terminal ZFs to engage in sequence-specific DNA binding (27, 28). Therefore, augmentation of the DNA binding affinity and stability of the C-terminal ZF domain is likely to increase its contribution to the overall DNA binding affinity of IK. Like IK, other transcription factors, such as the Myc-associated ZF protein MAZ with six C2H2-type ZF motifs, show enhanced DNA binding activity when phosphorylated on a regulatory S-residue in their C-terminal domain (29).
The structural basis of SYK activation is not fully understood because of the lack of a 3D anatomic structure of full-length SYK in active conformation. Based on the anatomic structure of the related ZAP70 kinase, a model of immunoreceptor tyrosine-based activation motif (ITAM)-based signaling has been proposed to explain BCR-mediated SYK activation (30, 31). However, elevation of SYK enzymatic activity is induced by a variety of BCR-independent signals, including oxidative stress that cannot be explained by recruitment of SYK to ITAMs (32, 33). Furthermore, the X-ray structure of unphosphorylated kinase catalytic domain of SYK showed that the enzyme adopts a conformation of the activation loop typically seen only in activated Y-phosphorylated Y-kinases (34). In addition, single-particle electron microscopy studies suggested that the regulation of the activation of SYK might be modulated by subtle or minor changes in the positioning of the regulatory domains (SH2-SH2 region) rather than full opening mechanisms proposed for Src kinases (35). It has also been shown that numerous signaling events such as phosphorylation by Src-family kinases, autophosphorylation, and substrate-binding to the SH2 domains can equally activate SYK (36). The CD19 receptor plays a critical role in initiation of SYK-dependent signaling events in both immature and mature B-lineage lymphoid cells (37–39) (SI Text). The present study demonstrates that activation of SYK after CD19 receptor engagement in a BCR-negative human pro–B-cell line is associated with increased S-phosphorylation of native IK, and SYK is directly responsible for this BCR-independent S-phosphorylation of native IK after CD19-receptor engagement. Hence, there are a multitude of signals that contribute to sustained baseline SYK activity in cells and this activity appears to be important for the optimal transcription factor function of IK. Whether or not the kinase-ligand interactions between SYK and IK can cause conformational changes reshaping the active site of SYK via the fairly common process of “induced fit” (40), and thereby further promote SYK-induced S-phosphorylation of IK, will require a 3D structure determination of full-length SYK in a complex with IK using X-ray crystallography or NMR spectroscopy. Similarly, the elucidation of the structural basis of IK activation by SYK-induced S-phosphorylation will require a 3D structure determination of IK at atomic resolution before and after phosphorylation.
Materials and Methods
Standard Biochemical, Imaging, and Transfection Methods.
Confocal laser scanning microscopy, coimmunoprecipitations, Western blot analyses, and EMSA were performed as per previously described standard procedures (SI Text). RT-PCR was used to evaluate the expression levels of IK target genes.
Mass Spectrometry.
We used MS to identify the SYK phosphorylation sites of IK. MS was performed in the University of Southern California Proteomics Core by using the NanoLC system from Eksigent, a nano-LC-MS/MS proteomics system that employs Eksigent’s Microfluidic Flow Control technology (SI Text).
Site-Directed Mutagenesis.
The full-length mouse IK cDNA (NM_001025597) was subcloned into the pCMV6-Entry precision shuttle vector (Cat# PS100001; Origene) at the restriction sites SgfI and MluI to generate the pCMV6-mIK mammalian cell-expression vector. The pCMV6-mIK construct was then used as a backbone vector to generate the mIKS358A_S361A and mIKS358D_S361D mutant vectors encoding IK proteins with A- or phosphomimetic D-mutations at the SYK-phosphorylation sites S358 and S361 using the QuikChange II Site-Directed Mutagenesis Kit from Agilent Technologies (SI Text).
Bioinformatics.
In analyses of gene-expression profiles of lymphocyte precursors with high vs. low SYK expression levels, we focused our analysis on validated IK target genes (SI Text). The Gene Pattern Web-based software (www.broadinstitute.org/cancer/software/genepattern) was used to extract expression values from the National Center for Biotechnology Information Gene Expression Omnibus database to compile gene expression profiles of 1,104 primary leukemia specimens from newly diagnosed or relapsed ALL patients (SI Text).
Supplementary Material
Acknowledgments
The authors thank Ernesto Barron of the University of Southern California Norris Comprehensive Cancer Center Cell and Tissue Imaging Core and Dr. Guangyu Zhang of the University of Southern California Proteomics Core for their technical assistance. This study was supported in part by Department of Health and Human Services Grants P30CA014089, U01-CA-151837, R01CA-154471, and R21-CA-164098 (to F.M.U.) and R01 HL095120 (to S.D.); a 2011 V-Foundation Translational Research Award (to F.M.U.); the Ronald McDonald House Charities of Southern California (F.M.U.); the Couples Against Leukemia Foundation (F.M.U.); a William Lawrence and Blanche Hughes Foundation grant (to F.M.U.); Nautica Triathalon and its producer Michael Epstein (F.M.U.); a St. Baldrick’s Foundation Career Development Award (to S.D.); and the Four Diamonds Fund of the Pennsylvania State University, College of Medicine (S.D.). J.Z was supported by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE18798)
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209828109/-/DCSupplemental.
References
- 1.Yoshida T, Ng SY, Georgopoulos K. Awakening lineage potential by Ikaros-mediated transcriptional priming. Curr Opin Immunol. 2010;22(2):154–160. doi: 10.1016/j.coi.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobb BS, et al. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev. 2000;14(17):2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson EC, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26(3):335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 2010;584(24):4910–4914. doi: 10.1016/j.febslet.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 5.Bottardi S, et al. Ikaros interacts with P-TEFb and cooperates with GATA-1 to enhance transcription elongation. Nucleic Acids Res. 2011;39(9):3505–3519. doi: 10.1093/nar/gkq1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, et al. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30(17):4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13(1):86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dovat S, Song C, Payne KJ, Li Z. Ikaros, CK2 kinase, and the road to leukemia. Mol Cell Biochem. 2011;356(1-2):201–207. doi: 10.1007/s11010-011-0964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, et al. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 1999;96(2):680–685. doi: 10.1073/pnas.96.2.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mullighan CG, et al. Children’s Oncology Group Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15(19):5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 12.Gurel Z, et al. Recruitment of Ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J Biol Chem. 2008;283(13):8291–8300. doi: 10.1074/jbc.M707906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gómez-del Arco P, Maki K, Georgopoulos K. Phosphorylation controls Ikaros’s ability to negatively regulate the G(1)-S transition. Mol Cell Biol. 2004;24(7):2797–2807. doi: 10.1128/MCB.24.7.2797-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popescu M, et al. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J Biol Chem. 2009;284(20):13869–13880. doi: 10.1074/jbc.M900209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng AM, et al. Syk tyrosine kinase required for mouse viability and B-cell development. Nature. 1995;378(6554):303–306. doi: 10.1038/378303a0. [DOI] [PubMed] [Google Scholar]
- 16.Mócsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat Rev Immunol. 2010;10(6):387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner M, et al. Syk tyrosine kinase is required for the positive selection of immature B cells into the recirculating B cell pool. J Exp Med. 1997;186(12):2013–2021. doi: 10.1084/jem.186.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uckun FM, Qazi S. Spleen tyrosine kinase as a molecular target for treatment of leukemias and lymphomas. Expert Rev Anticancer Ther. 2010;10(9):1407–1418. doi: 10.1586/era.10.112. [DOI] [PubMed] [Google Scholar]
- 19.Zhou F, Hu J, Ma H, Harrison ML, Geahlen RL. Nucleocytoplasmic trafficking of the Syk protein tyrosine kinase. Mol Cell Biol. 2006;26(9):3478–3491. doi: 10.1128/MCB.26.9.3478-3491.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koretzky GA, Abtahian F, Silverman MA. SLP76 and SLP65: Complex regulation of signalling in lymphocytes and beyond. Nat Rev Immunol. 2006;6(1):67–78. doi: 10.1038/nri1750. [DOI] [PubMed] [Google Scholar]
- 21.Heizmann B, Reth M, Infantino S. Syk is a dual-specificity kinase that self-regulates the signal output from the B-cell antigen receptor. Proc Natl Acad Sci USA. 2010;107(43):18563–18568. doi: 10.1073/pnas.1009048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uckun FM, Qazi S, Ma H, Tuel-Ahlgren L, Ozer Z. STAT3 is a substrate of SYK tyrosine kinase in B-lineage leukemia/lymphoma cells exposed to oxidative stress. Proc Natl Acad Sci USA. 2010;107(7):2902–2907. doi: 10.1073/pnas.0909086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Devarajan E, He J, Reddy SP, Dai JL. Transcription repressor activity of spleen tyrosine kinase mediates breast tumor suppression. Cancer Res. 2005;65(22):10289–10297. doi: 10.1158/0008-5472.CAN-05-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue L, et al. Sensitive kinase assay linked with phosphoproteomics for identifying direct kinase substrates. Proc Natl Acad Sci USA. 2012;109(15):5615–5620. doi: 10.1073/pnas.1119418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodman PA, Wood CM, Vassilev A, Mao C, Uckun FM. Spleen tyrosine kinase (Syk) deficiency in childhood pro-B cell acute lymphoblastic leukemia. Oncogene. 2001;20(30):3969–3978. doi: 10.1038/sj.onc.1204515. [DOI] [PubMed] [Google Scholar]
- 26.Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14(12):8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258(5083):808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 28.Payne MA, et al. Zinc finger structure-function in Ikaros. World J Biol Chem. 2011;2(6):161–166. doi: 10.4331/wjbc.v2.i6.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsutsui H, et al. The DNA-binding and transcriptional activities of MAZ, a myc-associated zinc finger protein, are regulated by casein kinase II. Biochem Biophys Res Commun. 1999;262(1):198–205. doi: 10.1006/bbrc.1999.1130. [DOI] [PubMed] [Google Scholar]
- 30.Deindl S, et al. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell. 2007;129(4):735–746. doi: 10.1016/j.cell.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 31.deCastro RO. Regulation and function of Syk tyrosine kinase in mast cell signaling and beyond. J Signal Transduct. 2011 doi: 10.1155/2011/507291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sada K, Takano T, Yanagi S, Yamamura H. Structure and function of Syk protein-tyrosine kinase. J Biochem. 2001;130(2):177–186. doi: 10.1093/oxfordjournals.jbchem.a002970. [DOI] [PubMed] [Google Scholar]
- 33.Uckun FM, Ozer Z, Qazi S, Tuel-Ahlgren L, Mao C. Polo-like-kinase 1 (PLK1) as a molecular target to overcome SYK-mediated resistance of B-lineage acute lymphoblastic leukaemia cells to oxidative stress. Br J Haematol. 2010;148(5):714–725. doi: 10.1111/j.1365-2141.2009.07983.x. [DOI] [PubMed] [Google Scholar]
- 34.Atwell S, et al. A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase. J Biol Chem. 2004;279(53):55827–55832. doi: 10.1074/jbc.M409792200. [DOI] [PubMed] [Google Scholar]
- 35.Arias-Palomo E, Recuero-Checa MA, Bustelo XR, Llorca O. Conformational rearrangements upon Syk auto-phosphorylation. Biochim Biophys Acta. 2009;1794(8):1211–1217. doi: 10.1016/j.bbapap.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsang E, et al. Molecular mechanism of the Syk activation switch. J Biol Chem. 2008;283(47):32650–32659. doi: 10.1074/jbc.M806340200. [DOI] [PubMed] [Google Scholar]
- 37.DeFranco AL. The two-headed antigen. B-cell co-receptors. Curr Biol. 1996;6(5):548–550. doi: 10.1016/s0960-9822(02)00539-0. [DOI] [PubMed] [Google Scholar]
- 38.Uckun FM, et al. Targeting human B precursor acute lymphoblastic leukemia cells with recombinant human CD19 ligand. 2010. Available at https://ash.confex.com/ash/2010/webprogram/Paper32270.html. Accessed December 4, 2010.
- 39.Depoil D, et al. CD19 is essential for B cell activation by promoting B cell receptor-antigen microcluster formation in response to membrane-bound ligand. Nat Immunol. 2008;9(1):63–72. doi: 10.1038/ni1547. [DOI] [PubMed] [Google Scholar]
- 40.Zhong H, Tran LM, Stang JL. Induced-fit docking studies of the active and inactive states of protein tyrosine kinases. J Mol Graph Model. 2009;28(4):336–346. doi: 10.1016/j.jmgm.2009.08.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.