Abstract
Malignant gliomas are the most common tumors originating within the central nervous system and account for over 15,000 deaths annually in the United States. The median survival for glioblastoma, the most common and aggressive of these tumors, is only 14 months. Therapeutic strategies targeting glioma cells migrating away from the tumor core are currently hampered by the difficulty of reproducing migration in the neural parenchyma in vitro. We utilized a tissue engineering approach to develop a physiologically relevant model of glioma cell migration. This revealed that glioma cells display dramatic differences in migration when challenged by random versus aligned electrospun poly-ɛ-caprolactone nanofibers. Cells on aligned fibers migrated at an effective velocity of 4.2 ± 0.39 μm/h compared to 0.8 ± 0.08 μm/h on random fibers, closely matching in vivo models and prior observations of glioma spread in white versus gray matter. Cells on random fibers exhibited extension along multiple fiber axes that prevented net motion; aligned fibers promoted a fusiform morphology better suited to infiltration. Time-lapse microscopy revealed that the motion of individual cells was complex and was influenced by cell cycle and local topography. Glioma stem cell–containing neurospheres seeded on random fibers did not show cell detachment and retained their original shape; on aligned fibers, cells detached and migrated in the fiber direction over a distance sixfold greater than the perpendicular direction. This chemically and physically flexible model allows time-lapse analysis of glioma cell migration while recapitulating in vivo cell morphology, potentially allowing identification of physiological mediators and pharmacological inhibitors of invasion.
Introduction
Malignant gliomas are the most common tumors originating within the central nervous system and account for over 15,000 deaths annually in the United States. The median survival for glioblastoma, the most common and aggressive of these tumors, is only 14 months.1 This bleak prognosis is due in large part to the uniquely invasive ability of glioma cells, which allows them to infiltrate normal brain tissue, evade immunodetection, and resist cytotoxic therapies.2,3 Tumor cells exhibit a diffuse migration around the tumor core and disperse much further along anatomical fiber-like and tube-like structures of the brain, such as white matter tracts and blood vessels.1 This dispersion is critical because it prevents complete surgical removal and contributes to tumor recurrence followed by a rapid, lethal outcome.
Current therapeutic strategies focus preferentially on targeting cell proliferation or angiogenesis to eliminate residual foci of the growing tumor after surgical resection of the neoplastic mass.4 However, there are no strategies effectively formulated to act against the slowly dividing, migratory cells that have colonized the neural parenchyma.2,5 Indeed, current evidence suggests that radiotherapy and anti-angiogenic approaches may cause an increase in invasion, thus eventually promoting a selection of migratory cells that eventually thwart the efficacy of these therapies.6 The development of effective anti-invasive approaches for malignant gliomas has been largely hampered by the difficulty in modeling glioma cell migration appropriately in vitro.1 A number of in vitro migration and invasion assays are in common use, but significant limitations restrict their potential to predict cell behavior in vivo.
The most common model of glioma cell migration through an extracellular matrix (ECM) utilizes type-I collagen or collagen-based mixtures (e.g., Matrigel™) to form a gel in which cell aggregates (or spheroids) are seeded and allowed to disperse over several days. This model has been used to characterize molecular factors that promote glioma invasion, such as pericellular proteases.7 Importantly, the elongated shape of the migratory cells in collagen is similar to the typical morphology of infiltrating tumor cells in vivo.8,9 However, there are limitations in design that make this model less than optimal: (1) the collagen matrix is uniform, abolishing directional mechanical stimuli, and is difficult to manipulate structurally, (2) the matrix degrades over the culture period, preventing the study of secondary foci formed from dispersed cells, and (3) downstream assays such as Western blotting or immunostaining are difficult to perform due to the embedded nature of the cells. More importantly, fibrillar collagen-I is absent from the adult neural parenchyma (i.e., neuropil and white matter),10 and the collagen located in the basal lamina of brain blood vessels is not degraded by glioma cells in vivo,11 thus calling into question the relevance of collagen invasion assays as predictors of brain infiltration.
Other in vitro assays commonly used to study glioma migration have additional limitations. The traditional wound healing and the Boyden chamber (Transwell, Corning Incorporated, Corning, NY) assays, which measure motility on flat surfaces, are quantitative and informative but rely on cells arranged in flat monolayers on rigid substrates. This condition forces the cells to adopt a fibroblast-like morphology and motile behavior,12 quite different from that in the brain.13 At the other end of the spectrum is the slice assay in which glioma cells are seeded on live brain slices supported by appropriate culture medium.14,15 While this model challenges glioma cells with a true neural cytoarchitecture and has predictive power,16,17 it is laborious, is difficult to reproduce, and includes poorly controlled variables such as the rapid degradation of myelin fibers and the death of neural cells in the brain slice during the culture period.
Glioma infiltration in the brain parenchyma involves both short-range, nonoriented dispersion through the ECM of the neuropil as well as long-range migration along the major axes of white matter fibers and blood vessels, which act as highways for cell dispersion.18,19 Novel models of glioma migration should be able to reproduce the different modalities of individual cell motion and, in particular, the influence of the substrate topography on motility. Most available models use homogeneous matrices and partially reproduce short-range motion, but fail to mimic oriented motility along elongated anatomical structures. To design a more realistic assay for the study of glioma cell migration, we investigated the potential of a common tissue engineering matrix, electrospun nanofibers, precisely engineered to produce aligned and random structures.
Such biocompatible scaffolds have previously been used to study various aspects of cancer cell biology. For example, using synthetic substrates, Song et al. cultured spheroids of cancer cell lines in a high aspect rotating wall bioreactor20 and demonstrated that proliferation influenced environmental sensitivity and susceptibility to chemotherapeutic agents. Liu et al. used injectable hyaluronic acid21 to show that scaffolds can improve the in vivo behavior of seeded tumors in a murine model.
For our model a biocompatible polymeric substrate, poly-ɛ-caprolactone (PCL), was electrospun to form either randomly organized or tightly aligned submicron fibers. On PCL scaffolds, the migratory behavior of glioma cells displays clear, reproducible, and quantifiable differences when challenged by aligned versus randomly oriented topographic cues, in agreement with the observed behavior of these cells in vivo. These chemically and physically flexible assays underscore the relevance of tissue engineering scaffolds as a valuable tool that models glioma motility in neural-like structures, with potential to become the basis for tailored bioassays of tumor biopsies. In contrast to standard approaches utilizing electrospun matrices for reconstructive purposes, here electrospinning is used to develop a realistic in vitro system simulating brain tumor migration while retaining compatibility with downstream analytical techniques such as Western blotting, immunostaining, and RT-PCR.22 A key aspect of this work uniting it with tissue engineering is the basic question of how far and how fast mammalian neural cells can move along such nanoscaled structures to establish viable internal populations of seeded cells.23 In addition, scaffolds that more adequately recapitulate key aspects of brain structure simulating tumor cell migration by extension provide considerable guidance to related regenerative approaches.24,25
Materials and Methods
Electrospun nanofibers
Random fiber preparation
An 18 wt% solution of PCL (MW = 65,000; Sigma-Aldrich, St. Louis, MO) in acetone (Mallinckrodt Chemicals, Hazelwood, MO) was prepared by heating acetone to 50°C followed by continuous stirring to dissolve the PCL. After cooling to room temperature, the solution was placed in a 60 cc syringe with a 20 gauge blunt tip needle and electrospun using a high voltage DC power supply (Glassman High Voltage, Inc., High Bridge, NJ) set to 24 kV, a 20 cm tip-to-substrate distance, and a 16 mL/h flow rate. A 3 × 3″ (7.6 × 7.6 cm) sheet approximately 100 μm in thickness was deposited onto aluminum foil in 2 min. The PCL sheets were then placed in a vacuum overnight to ensure removal of residual acetone. High-resolution electrospray ionization ESI analysis (Esquire, Billerica, MA) was used to establish that the resulting acetone content is beneath our ability to detect it (less than 10 ppm).26 Scanning electron microscopy (SEM) was used to examine the morphology of the as-deposited fiber and to allow quantification of fiber diameter taken from measurements of 100 fibers.
Aligned fiber preparation
Alignment of the otherwise random structure produced by normal electrospinning has utilized many techniques, but the requirements for actively exploring cell migration called for enhanced alignment efficiency combined with levels of production sufficient to match the statistical requirements of cell culture. A method known as the split ground technique,27 in which fiber deposition rapidly alternates between two separate grounding plates, was found to provide relatively efficient alignment. The same polymer solution used to produce random fiber was electrospun using 10 kV, a horizontal tip-to-substrate distance of 20 cm, and a flow rate of 3 mL/h. Before spinning, fluorescein isothiocynate isomer I (FITC; Fluka BioChemika, Steinheim, Germany) was added to cooled solution at 10 mg/mL of polymer solution while stirring continuously. During electrospinning, fibers were deposited for 1 min onto a glass disc having two separate grounds at a 5 mm separation to a thickness of approximately 50 μm. These aligned electrospun samples were again placed in a vacuum overnight, a process proven to ensure removal of residual acetone.26 The samples were then sealed in zip-lock polyethylene bags. The bags were submerged in a 45°C water bath for 10 min. This acted as a stress anneal for the aligned fibers and prevented wrinkling otherwise observed during cell culture. As before, SEM was used to examine the morphology of the as-deposited fiber and to allow quantification of fiber diameter (n = 100).
Dispersed glioma culture and analysis of migration
Cell culture
The human glioma cell line U251 was routinely cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin 50 IU/mL, and streptomycin 50 μg/mL. To provide stable fluorescently labeled nuclei, resuspended cells were infected with lentiviruses carrying a red fluorescent protein (RFP) fused to the nuclear histone 2-B (vector pLenti-H2B-RFP, kindly provided by Dr. Yoshinaga Saeki, The Ohio State University) and a cytoplasmic green fluorescent protein (GFP), using standard transduction protocols. Several experiments were also replicated with the human primary glioma cells X12,28 which were maintained by flank passage in immunodeficient (nude) mice and similarly transduced for stable expression of fluorescent proteins.
For initial migration studies, fluorescently labeled U251 or X12 glioma cells were dissociated and seeded on nanofiber substrates at an initial density of ∼7 × 104 cells/mL. The cells were dispersed homogeneously on 12-mm-diameter discs of random or aligned nanofibers that had been attached to the bottom of 35 mm culture dishes, and subsequently tracked over 24 h using time-lapse microscopy using a confocal microscope (LSM 510; Zeiss, Minneapolis, MN) fitted with a culture chamber to provide normal temperature and humidity conditions. The cells were identified by both GFP (U251) and RFP (U251 and X12) fluorescence, and a 50-μm-thick Z-stack was captured every 7 min. Subsequently, cells were fixed with 1% glutaraldehyde in PBS solution for 0.5 h. Dehydration was performed by washing the cells with solutions of increasing ethanol concentration (40%, 50%, 75%, 95%, 100%, and 100%, 5 min each), followed by rapid air-drying. Samples were mounted on aluminum pin mounts and coated with 15 nM of osmium for SEM.
Proliferation
Cell growth was monitored in 12-well dishes seeded on day 0 at 10,000 cells per well. Two separate experiments were conducted each of which involved three wells per fiber sample. The medium was changed every 24 h, and cell number determined through fluorimetric analysis of spent medium using Alamar blue (Invitrogen, Carlsbad, CA).
Migration analysis
Time-lapse confocal images were processed using ImageJ software to produce a maximal-intensity Z-projection at each captured time point. These images were then concatenated and their illumination was normalized to produce movies that were further analyzed by particle-tracking analysis. Using ImageJ, individual cells were manually identified and tracked throughout the entire duration (24–36 h) of the experiment. Total distance traveled, and average and individual cell velocities were then quantified and plotted over time.
Neurosphere glioma culture and analysis of migration
Cell culture
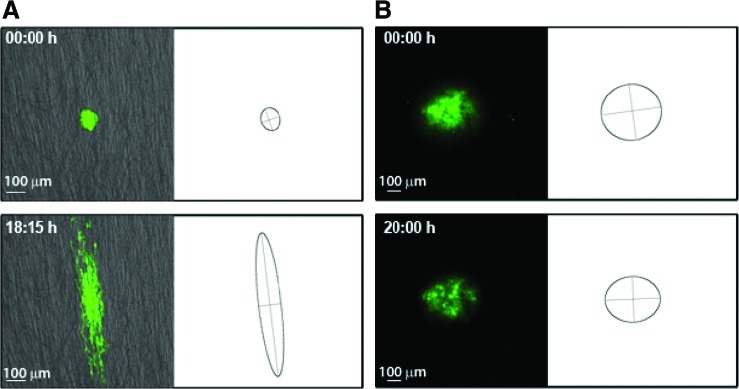
Fresh biopsy samples from high-grade gliomas were dissociated and cultured as previously described to enrich and maintain the population of glioma stem cells.29 Cells were maintained in Neurobasal culture medium (Invitrogen) supplemented with EGF 50 ng/mL, bFGF 50 ng/mL, LIF 10 ng/mL, and B27 supplement (Invitrogen), and grown in suspension as tumor aggregates known as neurospheres. Neurospheres were dissociated by trypsinization, and the isolated cells stably transduced for GFP expression with the lentiviral vector pCDH1-MCS-EF1-copGFP (System Biosciences, Mountain View, CA), following the manufacturer's recommendations. GFP-expressing neurospheres were seeded on discs of random or aligned PCL fibers (Fig. 8) and followed by time-lapse confocal microscopy, as described above. We allowed the cells to attach for 20 min before beginning cell tracking; this constitutes the minimum time needed for attachment and thus provides a real-time record of attachment and motion. Samples were subsequently fixed and processed for SEM using the procedures identical to those for dispersed cells.
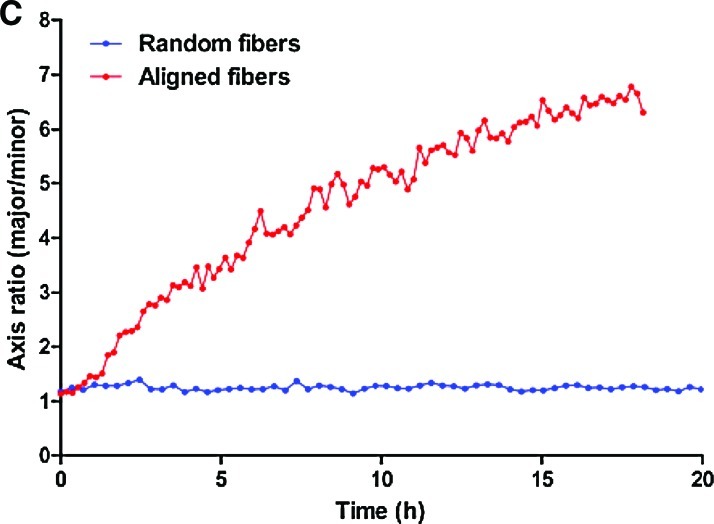
FIG. 8.
Representative frames showing cell dispersion from neurospheres seeded on aligned (A) and random (B) PCL fibers. The corresponding bounding ellipses were estimated by principal component analysis. The change in the ratio of the elliptic axes over time (i.e., a measure of anisotropic cell spread) revealed a sixfold increase in along-axis versus across-axis migration on aligned fibers (C). Color images available online at www.liebertonline.com/ten.
Migration analysis
Time-lapse confocal images were processed using ImageJ to produce movies of cell migration, as described above. The core mass of the neurospheres and their detached cells were analyzed as a population of scattered particles using principal component analysis. The major and minor axes of cell migration were calculated as eigenvectors of the covariance matrix of particle dispersion, and were further scaled to cover >95% of the original distribution of cells.
Results
Polycaprolactone fibers provide a substrate for glioma cell migration
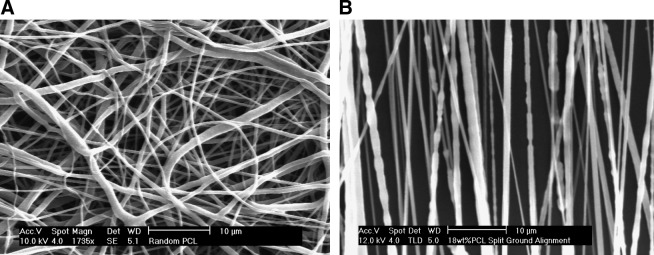
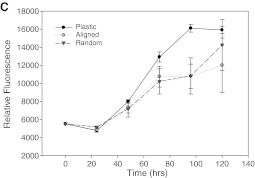
Electrospun PCL fibers alternating between the two ground plates showed a high degree of parallel alignment, in contrast to randomly deposited fibers (Fig. 1). Fiber diameters were nearly identical; the random fiber averaged 0.84 ± 0.04 μm, while the aligned fiber averaged 0.95 ± 0.05 μm. U251 glioma cells were seeded onto these fibers after covering them with appropriate culture medium. The cells rapidly adhered to both forms of the PCL fibers to exhibit strikingly different morphologies observed by high-magnification fluorescence microscopy (not shown) and SEM (Fig. 2). Cells adhered to parallel fibers essentially adopted a fusiform morphology and elongated along the axis of the single or few fibers to which they were attached (average length, 32 μm). In contrast, cells placed on randomly oriented fibers showed no preferential extension and spread similarly in all directions (average diameter, 15 μm). Adherence to PCL did not preclude cell motility or division, as analyzed below. Additionally, the cells did not show any evidence of toxicity-induced phenotypes or abnormal rates of cell death over culture periods of several days. Cell proliferation on the two types of fiber was identical both to each other and to tissue culture polystyrene (Fig. 3c), in agreement with prior uses of electrospun PCL with a wide variety of cell types.30–32
FIG. 1.
SEM of as-deposited, randomly oriented PCL fibers (A) and aligned PCL fibers (B). Scale bars: 10 μm.
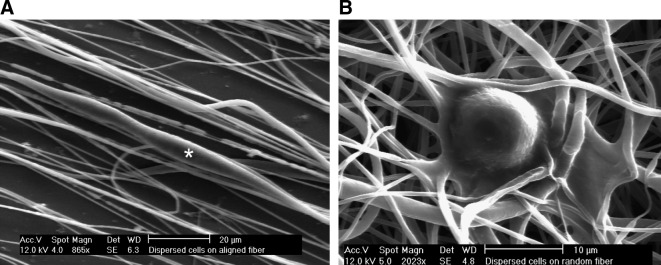
FIG. 2.
SEM showing examples of isolated U251 glioma cells migrating on aligned (A) or randomly oriented (B) PCL fibers. Note the cell in the center of (A) denoted with an asterisk. Cells on randomly oriented substrates interact with many fibers and do not show preferential pseudopodia extension. Conversely, cells on aligned substrates interact with relatively few nanofibers and are highly elongated in the direction of alignment.
FIG. 3.
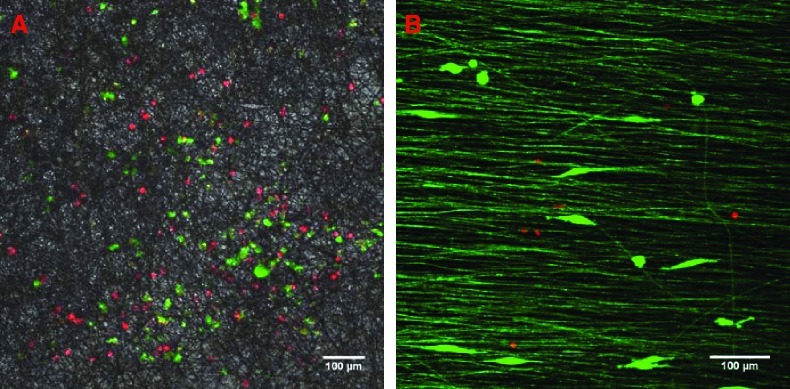
Confocal microscopy of U251 glioma cells detected by nuclear red and cytoplasmic green fluorescence on random (A) and aligned (B) PCL fibers. Random fibers in (A) are shown under phase contrast illumination; aligned fibers in (B) are shown under fluorescent illumination and appear green due to internal reflection of fluorescence. Scale bars: 100 μm. The growth curve of U251 glioma cells cultured on TCPS, aligned nanofibers, and random nanofibers is shown in (C). Color images available online at www.liebertonline.com/ten.
Glioma cells migrate differently on random and aligned fibers
We next analyzed migration of dispersed glioma cells on both types of scaffolds using time-lapse confocal fluorescence microscopy. Cells were engineered for stable expression of cytoplasmic GFP or nuclear RFP to facilitate detection and trajectory analysis.
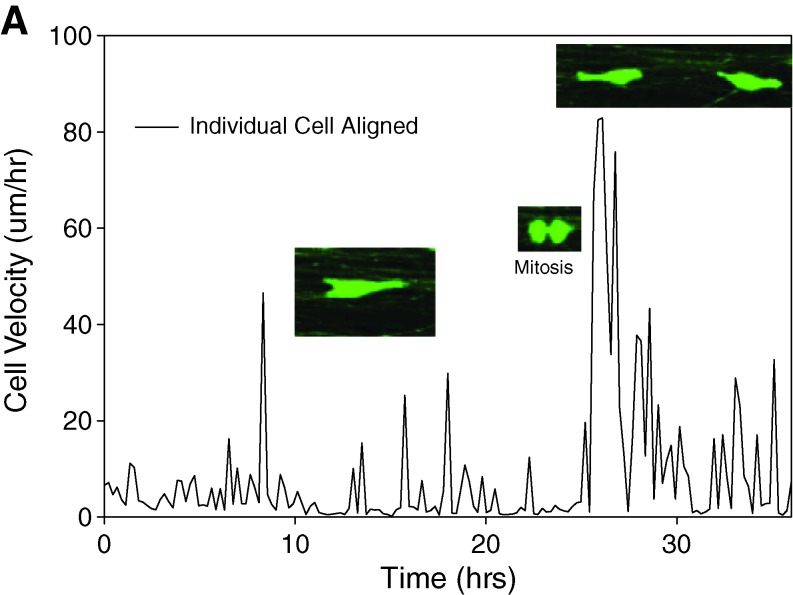
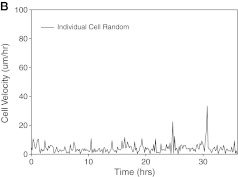
In agreement with the different morphologies observed in Figure 2, the motile behavior of the cells was also markedly different on randomly oriented versus aligned PCL fibers. Cells seeded on random fibers remained largely globular and contacted multiple fibers, displaying little progress in any direction while oscillating around major fiber intersections (Figs. 4A and 2B). In stark contrast, cells seeded on aligned fibers maintained an elongated morphology (except during division) and displayed decisive motion, that is, consistent motility along the fiber axis (Figs. 4B, 3B, and 2A). The results were identical when dispersed X12 glioma cells were assayed (not shown). Based on the detail inherent to our studies we define two distinct cell velocities: average velocity, or motion regardless of direction, and effective velocity reflecting the net distance traveled away from the cell's starting position along the fiber axes. The latter is significant in this context because it is more likely to represent invasion of a cell from one part of the brain to another rather than the random back-and-forth oscillation of a cell in the absence of an imposed chemotactic gradient.
FIG. 4.
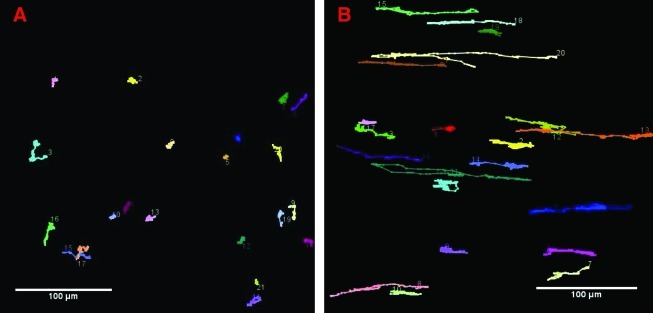
Motion cell-tracking of individual cells on as-deposited, random (A) and aligned (B) PCL nanofibers. The figure represents approximately 20 individual trajectories traced manually after a total tracking period of 36 h. Scale bars: 100 μm. Color images available online at www.liebertonline.com/ten.
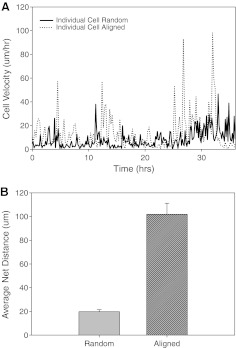
Average migration velocities calculated from individual U251 trajectories were 11.7 ± 0.7 μm/h (mean ± SEM) on aligned fibers versus 5.3 ± 0.3 μm/h on randomly oriented fibers. In contrast, the effective velocities are 4.2 ± 0.39 μm/h and 0.8 ± 0.08 μm/h, values that closely match experimental observations in vivo.33 Accordingly, the net distance traveled distance was approximately fivefold higher on aligned fibers than on random fibers.
Analysis of individual trajectories on aligned fibers disclosed that cell motility was characterized by periods of rapid motion interrupted by relatively short pauses (Figs. 5 and 6). The longest pauses corresponded, as expected, to periods of cell division, which were immediately followed by a burst of rapid motility of the daughter cells in opposite directions. Analysis of motility in relation to fiber alignment provided evidence that the efficiency of motion was highly influenced by small imperfections in alignment: cell encounters with even slightly misaligned fibers impeded migration, underscoring the strong effects of a directionally random substrate on migration.
FIG. 5.
Quantification of cell motion on nanofibers. (A) Point-to-point velocity of an individual cell seeded on random and aligned PCL fibers. (B) Net distance (mean ± SE) traveled by the cells on random and aligned PCL fibers over a 36 h period.
FIG. 6.
Representative motion of individual cells on aligned (A) and random (B) PCL nanofibers. The inset pictures show events of cell division, which were usually followed by a burst of motion of the daughter cells. Color images available online at www.liebertonline.com/ten.
Glioma neurospheres prepared as indicated in the methods section were able to adhere to PCL and, after seeding, showed better adhesion to this substrate than that usually observed on plastic culture dishes (Fig. 7). Time-lapse confocal tracking of cells migrating from the spheres revealed that cells on random fibers did not detach from the original aggregate, which retained its shape and dimensions over time (Fig. 8A). Conversely, cells readily detached from neurospheres seeded on aligned fibers and showed decisive motion away from the neurosphere along the fiber axes (Fig. 8B). Cell dispersion along the fibers was on average approximately sixfold higher than across the fibers (Fig. 8C).
FIG. 7.
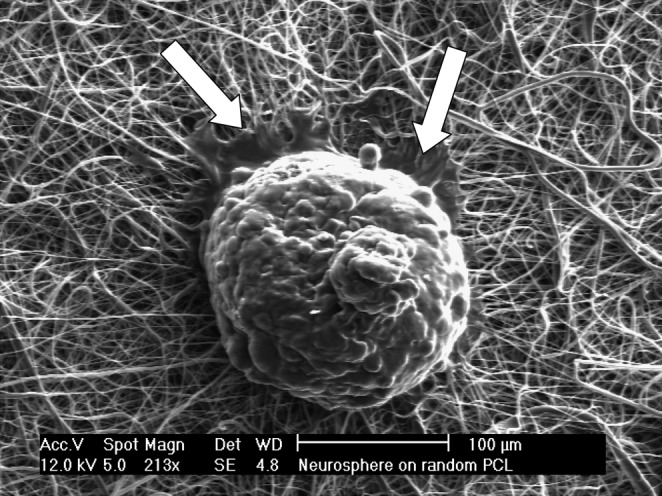
Representative image of a glioblastoma neurosphere seeded on a random PCL fiber scaffold. Notice the formation of adhesion lamellipodia (arrows) but predominant absence of cell detachment from the neurosphere. Scale bar: 100 μm.
Discussion
The major routes of glioma dispersion in the central nervous system are along the abluminal surface of blood vessels and the bundles of white matter fibers.18 These elongated structures provide the physical resistance and molecular anchors for the formation of focal adhesions and intracellular stress fibers necessary for cell traction.12 In addition, glioma cells transverse short distances through the gray matter neuropil, infiltrating the randomly organized, soft neural ECM formed by hyaluronic acid and associated proteoglycans.10 The increased dispersion of the tumor cells in white matter versus gray matter, which essentially contain the same ECM molecules, illustrates the influence of substrate topography on glioma migration in vivo, a condition that has not been intentionally mimicked in any previous in vitro model.
Here we show that differently electrospun PCL fibers can specifically reproduce this unique feature of glioma migration, namely, the effects of substrate orientation on cell motility. Electrospinning is an ideal method mimicking biological ECMs easily producing macroscopic quantities of nano- to microsized scaffolds with high relative porosity (70–90%) compatible with cell proliferation and motility. Natural biodegradable materials such as collagen, gelatin, elastin, chitosan, and hyaluronic acid as well as synthetic biodegradable polymers such as PCL and poly(glycolic) acid, poly(lactic) acid, and poly(lactic-glycolic) acid have been electrospun as scaffolds for culture of cardiac, chondral, and osseous cells.30–32 The split ground technique clearly yields highly aligned nanofibers compared with normal random deposition, resulting in easily reproducible, aligned substrates that mimic the topography of natural conduits for tumor cell dispersion in the brain.
A review of current progress shows that while 3D collagen matrices34 are actively utilized in cancer research, they do not allow the alignment inherent to white matter tracts in the brain. Biologically derived substrates have been used35–37 and have value but are similarly limited in structural and chemical flexibility. Schnell et al.24 utilized aligned PCL and PCL-gelatin to study both neurite outgrowth and noncancerous glial cell migration. Motion was monitored statically rather than in situ in contrast with our approach sampling the complete range of motion of migrating cells.
Cultured glioma cells on highly rigid flat surfaces (glass or plastic) exhibit typically flattened and distended morphologies (fan-like, teardrop-like, etc.) and random motion having little correlation to their migratory phenotype in vivo.13 In contrast, we observe that cells adhered to 3D electrospun fibers display either a spherical or a spindle-shaped morphology based primarily on the influence of substrate anisotropy. This is largely in agreement with morphologies observed in noninvasive versus infiltrating glioma cells in vivo.38,39 On aligned fiber the spindle-shaped morphology was maintained consistently and only modified significantly by cell division.
Our results also clearly show that glioma cells exhibit significant differences in either average or effective motion when presented with variable substrate alignment. Cells on aligned fibers exhibited active bidirectional motion along the axis of the fiber; perpendicular movement was observed only at fiber intersections. Conversely, cells on random fibers were continuously constrained by interactions with these fibers and showed greatly reduced effective motion away from their initial sites of seeding. Interestingly, their net movement was reduced even when compared with homogeneous substrates such as plastic culture dishes.
The effective velocity and ratio of glioma cell movement on aligned versus random fibers (approximately fivefold) closely matched the parameters observed for cell migration on white versus gray matter in vivo,16,40,41 suggesting that even in the absence of specific molecular signals from the ECM, the topography of white and gray matter could be enough to direct glioma migration. This difference in substrate presentation has been postulated as the underlying reason for the preferred dispersion of the tumors along white matter fibers that then act as pathways of least resistance even in the absence of preferred molecules for cell adhesion.42 It is interesting to note that the strong inhibition of glioma cell motility on random fibers was not observed with other cell types that we have previously tested on electrospun PCL,23 although those studies have been conducted over much longer time periods allowing far more proliferation. Work is in progress to characterize long-term glioma cultures on these PCL scaffolds.
Even outside of the benefits associated with highly detailed measurements of velocity, time-lapse microscopy of migratory cells on aligned fibers reveals that a novel form of data, the heterogeneity of motion, can be obtained from these scaffolds. Analysis of individual migratory cells on aligned fibers revealed that cells experienced bursts of motility followed by longer pauses that often coincided with fiber–fiber intersections even if the angle of misalignment was small (∼20°). For most cells, motility decreased to zero to allow cell division that was immediately followed by bursts of motility as the daughter cells separated in opposite directions following the same stereotyped pattern of migration as the mother cell. Mitosis and slight defects in fiber alignment were the two major influences on cell motility throughout the assays. To our knowledge, no such prior connection between cell cycle and migration exists in the literature. Further discoveries relating the physical and chemical influence of the microenvironment on cycle-based motion are expected and could provide detailed translation of less obvious cycle-based changes in relevant intracellular processes (e.g., cytoskeletal tensional homeostasis and RNA transcription) to their effects on migration.
The previous experiments demonstrated that individual glioma cells could migrate on PCL fibers in a topography-dependent manner. Next, to mimic the directional component of glioma cell migration from a central mass, we tested outward migration of glioma cells from aggregated spheres seeded on the PCL fibers, providing a novel application of aligned electrospun fibers in simulating migration away from a centralized tumor mass. For these experiments, we prepared aggregates of glioma cells from biopsy samples (glioma neurospheres), using protocols optimized for culture of tumor initiating cells. Several lines of evidence have indicated that these cells, also known as glioma stem cells, may be the key population that supports tumor growth and underlies its recurrence after spreading in the neural parenchyma.43,44 Thus, they represent a remarkably important population of clinically derived gliomas whose migratory behavior has not yet been analyzed. These neurospheres were prepared in conditions that maintain the stem cell component of the tumor45,46 and have been shown to produce specimens with a genetic makeup and phenotype closely matching the original tumor (compared to the rapidly drifting features of biopsies cultured in traditional, high-serum conditions). More importantly, glioma neurospheres may contain the essential population(s) to reproduce the original tumor, but the migratory abilities of these cells—a key property for tumor recurrence—have not been studied.
The influence of substrate alignment on motility out of neurospheres was plainly evident. After seeding on random fiber, not only did neurosphere-derived cells not spread but they also failed to even detach from the sphere. The absence of cell detachment suggested that cell–cell adhesion on/in the sphere largely predominated over cell–substrate attachment to the PCL. However, this phenomenon was restricted to the randomly aligned PCL; cells from neurospheres seeded on aligned PCL rapidly detached and migrated along the direction of the fibers, indicating that the strong cell–cell attachment was not an absolute property of the spheres but was instead modulated by the underlying topography. Interestingly, cell detachment on aligned fibers was largely parallel to the orientation of the fibers and little perpendicular detachment and migration were observed, indicating that after detachment the neurosphere cells behaved essentially as the previously analyzed dispersed cell lines. Thus, our model appears as an attractive option to study the migration of biopsy-derived glioma stem cells under the same conditions used for routine culture of floating neurospheres.
Properly fabricated, electrospun fiber scaffolds overcome several limitations of traditional cell cultures and provide significant advantages for glioma cell modeling. First, the electrospun fibers present all the advantages of a well-defined in vitro system coupled to the convenient monitoring of glioma cells in 3D using time-lapse microscopy. Second, because the PCL forms a resistant, self-sustaining 3D scaffold, this model does not require the addition of gelling molecules that may stimulate artificial cell behaviors. Third, additional techniques can produce useful postspinning modifications such as laser ablation to modify fiber alignment30,47,48 and inclusion of relevant ECM molecules as shells around the PCL fibers.49–51 These modifications, which we are actively exploring, can expand the model to more accurately mimic the molecular signatures of the terrains on which glioma cells disperse, that is, white matter, basal lamina of blood vessels, and the like. Thus, from a design standpoint, the electrospun fiber model can reach very high complexity while providing, at the same time, a degree of control found in simple in vitro assays.
Finally, it is important to remark upon the quantitative nature of these assays allowing accurate comparison of glioma cells and, more importantly, of neurospheres derived from clinical samples. Preparation of neurospheres from patient biopsies followed by rapid culture on conveniently modified PCL scaffolds is a technique amenable to reasonable throughput to result in rapid and quantitative analysis. We envision the use of these scaffolds as the basis for assays that quantify the migratory capacity of biopsy-derived cells and test anti-proliferative and anti-invasive compounds in an efficient and reproducible manner. Thus, these scaffolds could provide the basis for patient-tailored bioassays in which the most effective drug combinations are identified in a quantifiable manner against each specific glioma biopsy. The resulting clinically relevant information could then be used to improve treatment or prevent tumor recurrence. Novel scaffolds that recapitulate these key aspects of tumor invasion may, by extension, return considerable guidance to approaches for neural tissue regeneration.24,25
Acknowledgments
This work was partly supported by a research grant from the National Science Foundation under Grant No. EEC-0425626 (J.J. and J.J.L.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. Additional support was also provided by the American Brain Tumor Association (M.S.V. and M.O.N.) and the Dardinger Center Fund for Neuro-Oncology Research at the Ohio State University (M.S.V. and S.E.L.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Central Brain Tumor Registry of the United States. Statistical Report: Primary Brain Tumors in the United States 1998–2002. CBTRUS. 2005.
- 2.Holland E.C. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA. 2000;97:6242. doi: 10.1073/pnas.97.12.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fathallah-Shaykh H.M. Darts in the dark cure animal, but not human, brain tumors. Arch Neurol. 2002;59:721. doi: 10.1001/archneur.59.5.721. [DOI] [PubMed] [Google Scholar]
- 4.Salgaller M.L. Liau L.M. Current status of clinical trials for glioblastoma. Rev Recent Clin Trials. 2006;1:265. doi: 10.2174/157488706778250140. [DOI] [PubMed] [Google Scholar]
- 5.Tonn J.C. Goldbrunner R. Mechanisms of glioma cell invasion. Acta Neurochir. 2003;(Suppl 88):163. doi: 10.1007/978-3-7091-6090-9_22. [DOI] [PubMed] [Google Scholar]
- 6.Lamszus K. Kunkel P. Westphal M. Invasion as limitation to anti-angiogenic glioma therapy. Acta Neurochir. 2003;(Suppl 88):169. doi: 10.1007/978-3-7091-6090-9_23. [DOI] [PubMed] [Google Scholar]
- 7.Rao J.S. Molecular mechanisms of glioma invasiveness: the role of proteases. Nat Rev Cancer. 2003;3:489. doi: 10.1038/nrc1121. [DOI] [PubMed] [Google Scholar]
- 8.Levicar N. Nuttall R.K. Lah T.T. Proteases in brain tumour progression. Acta Neurochir (Wien) 2003;145:825. doi: 10.1007/s00701-003-0097-z. [DOI] [PubMed] [Google Scholar]
- 9.VanMeter T.E. Rooprai H.K. Kibble M.M. Fillmore H.L. Broaddus W.C. Pilkington G.J. The role of matrix metalloproteinase genes in glioma invasion: co-dependent and interactive proteolysis. J Neurooncol. 2001;53:213. doi: 10.1023/a:1012280925031. [DOI] [PubMed] [Google Scholar]
- 10.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6:489. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 11.Paulus W. Roggendorf W. Schuppan D. Immunohistochemical investigation of collagen subtypes in human glioblastomas. Virchows Arch A Pathol Anat Histopathol. 1988;413:325. doi: 10.1007/BF00783025. [DOI] [PubMed] [Google Scholar]
- 12.Georges P.C. Miller W.J. Meaney D.F. Sawyer E.S. Janmey P.A. Matrices with compliance comparable to that of brain tissue select neuronal over glial growth in mixed cortical cultures. Biophys J. 2006;90:3012. doi: 10.1529/biophysj.105.073114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beadle C. Assanah M.C. Monzo P. Vallee R. Rosenfeld S.S. Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19:3357. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohnishi T. Matsumura H. Izumoto S. Hiraga S. Hayakawa T. A novel model of glioma cell invasion using organotypic brain slice culture. Cancer Res. 1998;58:2935. [PubMed] [Google Scholar]
- 15.Jung S. Ackerley C. Ivanchuk S. Mondal S. Becker L.E. Rutka J.T. Tracking the invasiveness of human astrocytoma cells by using green fluorescent protein in an organotypical brain slice model. J Neurosurg. 2001;94:80. doi: 10.3171/jns.2001.94.1.0080. [DOI] [PubMed] [Google Scholar]
- 16.Palfi S. Swanson K.R. De B.S. Chretien F. Oliveira R. Gherardi R.K. Kros J.M. Peschanski M. Christov C. Correlation of in vitro infiltration with glioma histological type in organotypic brain slices. Br J Cancer. 2004;91:745. doi: 10.1038/sj.bjc.6602048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viapiano M.S. Hockfield S. Matthews R.T. BEHAB/brevican requires ADAMTS-mediated proteolytic cleavage to promote glioma invasion. J Neurooncol. 2008;88:261. doi: 10.1007/s11060-008-9575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louis D.N. Molecular pathology of malignant gliomas. Annu Rev Pathol Mech Dis. 2006;1:97. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 19.Bellail A.C. Hunter S.B. Brat D.J. Tan C. Van Meir E.G. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Song H. David O. Clejan S. Giordano C.L. Pappas-Lebeau H. Xu L. O'Connor K.C. Spatial composition of prostate cancer spheroids in mixed and static cultures. Tissue Eng. 2004;10:1266. doi: 10.1089/ten.2004.10.1266. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y.C. Shu X.Z. Prestwich G.D. Tumor engineering: orthotopic cancer models in mice using cell-loaded, injectable, cross-linked hyaluronan-derived hydrogels. Tissue Eng. 2007;13:1091. doi: 10.1089/ten.2006.0297. [DOI] [PubMed] [Google Scholar]
- 22.Nam J. Rath B. Knobloch T.J. Lannutti J.J. Agarwal S. Novel electrospun scaffolds allowing in vitro molecular analysis of chondrocytes experiencing dynamic compression. Tissue Eng. 2009;15:513. doi: 10.1089/ten.tea.2007.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam J. Huang Y. Agarwal S. Lannutti J. Improved cellular infiltration in electrospun fiber via engineered porosity. Tissue Eng. 2007;13:22249. doi: 10.1089/ten.2006.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnell E. Klinkhammer K. Balzer S. Brook G. Klee D. Dalton P. Mey J. Guidance of glial cell migration and axonal growth on electrospun nanofibers of poly-epsilon-caprolactone and a collagen/poly-epsilon-caprolactone blend. Biomaterials. 2007;28:3012. doi: 10.1016/j.biomaterials.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhang N. Yan H.H. Wen X.J. Tissue-engineering approaches for axonal guidance. Brain Res Rev. 2005;49:48. doi: 10.1016/j.brainresrev.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Nam J. Huang Y. Agarwal S. Lannutti J. Materials selection and residual solvent retention in biodegradable electrospun fibers. J Appl Poly Sci. 2008;107:1547. [Google Scholar]
- 27.Huang Z.M. Zhang Y.Z. Kotaki M. Ramakrishna S. A review on polymer nanofibers by electrospinning and their applications in nanocomposites. Composites Sci Technol. 2003;63:2223. [Google Scholar]
- 28.Giannini C. Sarkaria J.N. Saito A. Uhm J.H. Galanis E. Carlson B.L. Schroeder M.A. James C.D. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neurooncol. 2005;7:164. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J. Kotliarova S. Kotliarov Y. Li A. Su Q. Donin N.M. Pastorino S. Purow B.W. Christopher N. Zhang W. Park J.K. Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 30.Lannutti J. Reneker D. Ma T. Tomasko D. Farson D. Electrospinning for tissue engineering scaffolds. Mater Sci Eng: C. 2007;27:504. [Google Scholar]
- 31.Martins A. Araujo J.V. Reis R.L. Neves N.M. Electrospun nanostructured scaffolds for tissue engineering applications. Nanomedicine. 2007;2:929. doi: 10.2217/17435889.2.6.929. [DOI] [PubMed] [Google Scholar]
- 32.Pham Q.P. Sharma U. Mikos A.G. Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng. 2006;12:1197. doi: 10.1089/ten.2006.12.1197. [DOI] [PubMed] [Google Scholar]
- 33.Giese A. Loo M.A. Rief M.D. Tran N. Berens M.E. Substrates for astrocytoma invasion. Neurosurgery. 1995;37:294. doi: 10.1227/00006123-199508000-00015. [DOI] [PubMed] [Google Scholar]
- 34.Friedl P. Noble P.B. Walton P.A. Laird D.W. Chauvin P.J. Tabah R.J. Black M. Zanker K.S. Migration of coordinated cell clusters in mesenchymal and epithelial cancer explants in-vitro. Cancer Res. 1995;55:4557. [PubMed] [Google Scholar]
- 35.Katz B.Z. Romer L. Miyamoto S. Volberg T. Matsumoto K. Cukierman E. Geiger B. Yamada K.M. Targeting membrane-localized focal adhesion kinase to focal adhesions—roles of tyrosine phosphorylation and Src family kinases. 2003;278:29115. doi: 10.1074/jbc.M212396200. [DOI] [PubMed] [Google Scholar]
- 36.Li G.N. Hoffman-Kim D. Tissue-engineered platforms of axon guidance. Tissue Eng B Rev. 2008;14:33. doi: 10.1089/teb.2007.0181. [DOI] [PubMed] [Google Scholar]
- 37.Serebriiskii I. CastellÛ-Cros R. Lamb A. Golemis E.A. Cukierman E. Fibroblast-derived 3D matrix differentially regulates the growth and drug-responsiveness of human cancer cells. Matrix Biol. 2008;27:573. doi: 10.1016/j.matbio.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki S.O. Iwaki T. Dynamic analysis of glioma cells: looking into “movement phenotypes”. Neuropathology. 2005;25:254. doi: 10.1111/j.1440-1789.2005.00626.x. [DOI] [PubMed] [Google Scholar]
- 39.Caspani E.M. Echevarria D. Rottner K. Small J.V. Live imaging of glioblastoma cells in brain tissue shows requirement of actin bundles for migration. Neuron Glia Biol. 2006;2:105. doi: 10.1017/S1740925X06000111. [DOI] [PubMed] [Google Scholar]
- 40.Swanson K.R. Alvord E.C. Murray J.D. A quantitative model for differential motility of gliomas in grey and white matter. Cell Prolif. 2000;33:317. doi: 10.1046/j.1365-2184.2000.00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swanson K.R. Quantifying glioma cell growth and invasion in vitro. Math Compu Model. 2008;47:638. [Google Scholar]
- 42.Giese A. Westphal M. Glioma invasion in the central nervous system. Neurosurgery. 1996;39:235. doi: 10.1097/00006123-199608000-00001. [DOI] [PubMed] [Google Scholar]
- 43.Shih A.H. Holland E.C. Developmental neurobiology and the origin of brain tumors. J Neurooncol. 2004;70:125. doi: 10.1007/s11060-004-2746-3. [DOI] [PubMed] [Google Scholar]
- 44.Gilbertson R.J. Rich J.N. Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 45.Singh S.K. Hawkins C. Clarke I.D. Squire J.A. Bayani J. Hide T. Henkelman R.M. Cusimano M.D. Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 46.Singh S.K. Hawkins C. Clarke I.D. Squire J. Bayani J. Hide T. Cusimano M.D. Dirks P.B. Adult human glioma growth is exclusively maintained in vitro and in vivo by CD133 + cancer stem cells. Neurooncol. 2004;6:348. [Google Scholar]
- 47.Choi H.W. Johnson J.K. Nam J. Farson D.F. Lannutti J.J. Structuring electrospun polycaprolactone nanofiber tissue scaffolds by femtosecond laser ablation. J Laser Appl. 2007;19:225. [Google Scholar]
- 48.Farson D.F. Choi H.W. Zimmerman B. Steach J.K. Chalmers J.J. Olesik S.V. Lee L.J. Femtosecond laser micromachining of dielectric materials for biomedical applications. J Micromech Microeng. 2008;3:18. [Google Scholar]
- 49.Jiang H.L. Hu Y.Q. Li Y. Zhao P.C. Zhu K.J. Chen W.L. A facile technique to prepare biodegradable coaxial electrospun nanofibers for controlled release of bioactive agents. J Control Release. 2005;108:237. doi: 10.1016/j.jconrel.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Huang Z.M. Zhang Y.Z. Ramakrishna S. Double-layered composite nanofibers and their mechanical performance. J Polym Sci B Polym Phys. 2005;43:2852. [Google Scholar]
- 51.Yu J.H. Fridrikh S.V. Rutledge G.C. Production of submicrometer diameter fibers by two-fluid electrospinning. Adv Mater. 2004;16:1562. [Google Scholar]