Abstract
The anti-atherogenic cytokine, TGF-β, plays a key role during macrophage foam cell formation by modulating the expression of key genes involved in the control of cholesterol homeostasis. Unfortunately, the molecular mechanisms underlying these actions of TGF-β remain poorly understood. In this study we examine the effect of TGF-β on macrophage cholesterol homeostasis and delineate the role of Smads-2 and ‐3 during this process. Western blot analysis showed that TGF-β induces a rapid phosphorylation-dependent activation of Smad-2 and ‐3 in THP-1 and primary human monocyte-derived macrophages. Small interfering RNA-mediated knockdown of Smad-2/3 expression showed that the TGF-β-mediated regulation of key genes implicated in the uptake of modified low density lipoproteins and the efflux of cholesterol from foam cells was Smad-dependent. Additionally, through the use of virally delivered Smad-2 and/or Smad-3 short hairpin RNA, we demonstrate that TGF-β inhibits the uptake of modified LDL by macrophages through a Smad-dependent mechanism and that the TGF-β-mediated regulation of CD36, lipoprotein lipase and scavenger receptor-A gene expression was dependent on Smad-2. These studies reveal a crucial role for Smad signaling, particularly Smad-2, in the inhibition of foam cell formation by TGF-β through the regulation of expression of key genes involved in the control of macrophage cholesterol homeostasis.
Abbreviations: AcLDL, acetylated low density lipoprotein; ABCA-1, ATP-binding cassette transporter A-1; ABCG-1, ATP-binding cassette transporter G-1; ApoE, apolipoprotein E; ApoE−/−, apolipoprotein E deficient; CD36, cluster of differentiation 36; DiI, 1,1′-dioctadecyl-3,3,3′,3′-tetramethyllindocarbocyane perchlorate; HMDM, human monocyte-derived macrophages; LDL, low density lipoprotein; LPL, lipoprotein lipase; OxLDL, oxidized low density lipoprotein; shRNA, short hairpin RNA; SR-A, scavenger receptor A; THP-1, human acute monocytic leukemia cell line
Keywords: Foam cell, Atherosclerosis, Cholesterol, TGF-β, Macrophage
Highlights
► Anti-atherogenic cytokine TGF-β inhibits macrophage foam cell formation. ► The role of Smads in the control of macrophage cholesterol homeostasis was studied. ► Smads were found to play a key role in the TGF-β-mediated uptake of modified LDL. ► A dominant role of Smad2 was identified in the regulation of gene expression. ► The TGF-β-Smad axis may represent a powerful anti-foam cell therapeutic target.
1. Introduction
Atherosclerosis is a chronic inflammatory disease of the vasculature that accounts for nearly half of all mortalities in western society. This disease is governed by many risk factors including genetic predisposition and diet, and presents itself through the appearance of fibrotic plaques in the intima of the arterial wall, that upon rupture, result in thrombosis and ultimately myocardial infarction and stroke [1]. Atherosclerosis is initiated through the activation of the arterial endothelium by a number of risk factors leading to the recruitment of immune cells, particularly T lymphocytes and monocytes. The latter differentiate into macrophages, a process that is accompanied by increased expression of scavenger receptors, and then transform into lipid-loaded foam cells. Formation of macrophage foam cells via the uptake of modified LDL represents a critical step in atherosclerosis and is known to be tightly regulated by a multitude of both novel and classical cytokines such as interleukin (IL)-33, TNF-like protein 1A (TL1A), interferon-γ (IFN-γ), and transforming growth factor-β (TGF-β) [2–6].
TGF-β, an extensively studied pleiotropic cytokine, is widely recognized to exert athero-protective actions. Low circulating levels of TGF-β have been seen in patients with advanced atherosclerosis [6] while regions of the aorta with a high probability of lesion development, such as the dorsal descending thoracic aorta, display low levels of TGF-β expression, thereby suggesting an inverse correlation between TGF-β activity and the progression of the disease [7]. In addition, inhibition of TGF-β activity and/or expression in mouse models, achieved using neutralizing antibodies or targeted homologous gene disruption or expression of dominant negative forms of receptors, results in accelerated lesion development (in ApoE−/− mice) and an elevated inflammatory response [6,8,9]. Increased atherosclerosis is also observed when TGF-β signaling is specifically inhibited in T-cells [10]. Furthermore, numerous in vitro studies from our own and other laboratories have demonstrated that TGF-β inhibits the expression of several key genes implicated in the uptake of modified lipoproteins, such as scavenger receptor-A and -B1 (SR-A and SR-B1), CD36 and lipoprotein lipase (LPL) and, simultaneously, induces the expression of those involved in the efflux of cholesterol from macrophages such as apolipoprotein E (ApoE) and the ATP-binding cassette transporters -A1 and -G1 (ABCA1 and ABCG1) [5,6,11–20].
Due to its involvement in the pathogenesis of many diseases, such as arthritis, cancer and atherosclerosis, the TGF-β signal transduction pathway has also been the focus of intense research in recent years [6,21]. Briefly, TGF-β binds to its cognate heteromeric receptor complex, composed of two type I (TGF-βRI) and type II (TGF-βRII) receptors, on the cell surface. Phosphorylation of TGF-βRI, by TGF-βRII, on numerous serine and threonine residues results in a conformational change in the receptor architecture that facilitates the recruitment of Smad proteins. Aided by the Smad anchor for receptor activation (SARA), regulatory Smad-2 and Smad-3 become phosphorylated, complex with a common mediator Smad-4, and migrate to the nucleus where they can modulate the promoter activity of specific genes through interaction with Smad responsive elements and/or other transcription factors [22]. In addition, the TGF-β-mediated activation of other signaling pathways, such as mitogen-activated protein kinases, is required for the regulation of expression of some genes by this cytokine [23,24].
Both TGF-β and Smads are known to be highly expressed in both macrophages and foam cells of early-stage atherosclerotic lesions [25]. However, the role of Smad signaling in the TGF-β-mediated control of macrophage foam cell formation and associated changes in gene expression is unknown except for the inhibition of CD163 expression by this cytokine [15]. Further studies are required because of the existence of both Smad-dependent and ‐independent pathways in TGF-β signaling [26,27]. In addition, mechanistic studies examining the role of TGF-β signaling underlying modified LDL uptake, foam cell formation and associated changes in gene expression may ultimately open up promising novel therapeutic avenues. Therefore, the objective of this study was to investigate the roles of Smad-2 and Smad-3 in the TGF-β-mediated regulation of modified LDL uptake and changes in gene expression associated with foam cell formation.
2. Materials and methods
2.1. Reagents
All chemicals were purchased from Sigma-Aldrich (Poole, U.K.) unless otherwise stated. Recombinant human TGF-β1 was supplied by Peprotech (London, U.K.), and 1,1′-dioctadecyl-3,3,3′,3′-tetramethyllindocarbocyane perchlorate (DiI)-labeled acetylated LDL (DiI-AcLDL) and (DiI)-labeled oxidized LDL (DiI-OxLDL) were purchased from Intracel (Frederick, MD, USA).
2.2. Cell culture
Human monocyte-derived macrophages (HMDM) were differentiated from monocytes isolated from buffy coats supplied by the Welsh Blood service using Ficoll-Hypaque purification described elsewhere [2]. Ethical approval and informed consent for each donor was granted by the Welsh Blood Service for the use of human blood samples. Human acute monocytic leukemia cell line (THP-1) and HMDM were grown in complete RPMI-1640 supplemented with 10% (v/v) (THP-1) or 5% (v/v) (HMDM) (v/v) heat-inactivated FCS (heated to 56 °C for 30 minutes), penicillin (100 U/ml), streptomycin (100 μg/ml) and l-glutamine (2 mmol/l) at 37 °C in a humidified atmosphere containing 5% (v/v) CO2. THP-1 monocytes were differentiated into macrophages using 160 nM PMA for 24 hours to ensure high expression levels of scavenger receptors and other genes implicated in the control of macrophage foam cell formation [28]. In all experiments, unless otherwise stated, cells were incubated with TGF-β (30 ng/ml) for 48 hours. Recombinant human TGF-β was reconstituted in PBS/0.1% BSA that was subsequently used as a vehicle control.
2.3. LDL uptake assays
Cells were incubated for 24 or 48 hours with DiI-AcLDL or DiI-OxLDL (10 μg/ml; Intracel, Frederick, MD) in RPMI-1640 containing 0.2% (v/v) fatty acid free BSA at 37 °C. The uptake of modified LDL was analyzed by flow cytometry on a FACS Canto flow cytometer (BD Biosciences, Oxford, U.K.) with at least 10,000 events acquired for each sample. Data were represented as a percentage of the vehicle-treated control cells.
2.4. Real-time quantitative PCR
RNA extraction, reverse transcription and real-time quantitative PCR analysis were performed as described elsewhere [2]. Oligonucleotides sequences can be seen in supplementary Table I and were purchased from Sigma Aldrich (Poole, UK). Fold changes in expression were calculated using 2−(ΔCt1 − ΔCt2), where ΔCt represents the difference between the threshold cycle (CT) for each target gene and housekeeping mRNA transcript levels. Melting curve analysis was performed on each primer set to confirm amplification of a single product and all amplicons were sequenced to ensure reaction specificity (data not shown).
2.5. Western blotting
Total cell lysates were size-fractionated and analyzed by Western blotting as previously described [2,3,5,16,21]. Samples were subjected to electrophoresis alongside comparative molecular weight markers (GE Healthcare, WI, USA) to determine the size of the protein product. Antibodies specific to Phosho-Smad 2 (#3108), Phospho-Smad 3 (#9520), Total-Smad 2 (#5339), Total-Smad 3 (#9523) and Total-Smad 2/3 (#3102) were supplied by Cell Signaling Technologies (Danvers, MA, USA). Antibodies specific to ApoE (0650–1904), SR-A (sc-20660) and β-Actin were supplied by Biogenesis (Poole, U.K.), Santa Cruz Biotechnology (Santa Cruz, CA) and Sigma (Poole, UK) respectively.
2.6. Transfection of siRNA
THP-1 monocytes were transfected with validated small interfering (si)RNAs specific for Smad-2 and Smad-3 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (used as control; see supplementary Fig. 2) for a final concentration of 7.5 nM using INTERFERin™ in accordance with the manufacturer's protocol (PolyPlus Transfection). The cells were then left for 24 hours, differentiated into macrophages by incubation with 160 nM PMA for 24 hours as described above, and subsequently treated with TGF-β or vehicle for 24 hours. Gene silencing was measured by Western blotting.
2.7. Generation of adenovirus encoding Smad-2, Smad-3 and GAPDH shRNA
Adenovirus type 5 (Ad5) vectors were engineered to express Smad-2, Smad-3 or GAPDH short hairpin (sh)-RNA flanked by sequences from the mouse BIC gene (+ 134/+ 161 and + 221/+ 265, AY096003) to enable the shRNA to be processed by the endogenous microRNA (miRNA) cellular machinery as previously described [29]. Appropriate shRNAs, targeting the following sequences of the Smad-2 gene: AAGAGCAGCAAATTCCTGGTT (Rad-Smad 2 shRNA) or the Smad-3 gene: TCCATCTTCACTCAGGTAGCC (Rad-Smad 3 shRNA) or the GAPDH gene: AGAAGATGCGGCTGACTGTCG (Rad-GAPDH shRNA), were inserted into the Ad5 vector using recombineering technology as described elsewhere [30]. Briefly, two oligonucleotides (detailed in supplementary Table II) that overlap by 25 base pairs at their 3′ and 5′ ends respectively were designed to contain the appropriate shRNA sequence and arms of homology to facilitate homologous recombination into the Ad5 vector. Induced Escherichia coli SW102 containing the Ad5 vector genome in a modified bacterial artificial chromosome (BAC) were transformed with each oligonucleotide (100 ng) and appropriate recombinants were identified by sequence analysis. Adenoviruses were then amplified, purified and tittered as described elsewhere [30].
2.8. Adenoviral infection
THP-1 monocytes or HMDM were infected with Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA at a multiplicity of infection (MOI) of 100 for each virus in 0.5 ml RPMI-1640 medium for 2.5 hours at 37 °C (rocking) prior to addition of 1 ml RPMI-1640 medium (including 160 nM PMA for THP-1 monocytes to induce differentiation into macrophages) and incubation for a further 72 hours. Macrophages were then stimulated with TGF-β or vehicle control for 48 hours. An MOI of 100 was sufficient to infect > 90% THP-1 cells as measured by flow cytometry following infection with a GFP-expressing recombinant adenovirus (data not shown).
2.9. Statistical analysis
All data are presented as mean [± standard deviation (SD)] on the assigned number of independent experiments or, in experiments involving HMDM, experiments performed using samples from different donors. For single comparisons, values for p were calculated using the Student's t-test. For multiple comparisons, values of p were calculated using one-way ANOVA with Tukey's post-hoc analysis where homogeneity of variance was met (as determined by Levene's test of homogeneity of variances) or Welch's robust test of equality of means followed by Games–Howell post-hoc analysis. Values of p were considered significant below 0.05.
3. Results
3.1. TGF-β modulates the expression of key genes implicated in the uptake and efflux of cholesterol through a Smad-dependent mechanism
PMA differentiated THP-1 cells are commonly used as a model to delineate human macrophage cellular functions and gene expression associated with atherosclerosis in the light of their conserved responses with primary HMDM and in vivo evidence [2,3,31]. This cellular system was therefore employed to examine the potential role of Smad signaling in the TGF-β-mediated regulation of macrophage cholesterol homeostasis and foam cell formation in human macrophages. Key findings were confirmed in primary cultures of HMDM.
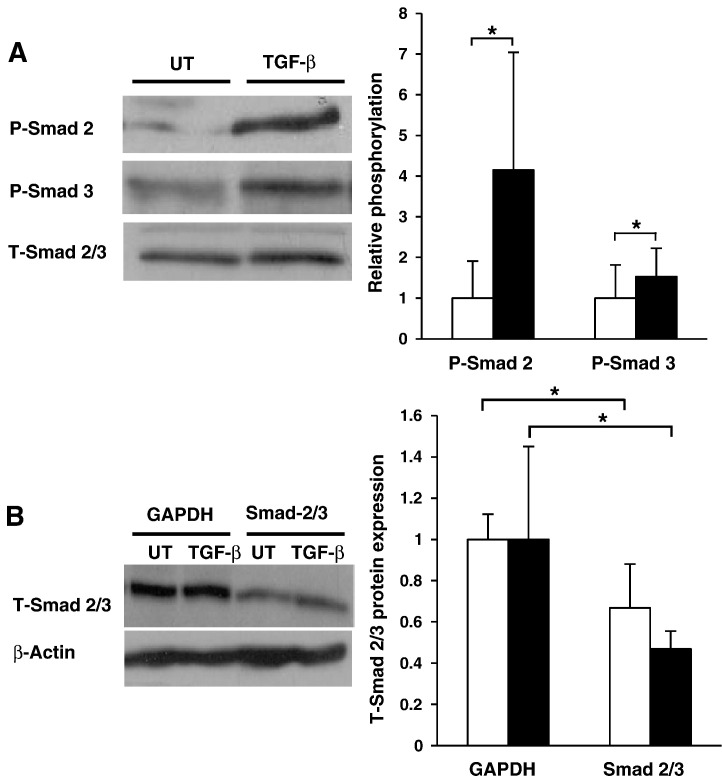
We first examined the ability of TGF-β to activate the Smad signaling cascade in human macrophages. Previous studies by our group have shown that 30 ng/ml of TGF-β induces maximal changes in macrophage gene expression [5,16,21]. We therefore performed all experiments using this previously established concentration. As shown in Fig. 1A, both Smad-2 and ‐3 were rapidly phosphorylated (and therefore activated) after 60 minutes incubation with TGF-β in HMDM. Supplementary Fig. 1 shows that this response was rapid and sustained, albeit at varying levels, for 3 hours and confirms that the TGF-β-mediated activation of Smad-2 and ‐3 was conserved in THP-1 macrophages.
Fig. 1.
Activation and silencing of Smad-2 and ‐3 in human macrophages. (A) Smad-2 phosphorylation or Smad-3 phosphorylation was determined in HMDM either left untreated (empty bars) or incubated with TGF-β (30 ng/ml; filled bars) for 60 minutes. Equal amounts of protein extracts were subjected to Western blot analysis using antisera against phosphorylated (P-)Smad 2, P-Smad 3 or total (T-)Smad 2/3. P-Smad 2 and P-Smad 3 levels were normalized to T-Smad 2/3 expression and displayed as a fold change compared to untreated controls (arbitrarily assigned as 1). Data represent the mean ± SD of 3 or 5 independent experiments (P-Smad 2 and P-Smad 3 respectively). (B) The expression of GAPDH or total (T-)Smad 2/3 protein was knocked down in untreated THP-1 macrophages (UT, empty bars) or those incubated with TGF-β (30 ng/ml; TGF-β, filled bars) using gene specific siRNA transfection. Equal amounts of protein extracts were subjected to Western blot analysis using antisera against T-Smad 2/3 or β-Actin. T-Smad 2/3 levels were normalized to β-actin expression and are displayed as a fold change compared to GAPDH siRNA controls (arbitrarily assigned as 1). Data represent the mean ± SD of 3 independent experiments. Statistical analysis was performed using the Student's t-test, * P < 0.05.
Many studies have reported a TGF-β-mediated reduction in the expression of key genes involved in the uptake of modified lipoproteins by macrophages, namely CD36, SR-A, SR-B1 and LPL [16–18], and the induction of those implicated in cholesterol efflux, such as ApoE, ABCA1 and ABCG1 [5,19,20]. Unfortunately, the role of Smad-2 and ‐3 in the TGF-β-mediated regulation of expression of these genes is poorly understood and was therefore investigated first using a dual-Smad2/3 siRNA-mediated knockdown assay in THP-1 macrophages. Comparisons were made with knockdown of GAPDH, which has been used as a control in numerous previous studies [32–36]. As shown in Fig. 1B, transfection of THP-1 cells with a combination of Smad-2 and ‐3 specific siRNAs resulted in a significant knockdown of total (T)-Smad 2/3 protein expression in both TGF-β treated and untreated cells when compared to control GAPDH siRNA transfected cells. In all cases, the TGF-β-mediated changes in the expression of genes studied were seen in cells transfected with GAPDH siRNA thus confirming the fidelity of our siRNA system. The decrease in GAPDH expression following siRNA-mediated knockdown of this gene was confirmed by RT-PCR (see supplementary Fig. 2).
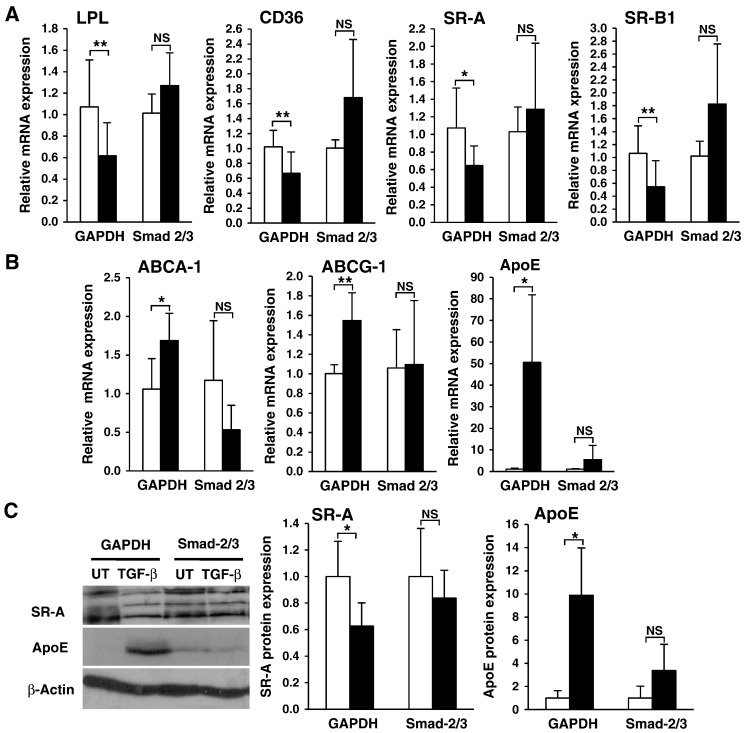
As shown in Fig. 2A and B, consistent with published literature [5,6,12–14,16–20], the expression of key genes involved in cholesterol uptake (LPL, CD36, SR-A and SR-B1) were significantly inhibited by TGF-β in GAPDH transfected THP-1 macrophages, while those involved in cholesterol efflux (ABCA1, ABCG1 and ApoE) were significantly induced. In contrast to this, the ability of TGF-β to replicate these responses was abolished in Smad-2/3 depleted cells as shown by the non-significant changes in transcript level observed for each gene compared to GAPDH siRNA transfected cells, thereby suggesting that Smad signal transduction plays an integral role in the regulation of expression of these genes. Smads are transcription factors and the changes in mRNA expression of downstream genes, such as LPL and ApoE, are reflective of their functional actions. Numerous studies by our group have consistently shown a direct correlation between the expression of LPL, SR-A, SR-B1, CD36, ApoE, ABCA1 and ABCG1 transcripts and their protein levels in both THP-1 macrophages and HMDMs suggesting that any changes in mRNA expression observed for these genes are likely to be conserved at protein levels [2,5,21]. Nevertheless, this was confirmed by Western blot analysis of selected proteins in an identical knockdown system. As shown in Fig. 2C, the TGF-β-mediated modulation of expression of SR-A (all 3 isoforms) and ApoE, was abrogated in response to Smad-2/3 knockdown.
Fig. 2.
TGF-β regulates the expression of key genes implicated in the control of cholesterol homeostasis in human macrophages through a Smad-dependent pathway. The expression of total (T-)Smad 2/3 or GAPDH protein in THP-1 macrophages was knocked down using gene specific siRNA transfection (as seen in Fig. 1B). The cells were then either left untreated (UT, empty bars) or stimulated (TGF-β, filled bars) with 30 ng/ml of TGF-β for 6 hours (ABCA1 and ABCG1) or 24 hours (other genes). Total RNA was subjected to real-time quantitative PCR using primers against (A) LPL, CD36, SR-A, SR-B1 or (B) ABCG-1, ABCA-1 and ApoE as indicated. The mRNA expression levels were calculated using comparative Ct method and normalized to 28S ribosomal (r)RNA levels (or RPL13A for ABCA-1 and ABCG-1) with untreated cells given an arbitrary value of 1. Data represent the mean ± SD of 3 independent experiments. (C) Equal amounts of protein extracts were subjected to Western blot analysis using antisera against SR-A, ApoE or β-actin as indicated. Protein expression, as determined by densitometric analysis, was normalized to β-actin and is displayed as a fold change compared to the untreated controls (arbitrarily assigned as 1). Data represent the mean ± SD of 3 independent experiments. Multiple immunoreactive polypeptides for SR-A represent different isoforms that are known to be produced by alternative splicing. In all cases, statistical analysis was performed using the Student's t-test, * P < 0.05; ** P < 0.01. NS indicates not significant.
3.2. TGF-β reduces modified LDL uptake in THP-1 macrophages through a Smad-dependent mechanism
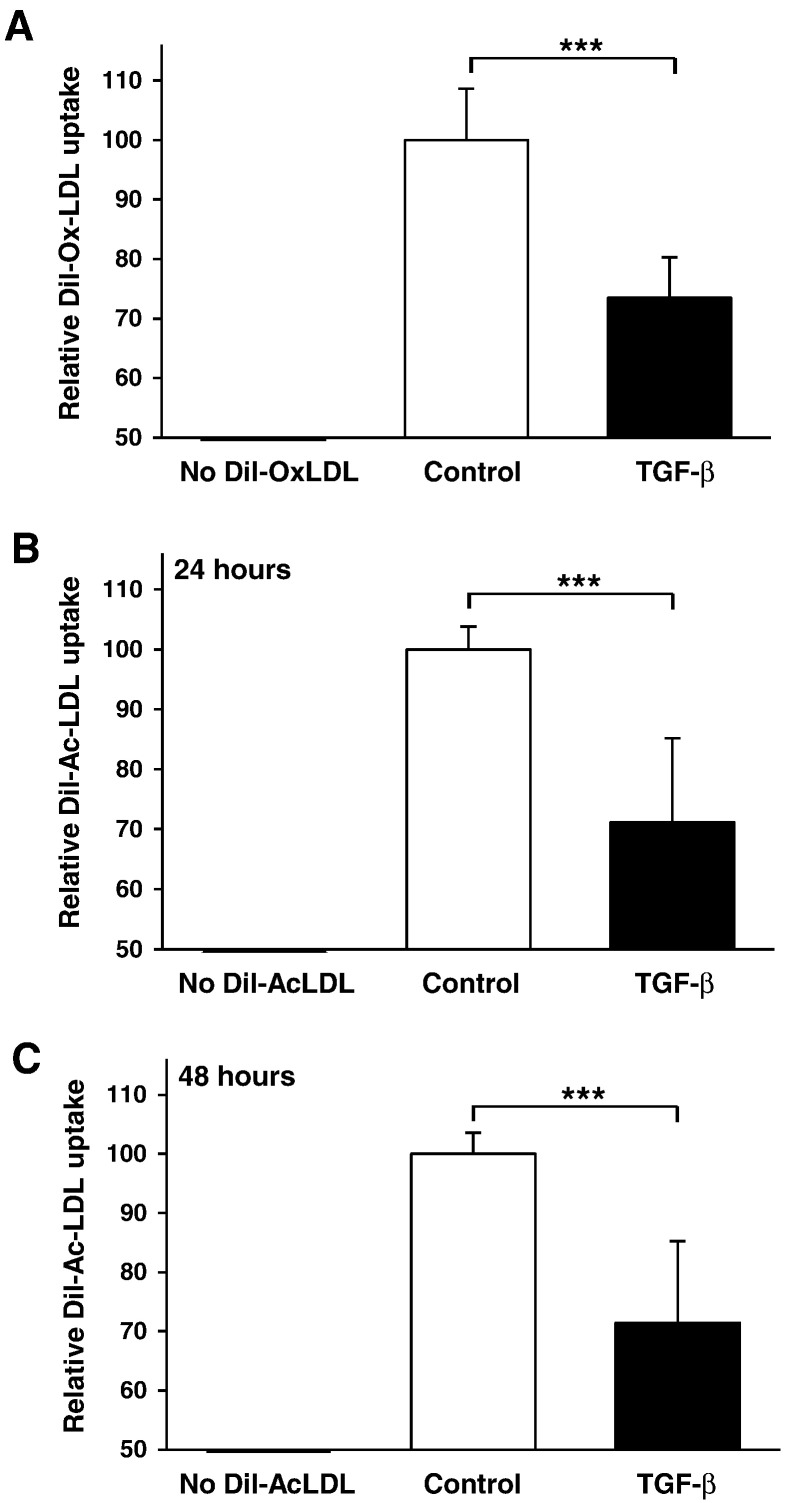
In order to determine if the changes observed in gene expression impart an observable change in cellular lipid uptake, we examined the effect of exogenous TGF-β stimulation on the uptake of OxLDL and AcLDL, both of which are commonly used to perform in vitro foam cell formation assays [2,3,37,38]. DiI-labeled LDLs were used at concentrations reflecting those employed in other published in vitro studies [2,3] and, similar to several previous publications [39–43], the effect of TGF-β on the uptake of modified LDL was represented relative to control cells. As shown in Fig. 3A, incubation of the cells with TGF-β for 24 hours produced an approximate 25% reduction in OxLDL uptake. Interestingly, both 24 and 48 hour TGF-β stimulation with AcLDL resulted in a slightly greater 30% reduction in DiI-AcLDL uptake (Fig. 3B and C respectively). The magnitude of these reductions exceeds that seen for other anti-atherogenic cytokines in identical systems, such as IL-33 [2], suggesting that TGF-β plays a key role in the inhibition of foam cell formation.
Fig. 3.
TGF-β inhibits modified LDL uptake by human macrophages. (A) DiI-OxLDL uptake is shown in THP-1 macrophages either left untreated (empty bars) or incubated with TGF-β (30 ng/ml: filled bars) for 24 hours. Data represent the mean ± SD of 4 independent experiments, or DiI-AcLDL uptake is shown in THP-1 macrophages either left untreated (empty bars) or incubated with TGF-β (30 ng/ml: filled bars) for (B) 24 hours or (C) 48 hours. Data represent the mean ± SD of 6 independent experiments. Relative modified LDL uptake is displayed as a % of untreated controls (arbitrarily assigned as 100%). Statistical analysis was performed using the Student's t-test; *** P < 0.001.
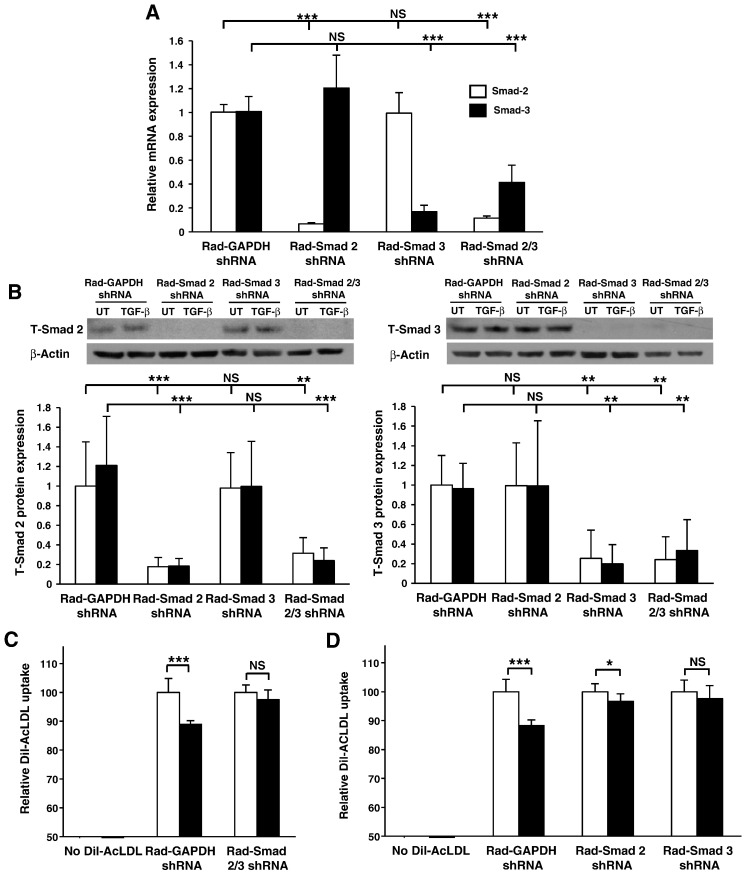
The involvement of Smad-2 and Smad-3 in this process was subsequently analyzed using THP-1 cells expressing adenoviral delivered shRNAs that facilitate the specific knockdown of Smad-2 or Smad-3 transcript levels; approximately 94% and 87% reductions respectively (Fig. 4A) translate into approximately 84% (T-Smad 2) and 88% (T-Smad 3) reductions in protein level (Fig. 4B). TGF-β stimulation had no significant effect on T-Smad protein levels. It should be noted that in all experiments hereafter a virally infected shRNA system was adopted due to the improved level of gene silencing observed compared to our siRNA system (Fig. 1B). In addition, in another study of a similar nature on death receptor 3 (DR3) we had shown that the changes in foam cell formation and the expression of downstream genes following knockdown of DR3 was similar to those seen using bone marrow-derived macrophages from DR3 knockout mice [3]. The duration of TGF-β treatment was increased to 48 hours in accordance with maximal AcLDL uptake in our virally infected system. As shown in Fig. 4C, comparison of cytokine treated and untreated cells shows that the TGF-β-mediated reduction in AcLDL uptake in the GAPDH shRNA infected control was attenuated in cells deficient in both Smad-2 and Smad-3 expression, and similarly, in cells deficient in Smad-2 or Smad-3 expression (Fig. 4D). It must be noted that a reduced magnitude in AcLDL uptake was observed in the GAPDH shRNA infected control cells (Fig. 4C and D) when compared to uninfected cells (Fig. 3C). Despite this slightly attenuated response, which could be attributed to the infection process, these data suggest that TGF-β inhibits the uptake of AcLDL and that this process was dependent on Smad-2 and Smad-3.
Fig. 4.
TGF-β inhibits modified LDL uptake by human macrophages through a Smad-dependent pathway. (A) Relative Smad-2 (empty bars) or Smad-3 (filled bars) mRNA expression was determined in THP-1 macrophages infected with either Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA or both Rad-Smad 2 shRNA and Rad-Smad 3 shRNA. Gene-specific mRNA expression levels were calculated using the comparative Ct method and normalized to RPL13A levels with untreated Rad-GAPDH shRNA infected cells given an arbitrary value of 1. Data represent the mean ± SD of 3 independent experiments. (B) T-Smad 2 or T-Smad 3 protein expression was determined in THP-1 macrophages infected with either Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA or both Rad-Smad 2 shRNA and Rad-Smad 3 shRNA in the absence (empty bars) or presence (filled bars) of 48 hours treatment with 30 ng/ml TGF-β. Protein expression, as determined by densitometric analysis, was normalized to β-actin and is displayed as a fold change compared to the untreated Rad-GAPDH shRNA infected cells (arbitrarily assigned as 1). Data represent the mean ± SD of 3 independent experiments. (C) DiI-AcLDL uptake is shown in THP-1 macrophages infected with Rad-GAPDH shRNA or Rad-Smad 2 shRNA and Rad-Smad 3 shRNA together. Data represent the mean ± SD of 3 independent experiments, or in (D) THP-1 macrophages infected with Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA individually in the absence (empty bars) or presence (filled bars) of 48 hour TGF-β (30 ng/ml) stimulation. Data represent the mean ± SD of 4 independent experiments. Relative modified LDL uptake is displayed as a % of untreated controls (arbitrarily assigned as 100%). Statistical analysis was performed using one-way ANOVA with Tukey's post hoc analysis where homogeneity of variance was met (Panel B), or Welch's test of equality of means with Games–Howell post-hoc analysis (Panel A), or Student's t-test (for single comparisons in panels C and D). * P < 0.05; ** P < 0.01; *** P < 0.001. NS indicates not significant.
3.3. TGF-β alters the expression of key genes involved in macrophage cholesterol uptake through a Smad-2-dependent mechanism
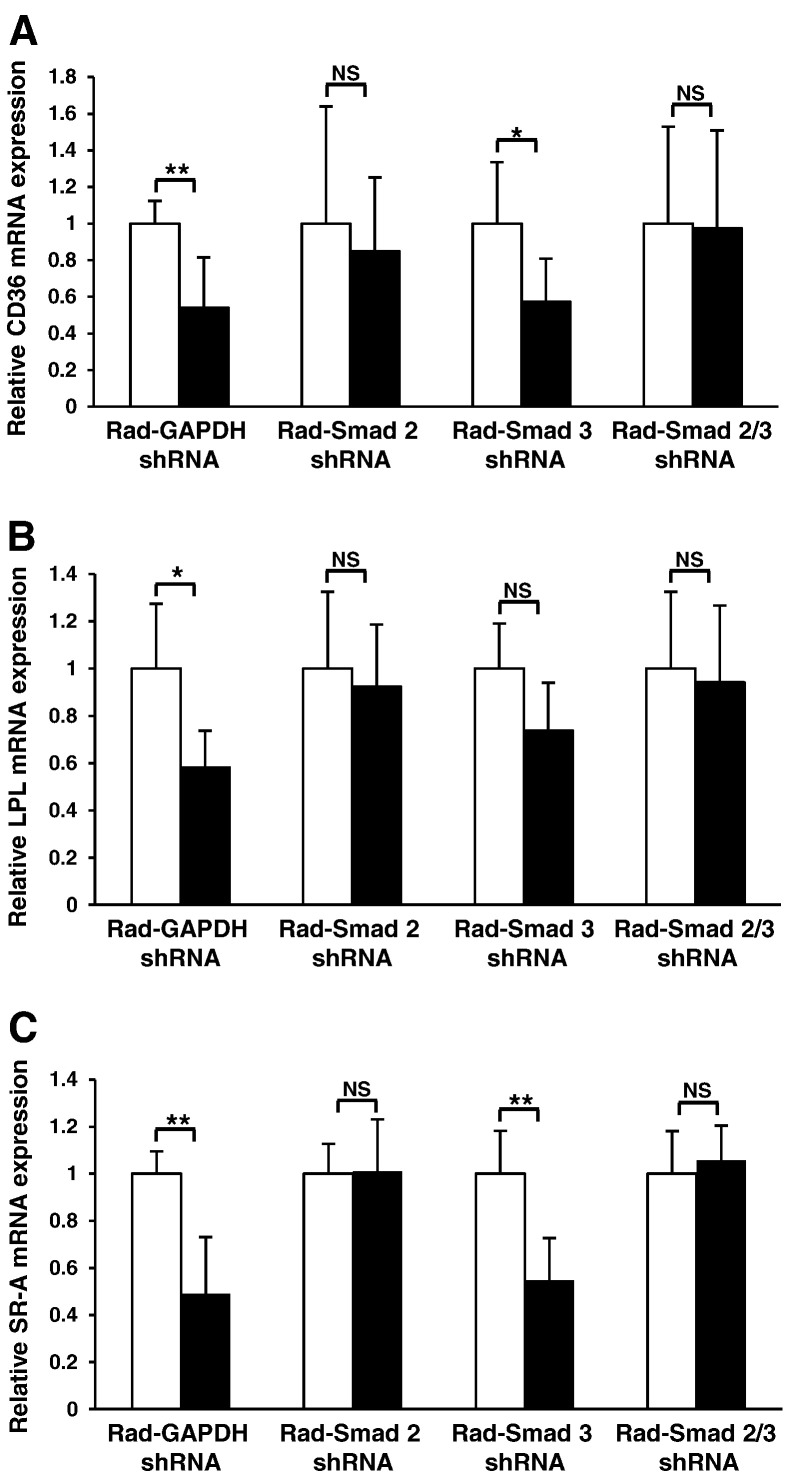
In order to confirm that the regulation of key genes involved in macrophage cholesterol uptake is Smad-dependent, and to determine if Smad-2 and Smad-3 play distinct roles in this process, we next examined the transcript levels of CD36, LPL and SR-A in our Smad depleted, TGF-β stimulated virally infected system. As already discussed, efficient silencing of Smad mRNA and protein expression was achieved using Rad-Smad shRNA infection of THP-1 cells (Fig. 4A and B). As shown in Fig. 5, a statistically significant reduction in the expression of CD36 (Fig. 5A), LPL (Fig. 5B) and SR-A (Fig. 5C) transcripts were observed in TGF-β treated, GAPDH shRNA infected cells when compared to the untreated GAPDH shRNA infected cells thus corroborating our previous findings. Furthermore, Smad-2 shRNA infection abolished the TGF-β responsiveness for all transcripts examined while Smad-3 depleted THP-1 macrophages maintained their ability to significantly down regulate CD36 and SR-A mRNA expression (Fig. 5A and C) in response to TGF-β. It should be noted that despite not reaching significance, TGF-β stimulation did induce a trend towards a decrease in LPL mRNA expression in Smad-3 depleted cells. Silencing of both Smad-2 and ‐3 in THP-1 macrophages also abolished the responsiveness of these genes to TGF-β-mediated inhibition (Fig. 5).
Fig. 5.
TGF-β regulates the expression of key uptake genes in THP-1 macrophages through a Smad-dependent pathway. The relative CD36 (A), LPL (B) or SR-A (C) mRNA expression was determined in THP-1 macrophages infected with either Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA or both Rad-Smad 2 shRNA and Rad-Smad 3 shRNA in the absence (empty bars) or presence (filled bars) of 48 hour TGF-β (30 ng/ml) stimulation. Data represent the mean ± SD of 3 independent experiments. Gene-specific mRNA expression levels were calculated using the comparative Ct method and normalized to RPL13A levels with untreated cells given an arbitrary value of 1. Statistical analysis was performed using the Student's t-test, * P < 0.05; ** P < 0.01. NS indicates not significant.
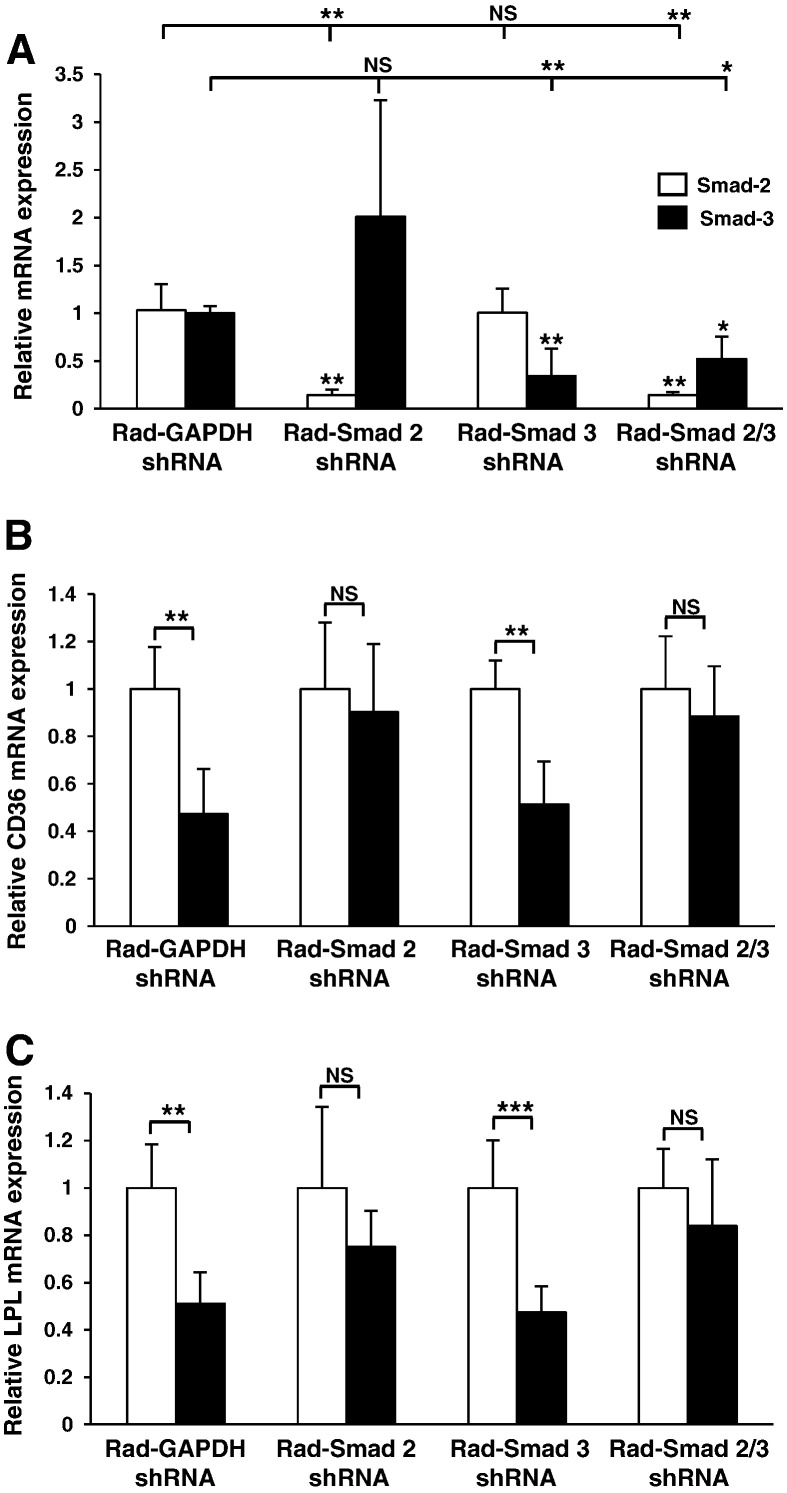
In order to determine if these responses were conserved in primary cells, representative experiments were performed in HMDM. Fig. 6A illustrates effective silencing of Smad-2 (86%) and Smad-3 (66%) gene expression in HMDM and, interestingly, in these cells Smad-2 knockdown can be seen to induce a non-significant 2-fold increase in Smad-3 expression. As in the case of THP-1 cells, TGF-β stimulation reduced the expression of CD36 and LPL in Rad-GAPDH shRNA infected cells (Fig. 6B and C). Likewise, knockdown of Smad-2 expression resulted in a marked attenuation of TGF-β responsiveness for both CD36 and LPL expression. In addition, a similar effect was seen following knockdown of both Smad-2 and Smad-3. Furthermore, a statistically significant reduction of CD36 and LPL mRNA expression was still observed when the expression of only Smad-3 was knocked down. Overall, these data confirm that TGF-β can down regulate the expression of key genes involved in the uptake of cholesterol and suggests that this process is mainly dependent on Smad-2 and not Smad-3 signaling.
Fig. 6.
TGF-β regulates the expression of key uptake genes in HMDMs through a Smad-dependent pathway. (A) Relative Smad-2 (empty bars) or Smad-3 (filled bars) mRNA expression was determined in HMDM infected with either Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA or both Rad-Smad 2 shRNA and Rad-Smad 3 shRNA. Data represent the mean ± SD of 3 independent experiments. (B) Relative CD36 or (C) LPL mRNA expression was analyzed in HMDM infected with either Rad-GAPDH shRNA or Rad-Smad 2 shRNA or Rad-Smad 3 shRNA or both Rad-Smad 2 shRNA and Rad-Smad 3 shRNA and then incubated for 48 hours in the absence (empty bars) or presence (filled bars) of TGF-β (30 ng/ml). Data represent the mean ± SD of 3 independent experiments. Gene-specific mRNA expression levels were calculated using the comparative Ct method and normalized to RPL13A levels with untreated cells given an arbitrary value of 1. Statistical analysis was performed using the Student's t-test (for single comparisons), or for multiple comparisons one-way ANOVA with Tukey's post hoc analysis where homogeneity of variance was met (panel A; Smad 2, empty bars); or Welch's test of equality of means with Games–Howell post-hoc analysis (A; Smad 3, filled bars), ** P < 0.01; *** P < 0.001. NS indicates not significant.
4. Discussion
The formation and accumulation of lipid engorged foam cells in the intima of the arterial wall is the key occurrence during the development of atherosclerosis [44]. It is understood that TGF-β can inhibit macrophage foam cell formation by regulating the expression of classic lipoprotein uptake and efflux genes [45]. However, the molecular mechanisms underlying such regulation is poorly understood and Smads have been shown to be required for the action of TGF-β on only a single gene, CD136 [15]. Further studies are necessary because of the existence of Smad-dependant and ‐independent mechanisms for the action of this cytokine [26,27]. In this study, for the first time, we clearly demonstrate that the Smad signal transduction pathway plays a critical role during TGF-β-mediated modulation of key cholesterol uptake (LPL, SR-A, SR-B1 and CD36) and efflux genes (ApoE, ABCA1 and ABCG1), and that this response is accompanied by a Smad-dependant reduction in AcLDL uptake in macrophages. These findings provide novel insights into the mechanisms underlying the TGF-β regulation of foam cell formation.
TGF-β, its receptors and Smads are all known to be expressed by macrophages in early stage atherosclerotic lesions present in human aortas [25]. Following arterial injury or activation, TGF-β is converted from a latent precursor to an activated form by a group of calcium-dependent proteases called furin-like proprotein convertases [46]. Active forms of Smad-2 and ‐3 have also been shown to exist in aortic lesions of atherosclerotic ApoE deficient mice [47], although it is unclear whether Smad activation occurs in response to arterial injury in humans. To this end, we have shown that Smad-2 and ‐3 are rapidly phosphorylated (and therefore activated) in response to TGF-β in both THP-1 macrophages and HMDM (Fig. 1A and supplementary data). The Smad signaling pathway has also been shown to be active in human atherosclerotic lesions suggesting that macrophages present in such lesions are TGF-β/Smad signaling sensitive [48].
Despite the establishment of Smads as classical transducers of the TGF-β signal, little is known about their role during the regulation of macrophage cholesterol homeostasis. Using siRNA knockdown of Smad-2 and ‐3 in THP-1 macrophages, we initially demonstrated that the Smad signaling pathway (Smad-2 and ‐3 together) was crucial for the TGF-β-induced expression of ApoE, ABCA1 and ABCG1 and also for the TGF-β-mediated inhibition of LPL, SR-A, CD36 and SR-B1 expression (Fig. 2). More detailed examination, using a more robust and specific viral shRNA system, demonstrated that the Smad signaling cascade plays a crucial role during the TGF-β-mediated inhibition of AcLDL uptake by macrophages (Fig. 4) and highlighted the Smad-2 dependency of CD36, SR-A and to some extent LPL gene expression in THP-1 macrophages and that of CD36 and LPL in HMDM (Figs. 5 and 6). Considering the involvement of Smad-2 during the TGF-β-mediated inhibition of CD36, a major scavenger receptor for OxLDL, these data also suggest that the TGF-β-mediated inhibition of OxLDL uptake (Fig. 3A) may also be mediated by a Smad-2 dependent mechanism.
Interestingly, as seen during the knockdown of Smad-2 expression in NIH/3T3 fibroblast cell line [49], depletion of Smad-2 expression in HMDM results in a non-significant increase in Smad-3 expression (Fig. 6A) suggesting that a compensatory mechanism may exist between Smad-2 and Smad-3 expression. Despite this, a TGF-β-mediated reduction in the expression of CD36 and LPL in HMDM was not significant in Smad-2 (or both Smad-2 and Smad-3) depleted cells (Fig. 6). In contrast, a statistically significant reduction in the expression of these genes was seen in Smad-3 depleted cells, thereby substantiating the dominant role of Smad-2.
The emergence of Smad-2 as the key player in the process of macrophage lipid accumulation may not appear surprising given the superior levels of activated Smad-2 compared to Smad-3 seen in both THP-1 macrophages and HMDM (Fig. 1 and supplementary data), although the majority of existing evidence does suggest that Smad-3 is the functionally dominant TGF-β signal transducer [50,51]. However, it has now become apparent that Smad-2 can act as the dominant component of the TGF-β signal transduction in many cellular systems [22]. One of the key differences between the two Smads is the ability of Smad-3 to bind directly to DNA through its MH1 domain while Smad-2 requires Smad-4 for DNA binding due to a 30 amino-acid insert in this domain [22]. This difference may be important in the regulation of genes involved in macrophage cholesterol homeostasis and foam cell formation.
5. Conclusion
In conclusion, we have demonstrated for the first time that the Smad signaling pathway, particularly Smad-2, plays a crucial role in the control of macrophage cholesterol uptake and expression of key genes implicated in this process by TGF-β. Our findings, along with the ability of TGF-β to increase the expression of key macrophage cholesterol efflux genes (Fig. 2) and cellular cholesterol efflux, as shown elsewhere [18–20], indicate that TGF-β has substantial anti-foam cell properties and highlight its potential as a powerful anti-foam cell therapeutic tool for the future.
Acknowledgements
This work was supported by the British Heart Foundation (PG/08/073/25520).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbadis.2012.06.002.
Appendix A. Supplementary data
Supplementary materials.
References
- 1.McLaren J.E., Michael D.R., Ashlin T.G., Ramji D.P. Cytokines, macrophage lipid metabolism and foam cells: implications for cardiovascular disease therapy. Prog. Lipid Res. 2011;50:331–347. doi: 10.1016/j.plipres.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 2.McLaren J., Michael D., Salter R., Ashlin T., Calder C., Miller A., Liew F., Ramji D. IL-33 reduces macrophage foam cell formation. J. Immunol. 2010;185:1222–1229. doi: 10.4049/jimmunol.1000520. [DOI] [PubMed] [Google Scholar]
- 3.McLaren J., Calder C., McSharry B., Sexton K., Salter R., Singh N., Wilkinson G., Wang E., Ramji D. The TNF-like protein 1A-death receptor 3 pathway promotes macrophage foam cell formation in vitro. J. Immunol. 2010;184:5827–5834. doi: 10.4049/jimmunol.0903782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLaren J., Ramji D. Interferon gamma: a master regulator of atherosclerosis. Cytokine Growth Factor Rev. 2009;20:125–135. doi: 10.1016/j.cytogfr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Singh N., Ramji D. Transforming growth factor-beta-induced expression of the apolipoprotein E gene requires c-Jun N-terminal kinase, p38 kinase, and casein kinase 2. Arterioscler. Thromb. Vasc. Biol. 2006;26:1323–1329. doi: 10.1161/01.ATV.0000220383.19192.55. [DOI] [PubMed] [Google Scholar]
- 6.Singh N., Ramji D. The role of transforming growth factor-beta in atherosclerosis. Cytokine Growth Factor Rev. 2006;17:487–499. doi: 10.1016/j.cytogfr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Borkowski P., Robinson M., Kusiak J., Borkowski A., Brathwaite C., Mergner W. Studies on TGF-beta 1 gene expression in the intima of the human aorta in regions with high and low probability of developing atherosclerotic lesions. Mod. Pathol. 1995;8:478–482. [PubMed] [Google Scholar]
- 8.Shull M., Ormsby I., Kier A., Pawlowski S., Diebold R., Yin M., Allen R., Sidman C., Proetzel G., Calvin D. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mallat Z., Gojova A., Marchiol-Fournigault C., Esposito B., Kamaté C., Merval R., Fradelizi D., Tedgui A. Inhibition of transforming growth factor-beta signaling accelerates atherosclerosis and induces an unstable plaque phenotype in mice. Circ. Res. 2001;89:930–934. doi: 10.1161/hh2201.099415. [DOI] [PubMed] [Google Scholar]
- 10.Robertson A., Rudling M., Zhou X., Gorelik L., Flavell R., Hansson G. Disruption of TGF-beta signaling in T cells accelerates atherosclerosis. J. Clin. Invest. 2003;112:1342–1350. doi: 10.1172/JCI18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tall A.R., Yvan-Charvet L., Terasaka N., Pagler T., Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Moore K., Freeman M. Scavenger receptors in atherosclerosis: beyond lipid uptake. Arterioscler. Thromb. Vasc. Biol. 2006;26:1702–1711. doi: 10.1161/01.ATV.0000229218.97976.43. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y.W., Wang Q., Ma X., Li X.X., Liu X.H., Xiao J., Liao D.F., Xiang J., Tang C.K. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells. J. Atheroscler. Thromb. 2010;17:493–502. doi: 10.5551/jat.3152. [DOI] [PubMed] [Google Scholar]
- 14.Han J., Hajjar D.P., Tauras J.M., Feng J., Gotto A.M., Nicholson A.C. Transforming growth factor-beta1 (TGF-beta1) and TGF-beta2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-gamma. J. Biol. Chem. 2000;275:1241–1246. doi: 10.1074/jbc.275.2.1241. [DOI] [PubMed] [Google Scholar]
- 15.Pioli P., Goonan K., Wardwell K., Guyre P. TGF-beta regulation of human macrophage scavenger receptor CD163 is Smad3-dependent. J. Leukoc. Biol. 2004;76:500–508. doi: 10.1189/jlb.1203617. [DOI] [PubMed] [Google Scholar]
- 16.Irvine S., Foka P., Rogers S., Mead J., Ramji D. A critical role for the Sp1-binding sites in the transforming growth factor-beta-mediated inhibition of lipoprotein lipase gene expression in macrophages. Nucleic Acids Res. 2005;33:1423–1434. doi: 10.1093/nar/gki280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bottalico L., Wager R., Agellon L., Assoian R., Tabas I. Transforming growth factor-beta 1 inhibits scavenger receptor activity in THP-1 human macrophages. J. Biol. Chem. 1991;266:22866–22871. [PubMed] [Google Scholar]
- 18.Draude G., Lorenz R. TGF-beta1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H1042–H1048. doi: 10.1152/ajpheart.2000.278.4.H1042. [DOI] [PubMed] [Google Scholar]
- 19.Argmann C., Van Den Diepstraten C., Sawyez C., Edwards J., Hegele R., Wolfe B., Huff M. Transforming growth factor-beta1 inhibits macrophage cholesteryl ester accumulation induced by native and oxidized VLDL remnants. Arterioscler. Thromb. Vasc. Biol. 2001;21:2011–2018. doi: 10.1161/hq1201.099426. [DOI] [PubMed] [Google Scholar]
- 20.Panousis C., Evans G., Zuckerman S. TGF-beta increases cholesterol efflux and ABC-1 expression in macrophage-derived foam cells: opposing the effects of IFN-gamma. J. Lipid Res. 2001;42:856–863. [PubMed] [Google Scholar]
- 21.Salter R.C., Arnaoutakis K., Michael D.R., Singh N.N., Ashlin T.G., Buckley M.L., Kwan A.P., Ramji D.P. The expression of a disintegrin and metalloproteinase with thrombospondin motifs 4 in human macrophages is inhibited by the anti-atherogenic cytokine transforming growth factor-β and requires Smads, p38 mitogen-activated protein kinase and c-Jun. Int. J. Biochem. Cell Biol. 2011;43:805–811. doi: 10.1016/j.biocel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown K., Pietenpol J., Moses H. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J. Cell. Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 23.Woodward R.N., Finn A.V., Dichek D.A. Identification of intracellular pathways through which TGF-beta1 upregulates PAI-1 expression in endothelial cells. Atherosclerosis. 2006;186:92–100. doi: 10.1016/j.atherosclerosis.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 24.Deaton R.A., Su C., Valencia T.G., Grant S.R. Transforming growth factor-beta1-induced expression of smooth muscle marker genes involves activation of PKN and p38 MAPK. J. Biol. Chem. 2005;280:31172–31181. doi: 10.1074/jbc.M504774200. [DOI] [PubMed] [Google Scholar]
- 25.Kalinina N., Agrotis A., Antropova Y., Ilyinskaya O., Smirnov V., Tararak E., Bobik A. Smad expression in human atherosclerotic lesions: evidence for impaired TGF-beta/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler. Thromb. Vasc. Biol. 2004;24:1391–1396. doi: 10.1161/01.ATV.0000133605.89421.79. [DOI] [PubMed] [Google Scholar]
- 26.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 27.Moustakas A., Heldin C.H. Non-Smad TGF-beta signals. J. Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 28.Grewal T., Priceputu E., Davignon J., Bernier L. Identification of a gamma-interferon-responsive element in the promoter of the human macrophage scavenger receptor A gene. Arterioscler. Thromb. Vasc. Biol. 2001;21:825–831. doi: 10.1161/01.atv.21.5.825. [DOI] [PubMed] [Google Scholar]
- 29.Chung K., Hart C., Al-Bassam S., Avery A., Taylor J., Patel P., Vojtek A., Turner D. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanton R., McSharry B., Armstrong M., Tomasec P., Wilkinson G. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques. 2008;45:659–662. doi: 10.2144/000112993. (664–658) [DOI] [PubMed] [Google Scholar]
- 31.Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- 32.Izawa D., Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nat. Cell Biol. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W.Y., Wang D.H., Yen R.C., Luo J., Gu W., Baylin S.B. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Yang S.H., Sharrocks A.D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 35.Larigauderie G., Furman C., Jaye M., Lasselin C., Copin C., Fruchart J.C., Castro G., Rouis M. Adipophilin enhances lipid accumulation and prevents lipid efflux from THP-1 macrophages: potential role in atherogenesis. Arterioscler. Thromb. Vasc. Biol. 2004;24:504–510. doi: 10.1161/01.ATV.0000115638.27381.97. [DOI] [PubMed] [Google Scholar]
- 36.Chandok M.R., Okoye F.I., Ndejembi M.P., Farber D.L. A biochemical signature for rapid recall of memory CD4 T cells. J. Immunol. 2007;179:3689–3698. doi: 10.4049/jimmunol.179.6.3689. [DOI] [PubMed] [Google Scholar]
- 37.Geng Y., Hansson G. Interferon-gamma inhibits scavenger receptor expression and foam cell formation in human monocyte-derived macrophages. J. Clin. Invest. 1992;89:1322–1330. doi: 10.1172/JCI115718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein J., Ho Y., Basu S., Brown M. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl. Acad. Sci. U. S. A. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morawietz H., Rueckschloss U., Niemann B., Duerrschmidt N., Galle J., Hakim K., Zerkowski H.R., Sawamura T., Holtz J. Angiotensin II induces LOX-1, the human endothelial receptor for oxidized low-density lipoprotein. Circulation. 1999;100:899–902. doi: 10.1161/01.cir.100.9.899. [DOI] [PubMed] [Google Scholar]
- 40.Morawietz H., Duerrschmidt N., Niemann B., Galle J., Sawamura T., Holtz J. Augmented endothelial uptake of oxidized low-density lipoprotein in response to endothelin-1. Clin. Sci. (Lond.) 2002;103(Suppl. 48):9S–12S. doi: 10.1042/CS103S009S. [DOI] [PubMed] [Google Scholar]
- 41.Jagavelu K., Tietge U.J., Gaestel M., Drexler H., Schieffer B., Bavendiek U. Systemic deficiency of the MAP kinase-activated protein kinase 2 reduces atherosclerosis in hypercholesterolemic mice. Circ. Res. 2007;101:1104–1112. doi: 10.1161/CIRCRESAHA.107.156075. [DOI] [PubMed] [Google Scholar]
- 42.Araki N., Johnson M.T., Swanson J.A. A role for phosphoinositide 3-kinase in the completion of macropinocytosis and phagocytosis by macrophages. J. Cell Biol. 1996;135:1249–1260. doi: 10.1083/jcb.135.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corcoran M.P., Lichtenstein A.H., Meydani M., Dillard A., Schaefer E.J., Lamon-Fava S. The effect of 17beta-estradiol on cholesterol content in human macrophages is influenced by the lipoprotein milieu. J. Mol. Endocrinol. 2011;47:109–117. doi: 10.1530/jme-10-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Libby P., Ridker P., Hansson G. Inflammation in atherosclerosis: from pathophysiology to practice. J. Am. Coll. Cardiol. 2009;54:2129–2138. doi: 10.1016/j.jacc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramji D., Singh N., Foka P., Irvine S., Arnaoutakis K. Transforming growth factor-beta-regulated expression of genes in macrophages implicated in the control of cholesterol homoeostasis. Biochem. Soc. Trans. 2006;34:1141–1144. doi: 10.1042/BST0341141. [DOI] [PubMed] [Google Scholar]
- 46.Sluijter J., Verloop R., Pulskens W., Velema E., Grimbergen J., Quax P., Goumans M., Pasterkamp G., de Kleijn D. Involvement of furin-like proprotein convertases in the arterial response to injury. Cardiovasc. Res. 2005;68:136–143. doi: 10.1016/j.cardiores.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Nachtigal P., Vecerova L., Pospisilova N., Micuda S., Brcakova E., Navarro Hernandez E., Pospechova K., Semecky V. Endoglin co-expression with eNOS, SMAD2 and phosphorylated SMAD2/3 in normocholesterolemic and hypercholesterolemic mice: an immunohistochemical study. Histol. Histopathol. 2009;24:1499–1506. doi: 10.14670/HH-24.1499. [DOI] [PubMed] [Google Scholar]
- 48.Bot P., Hoefer I., Sluijter J., van Vliet P., Smits A., Lebrin F., Moll F., de Vries J., Doevendans P., Piek J., Pasterkamp G., Goumans M. Increased expression of the transforming growth factor-beta signaling pathway, endoglin, and early growth response-1 in stable plaques. Stroke. 2009;40:439–447. doi: 10.1161/STROKEAHA.108.522284. [DOI] [PubMed] [Google Scholar]
- 49.Zheng R., Xiong Q., Zuo B., Jiang S., Li F., Lei M., Deng C., Xiong Y. Using RNA interference to identify the different roles of SMAD2 and SMAD3 in NIH/3T3 fibroblast cells. Cell Biochem. Funct. 2008;26:548–556. doi: 10.1002/cbf.1464. [DOI] [PubMed] [Google Scholar]
- 50.Dong Y., Tang L., Letterio J., Benveniste E. The Smad3 protein is involved in TGF-beta inhibition of class II transactivator and class II MHC expression. J. Immunol. 2001;167:311–319. doi: 10.4049/jimmunol.167.1.311. [DOI] [PubMed] [Google Scholar]
- 51.Frederick J., Liberati N., Waddell D., Shi Y., Wang X. Transforming growth factor beta-mediated transcriptional repression of c-myc is dependent on direct binding of Smad3 to a novel repressive Smad binding element. Mol. Cell. Biol. 2004;24:2546–2559. doi: 10.1128/MCB.24.6.2546-2559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.