Abstract
Cat's claw (Doxantha unguis-cati L.) vine accumulates nearly 80% palmitoleic acid (16:1Δ9) plus cis-vaccenic acid (18:1Δ11) in its seed oil. To characterize the biosynthetic origin of these unusual fatty acids, cDNAs for acyl-acyl carrier protein (acyl-ACP) desaturases were isolated from developing cat's claw seeds. The predominant acyl-ACP desaturase cDNA identified encoded a polypeptide that is closely related to the stearoyl (Δ9–18:0)-ACP desaturase from castor (Ricinis communis L.) and other species. Upon expression in Escherichia coli, the cat's claw polypeptide functioned as a Δ9 acyl-ACP desaturase but displayed a distinct substrate specificity for palmitate (16:0)-ACP rather than stearate (18:0)-ACP. Comparison of the predicted amino acid sequence of the cat's claw enzyme with that of the castor Δ9–18:0-ACP desaturase suggested that a single amino acid substitution (L118W) might account in large part for the differences in substrate specificity between the two desaturases. Consistent with this prediction, conversion of leucine-118 to tryptophan in the mature castor Δ9–18:0-ACP desaturase resulted in an 80-fold increase in the relative specificity of this enzyme for 16:0-ACP. The alteration in substrate specificity observed in the L118W mutant is in agreement with a crystallographic model of the proposed substrate-binding pocket of the castor Δ9–18:0-ACP desaturase.
The seed oils of higher plants contain many different fatty acids that determine the value of a particular oil for human nutrition or as a source of industrial chemicals (Murphy, 1994; Ohlrogge, 1994). For this reason, there is considerable interest in bringing about useful changes in the fatty acid composition of oilseed crops. One of the most promising areas of research in this regard relates to protein engineering of acyl-ACP desaturases (Cahoon et al., 1997b). The acyl-ACP desaturases are a family of closely related, soluble enzymes that catalyze insertion of the first double bond in a saturated fatty acyl chain (Cahoon et al., 1997a). The most widely occurring member of the family is the Δ9–18:0-ACP desaturase, which is responsible for 18:1 synthesis in plants (Nagai and Bloch, 1968; Shanklin and Somerville, 1991; Thompson et al., 1991). In addition, several other acyl-ACP desaturases have been described: the Δ4–16:0-ACP desaturase of coriander seed (Cahoon et al., 1992), the Δ6–16:0-ACP desaturase from blacked-eyed Susan (Thunbergia alata) seed (Cahoon et al., 1994a), and the Δ9–14:0-ACP desaturase of geranium trichomes (Schultz et al., 1996). The characterization of these variant enzymes and the availability of a crystal structure for the castor Ricinus communis L. Δ9–18:0-ACP desaturase (Lindqvist et al., 1996) raises the possibility that protein engineering can be used to manipulate the double-bond position and substrate specificities of these enzymes. Unusual monounsaturated fatty acids produced by redesigned desaturases may be useful for generating seed oils with altered physical properties and new commercial applications.
Based on previous studies (Cahoon et al., 1992, 1994a; Cahoon and Ohlrogge, 1994b; Schultz et al., 1996), seeds and other plant tissues that accumulate large amounts of unusual monounsaturated fatty acids represent potential sources of variant forms of acyl-ACP desaturases. The seed oils of several plant species are rich sources of 16:1Δ9 and its elongation product, 18:1Δ11 (Chisholm and Hopkins, 1965). These unusual fatty acids can account for 25 to 80% of the seed oils of such species. A likely route of synthesis of 16:1Δ9 and 18:1Δ11 would initially involve the Δ9 desaturation of 16:0-ACP by an acyl-ACP desaturase with enhanced specificity for this substrate relative to known Δ9–18:0-ACP desaturases. In this regard, a diverged acyl-ACP desaturase was recently identified in milkweed seed (Cahoon et al., 1997a), a tissue that accumulates 10% 16:1Δ9 and 15% 18:1Δ11. Although this enzyme displayed elevated activity with 16:0-ACP compared with known Δ9–18:0-ACP desaturases, it was most active in vitro with 18:0-ACP. A similar substrate-specificity profile was also observed when acyl-ACP desaturase activity was assayed in crude homogenates of developing milkweed seed.
Because 16:1Δ9 is found in the seed oils of very divergent species, it is likely that Δ9–16:0-ACP desaturases have evolved separately several times. This suggests the possibility that distinct molecular changes were involved in producing different Δ9–16:0-ACP desaturases from the ancestral Δ9–18:0-ACP enzyme. As an alternative source of a Δ9–16:0-ACP desaturase, we have examined acyl-ACP desaturase cDNAs in developing seed of cat's claw (Doxantha unguis-cati L.). This species is unrelated to milkweed and accumulates nearly 80% 16:1Δ9 plus 18:1Δ11 in its seed oil (Chisholm and Hopkins, 1965). In this report we describe the cloning of a cat's claw cDNA that encodes a 16:0-specific Δ9-ACP desaturase. By comparing the amino acid sequence of this enzyme with that of the castor Δ9–18:0-ACP desaturase, we were able to identify a single amino acid substitution located at the base of the substrate cleft, which, when introduced into the castor enzyme, dramatically increased its relative specificity for 16:0- versus 18:0-ACP.
MATERIALS AND METHODS
Immature fruits of cat's claw (Doxantha unguis-cati L.) were harvested from plants on the campus of Louisiana State University (Baton Rouge). Developing seeds were extracted from fruit pods, frozen in liquid N2, and stored at −80°C until they were used for RNA isolation.
RNA Preparation and Northern Blotting
Approximately 1 g of plant tissue was ground in liquid N2 in a precooled mortar and pestle. The fine powder was transferred to 5 mL of a 1:1 mixture of phenol and extraction buffer (0.1 m Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS) at 80°C. This suspension was mixed for 1 min. Chloroform:isoamyl alcohol (24:1, v/v) was then added and the suspension was vortexed again. The sample was briefly centrifuged to separate the phases, and the aqueous phase was recovered and mixed with an equal volume of 4 m LiCl. The RNA was then allowed to precipitate overnight at 4°C. The RNA pellet obtained was dissolved in water after centrifugation and precipitated again with 0.1 volume of 3 m sodium acetate, pH 5.2, and 2 volumes of ethanol. The pellet was washed with 70% alcohol, dried, dissolved in water, and stored at −80°C.
Total RNA was run in a formaldehyde-formamide-denaturing agarose gel and transferred to a nylon membrane with 20× SSC (Sambrook et al., 1989). For hybridization, DNA was labeled with [32P]dCTP to a specific activity of 3 × 108 dpm/mg DNA using random hexanucleotide primers. Hybridization and washing of the blot were done at 65°C as described previously (Shah et al., 1997).
Construction and Screening of a cDNA Library
mRNA was isolated from total RNA by adding biotinylated oligo(dT) to the RNA sample and mixing thoroughly. The mRNA-oligo(dT) hybrid molecules were captured with streptavidin-coated paramagnetic particles in a magnetic separation strand (PolyATract mRNA isolation system, Promega). Double-stranded cDNA was synthesized from the mRNA using a Superscript cDNA synthesis kit obtained from GIBCO-BRL. Size-fractionated cDNA was ligated to the EcoRI arms of Lambda-Ziplox (GIBCO-BRL) and packaged using Gigapack III gold-packaging extract (Stratagene).
The cDNA library was screened by hybridization with DNA probes labeled with [32P]dCTP. Blots were hybridized and washed at 57°C. Plasmids carrying positive cDNA inserts were released from the Lambda-Ziplox DNA by cre-loxP-mediated recombination in Escherichia coli strain DH10BZIP according to the manufacturer's protocol. Both strands of cDNA were sequenced by fluorescent dideoxy-termination using a DNA sequencer (model 373A, Applied Biosystems). Sequence information was analyzed with Genetics Computer Group (Madison, WI) programs using default settings for parameters unless otherwise indicated.
Desaturase Expression in E. coli
One of the positive cDNA clones isolated from the library pDU1 was amplified by PCR. The 5′ primer corresponded to amino acids 30 to 38 of the polypeptide encoded by pDU1 and contained a flanking SphI site. The 3′ primer was designed according to the sequence immediately downstream of the stop codon, and a terminal HindIII site was included. The PCR product was cloned in the SphI-HindIII site of the E. coli expression vector pQE-32 (Qiagen, Chatsworth, CA), which carries six His amino acid residues as a tag at the N terminus. Ligation in the correct reading frame with the His tag was confirmed by sequencing. The recombinant plasmid was introduced and expressed in E. coli strain DH10B, which also carries the repressor plasmid pREP4.
Six liters of E. coli DH10B cells harboring recombinant plasmids carrying cat's claw cDNA c566 was grown to an A600 = 0.4 at 37°C in BTNa medium (10 g/L bacto-tryptone and 5 g/L NaCl), then induced by addition of isopropylthio-β-galactoside (0.4 mm), and grown for an additional 4 h at 30°C. Bacterial cells were harvested by centrifugation and resuspended in 75 mL of a buffer consisting of 6.7 mm Mes/6.7 mm Hepes/6.7 mm Mops (pH 7.0) and 1 mm PMSF. The cells were lysed using a French pressure cell at 100 mPa and the extract was centrifuged at 100,000g. The resulting soluble protein was loaded onto a 1.7-mL Poros 20 HS cation-exchange column (PerSeptive Biosystems, Inc., Framingham, MA) interfaced with a Biocad Sprint HPLC system (PerSeptive Biosystems). Protein was loaded in cell lysis buffer lacking PMSF and eluted over a gradient of 20 column-volumes from 0 to 600 mm NaCl in the Mes/Hepes/Mops buffer described above. The cat's claw acyl-ACP desaturase was recovered at approximately 90% purity, as judged by SDS-PAGE using Coomassie blue staining. The enriched enzyme was exchanged into a buffer consisting of 40 mm Tris (pH 7.5), 50 mm NaCl, and 10% glycerol by gel filtration using a PD-10 column (Pharmacia). The protein was stored in aliquots at −70°C until use in enzyme assays.
In Vitro Assay
Activities of acyl-ACP desaturases were measured as described elsewhere (Cahoon et al., 1994a) with minor modifications. Anabaena nidulans vegetative Fd (22 μg/150 μL assay) and maize root Fd:NADP+ oxidoreductase (400 milliunits/assay) were used in place of spinach Fd and Fd reductase. Both cofactors were purified from E. coli expressing the corresponding cDNA (Ritchie et al., 1994; Cheng et al., 1995). No attempt was made to optimize assay conditions for the cat's claw desaturase. However, the conditions used are likely to be suitable because the cat's claw and castor (Ricinus communis L.) enzymes both operate in the stroma of seed plastids.
[14C]Acyl-ACPs were synthesized according to the method of Rock and Garwin (1979) using spinach ACP-I purified from recombinant sources. [1-14C]Myristic, palmitic, and stearic acids (American Radiolabeled Chemicals, Inc., St. Louis, MO) were used for the synthesis of acyl-ACPs, and the specific activity of each was 55 mCi/mmol.
Assay products were separated from the unreacted substrate by argentation TLC as previously described (Cahoon and Ohlrogge, 1994b). Radiation on TLC plates was detected using a phosphor imager (Molecular Dynamics, Sunnyvale, CA), and the distribution of radiolabel between the product and the unreacted substrate was determined using ImageQuant software (Molecular Dynamics). Double-bond positions were determined by the mobility of methyl esters of desaturation products on argentation TLC relative to authentic standards.
Preparation of Mutant L118W of the Castor Δ9–18:0-ACP Desaturase
Mutation L118W was introduced into the castor Δ9–18:0-ACP desaturase by overlap-extension PCR (Ho et al., 1989). Two separate PCR reactions were conducted using the coding sequence of the mature wild-type castor Δ9–18:0-ACP desaturase in the vector pET9d (Novagen, Madison, WI) as the template. One reaction was performed using the primer combination T7 primer and 5′-CCGAACTCCATCCCAGGTATTCAGCA-3′ (the mutation is underlined). The second reaction was conducted with the primer combination 5′-TGCTGAATACCTGGGATGGAGTTCGG-3′ and 5′-GCAAAAGCCAAAACGGTACCATCAGGATCA-3′ (primer 1). Following agarose-gel purification, the products of the two reactions were combined and amplified in a third PCR reaction using the T7 primer and primer 1 (without added template). The product of this reaction was gel purified, digested with XbaI and BamHI, and inserted in place of the corresponding portion of the mature wild-type castor Δ9–18:0-ACP desaturase cDNA in pET9d. The presence of the desired mutation was confirmed by DNA sequencing of the final construct. The activity of the L118W mutant with radiolabeled acyl-ACP substrates was determined following expression of this polypeptide in E. coli and subsequent purification by cation-exchange chromatography using the methods described above for analysis of the recombinant cat's claw acyl-ACP desaturase.
The methods used to map residues lining the substrate-binding pocket have been described (Lindqvist et al., 1996; Cahoon et al., 1997b). The projection of eight residues of the castor enzyme followed that used by Cahoon et al. (1997b). The substituted Trp in the L118W mutant was drawn with the bulky side group extending into the substrate-binding pocket.
RESULTS AND DISCUSSION
Isolation and Sequence Analysis of an Acyl-ACP Desaturase from Cat's Claw Seed
The seed oil of cat's claw contains more than 75% 16:1Δ9 and 18:1Δ11 fatty acids (Chisholm and Hopkins, 1965). To assess the possible role of an acyl-ACP desaturase in the biosynthesis of these unusual fatty acids, cDNAs for acyl-ACP desaturases from immature cat's claw seeds were characterized. Initially, a 32P-labeled probe derived from a cDNA of the castor Δ9–18:0-ACP desaturase was used to screen approximately 30,000 plaques from a cDNA library derived from developing cat's claw seeds. Twenty randomly selected, positive clones were converted to plasmids and subjected to restriction-enzyme mapping. All 20 clones shared a common set of restriction fragments, indicating that they represented cDNAs from a single gene (data not shown). The inserts from 8 of these plasmids were approximately the same size and longer than the remaining 12. One of these, designated pDU1, was chosen for further analysis.
The insert in pDU1 was fully sequenced in both directions, and this sequence has been deposited in the GenBank database under accession no. AF051134. Sequence analysis revealed an ORF of 1188 bp flanked by 19 bp of 5′-untranslated sequence and 300 bp of 3′-untranslated sequence plus a poly(A+) tail. The sequence of nucleotides flanking the proposed translation initiation site, AAAATGGC, differs from the eukaryotic consensus sequence ACAATGGC (Joshi, 1987) by only one nucleotide and is identical to the corresponding sequences in cDNAs encoding the 16:0-ACP desaturases of black-eyed Susan and coriander (Cahoon and Ohlrogge, 1994b; Cahoon et al., 1994a). No in-frame stop codon is present upstream of the ATG, but alignment of this ATG with the start codons in other acyl-ACP desaturases support its identification as the correct translation start.
The translated ORF of pDU1 corresponded to a polypeptide of 396 amino acids. Based on similarities with other acyl-ACP desaturases, the first 33 amino acids likely represent a plastid transit peptide. The calculated molecular mass of the mature protein was 41.7 kD. When compared with sequences available in the GenBank database, the predicted protein shares considerable sequence similarity with all of the acyl-ACP desaturases. The desaturases from coriander, geranium, milkweed, and black-eyed Susan all show deletions (from 6 to 22 residues) relative to the castor sequence near the amino terminus of the mature protein (Cahoon et al., 1997a). By contrast, the cat's claw protein had no deletions and was in fact entirely colinear with the castor Δ9–18:0-ACP desaturase (Fig. 1). The cat's claw and castor sequences were 85% identical (90% similar with conservative amino acid substitutions). This remarkably high degree of homology compares with 62 to 65% identity (72–74% similarity) for pairwise comparisons between the cat's claw or castor sequence and the enzyme from milkweed.
Figure 1.
Comparison of the deduced amino acid sequences of the cat's claw, castor, and milkweed acyl-ACP desaturases. Identical and similar residues are shown on backgrounds of black and gray, respectively. The cleavage site for the plastid transit peptide of the castor enzyme is indicated by the arrowhead. Eight residues that lie near the bottom of the substrate-binding pocket in the crystal structure of the castor enzyme are indicated by asterisks. The GenBank accession numbers for the sequences are: cat's claw, AF051134; castor, M59857; and milkweed, U60277.
Characterization of the Seed-Specific Δ9–16:0-ACP Desaturase
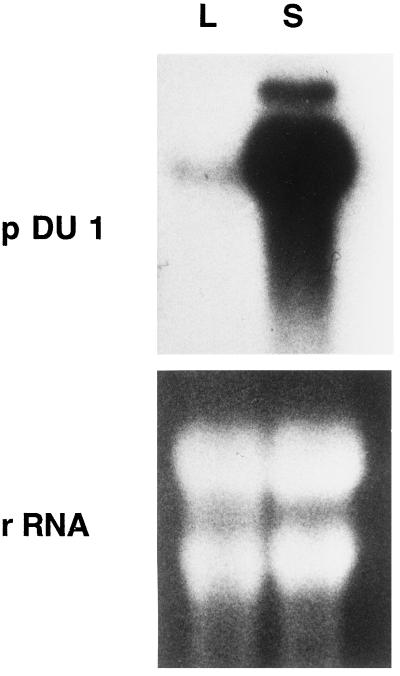
A gel blot containing total RNA from seed and leaf tissue of cat's claw was hybridized with a probe derived from the insert of pDU1. A transcript hybridizing to the probe was extremely abundant in seed tissue but very little transcript was present in the leaf sample (Fig. 2). We cannot exclude the possibility that the gene corresponding to the pDU1 cDNA is expressed at a low level in leaves, but, given the high amino acid sequence similarity within the acyl-ACP desaturase family, it is also possible that the pDU1 probe hybridized to some extent to a related transcript that in leaves would be expected to encode a chloroplast Δ9–18:0-ACP desaturase. The results shown in Figure 2, and the fact that the 20 clones analyzed from the cat's claw seed library represented the same gene, demonstrate that there is a single, dominant acyl-ACP desaturase in these seeds. The fact that fatty acids derived from the Δ9–18:0-ACP desaturase (18:1Δ9 plus 18:2) represent only 8% of the fatty acids in cat's claw oil (Chisholm and Hopkins, 1965) suggests that pDU1 might encode a relatively specific Δ9–16:0-ACP desaturase.
Figure 2.
Gel-blot analysis of acyl-ACP transcript levels in developing seeds (S) and leaves (L) of cat's claw. Equal amounts (10 μg) of total RNA from seeds and leaves were run in each lane and probed with the insert from pDU1. Ethidium bromide staining of the major rRNA bands was used to confirm equal loading of total RNA.
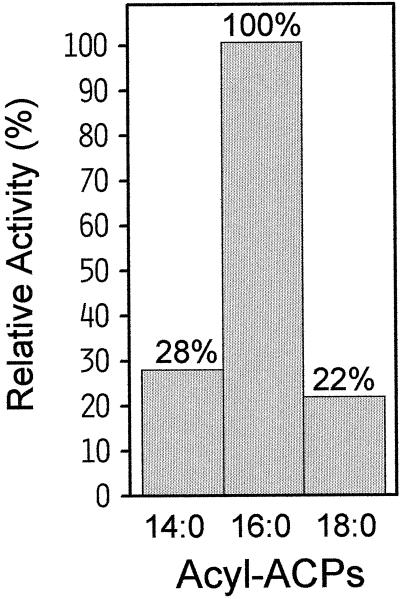
To investigate the substrate specificity of the cat's claw acyl-ACP desaturase, the ORF from pDU1 was expressed in E. coli DH10B cells using the vector pQE32. The recombinant protein produced in these cells contained six additional His residues at its N terminus that were derived from the vector. The desaturase was partially purified from bacterial lysates by cation-exchange chromatography and assayed for activity with acyl-ACPs of different chain lengths. The enzyme functioned primarily as a Δ9–16:0-ACP desaturase (Fig. 3), with approximately 4-fold lower relative activities when 14:0-ACP or 18:0-ACP were used as the substrates. In contrast to these results, the milkweed acyl-ACP desaturase showed a 12-fold higher activity with 18:0-ACP compared with that detected with 16:0-ACP (Cahoon et al., 1997b). It is interesting that two independent attempts to express the cat's claw cDNA in pET vectors (Novagen) lacking the His tag resulted in the accumulation of little if any recombinant protein, as determined by Coomassie blue staining of SDS-PAGE gels of crude bacterial extracts.
Figure 3.
Substrate specificities of the recombinant cat's claw acyl-ACP desaturase assayed with 14:0-, 16:0-, and 18:0-ACPs. Partially purified enzyme from E. coli lysate was assayed as described in Methods. The reaction rate with 16:0-ACP was 0.47 nmol min−1 mg−1 protein.
A Mutant Form of the Castor Enzyme Favors Δ9 Desaturation of 16:0-ACP
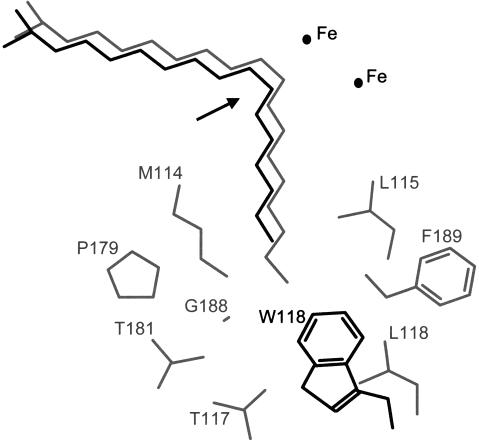
The amino acid sequences of all of the acyl-ACP desaturases are highly homologous and are colinear over most of their length. This suggests that the three-dimensional structure determined for the castor Δ9–18:0-ACP desaturase (Lindqvist et al., 1996) provides an excellent model for predicting the structure of the other enzymes. These predictions and site-specific mutagenesis studies have been used to identify eight residues located near the bottom of the substrate-binding pocket that help to determine the chain-length specificity of the enzymes (Cahoon et al., 1997b). The first of these residues, M114 of the mature castor protein, corresponds to M167 of the cat's claw sequence and M133 of the milkweed sequence shown in Figure 1, and the sequences are colinear throughout the remaining seven residues. Three of the eight residues are altered in the milkweed enzyme relative to the castor sequence (L115I, T117R, and P179T) but only one change occurs in the cat's claw protein L118W. A model of 16:0 and 18:0 bound to the active site is shown in Figure 4 along with stick-model representations of the eight previously identified residues. Clearly, the more bulky side chain of Trp at residue 118 can be predicted to reduce the depth of the substrate pocket and to favor the binding of 16:0-ACP over the longer substrate, 18:0-ACP.
Figure 4.
A model of 18:0 (gray) and 16:0 (black) bound to the active site of acyl-ACP desaturases. The structure of seven common residues lining the substrate pocket of the castor and cat's claw enzymes and L118 of the castor enzyme are shown in gray. The W118 residue of the cat's claw enzyme and the L118W castor mutant is shown in black. The position of the catalytic di-iron center is indicated and the arrow shows the position of double-bond insertion. Amino acid numbering is given with respect to the sequence of the mature castor Δ9–18:0-ACP desaturase.
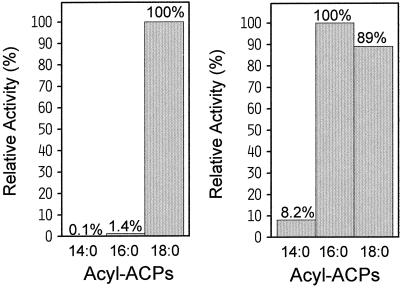
To test this hypothesis, we used site-specific mutagenesis to engineer the L118W variant of the Δ9–18:0-ACP castor desaturase. When this enzyme was assayed with acyl-ACP substrates of different chain lengths, its activity with 16:0-ACP was 115% relative to its activity with 18:0-ACP (Fig. 5). By contrast, the wild-type castor desaturase was more than 70 times more active with 18:0-ACP than with 16:0-ACP. Thus, the substitution of a single residue in the archetypal Δ9–18:0-ACP desaturase accounts for approximately one-half of the difference in relative substrate specificity between the castor and cat's claw enzymes (Fig. 3). Like the previously reported L118F/P179I mutant (Cahoon et al., 1997a), the L118W mutant of the castor Δ9-18:0-ACP desaturase is most active with 16:0-ACP. However, the L118F/P179I mutant displays a greater relative specificity for 16:0- versus 18:0-ACP than does the L118W mutant. The ratio of specific activity with 16:0-ACP:18:0-ACP is 2.4 for the L118F/P179I mutant compared with 1.1 for the L118W mutant. This difference demonstrates that, although W118 is a major determinant of substrate specificity, other residues likely contribute to creating the substrate profile displayed by the cat's claw acyl-ACP desaturase, which is more similar to that of the L118F/P179I mutant.
Figure 5.
Substrate specificities of recombinant wild-type castor Δ9–18:0-ACP desaturase (left) and the mutant L118W (right) assayed with 14:0-, 16:0-, and 18:0-ACPs. Partially purified enzymes from E. coli were assayed as described in Methods. The maximum reaction rate (100%) was 823 nmol min−1 mg−1 protein for the wild-type enzyme and 28.8 nmol min−1 mg−1 protein for the L118W mutant.
Identification of the underlying structural bases for the substrate specificity in studies of variant acyl-ACP desaturases further validates the crystallographic model of desaturase specificity (Lindqvist et al., 1996; Cahoon et al., 1997a, 1997b). Cat's claw and milkweed acyl-ACP desaturases have different amino acid substitutions that occlude the base of the substrate-binding cavity, resulting in similar reductions in chain-length specificity. This highlights the plasticity of protein structure-function relationships in this class of enzymes and supports the idea that acyl-ACP desaturases represent an excellent target for protein engineering. Such engineered enzymes have the potential to form the basis for a new generation of crop plants containing unusual fatty acids.
ACKNOWLEDGMENTS
We wish to thank P. Vijayan for collecting developing cat's claw seeds and other tissues and for shipping them to us. We are grateful to Jay Shockey for advice and help with computer analysis of the sequences. We also thank Armin Dorner for technical assistance.
Abbreviations:
- ACP
acyl carrier protein
- ORF
open reading frameX:Y a fatty acyl group containing X carbon atoms and Y cis double bonds
- ΔZ
site of a double bond or of double-bond insertion as the Z carbons from the carboxyl end of the fatty acid chain
- 14:0
myristate
- 16:0
palmitate
- 16:1Δ9
palmitoleate
- 18:0
stearate
- 18:1
oleate
- 18:1Δ11
cis-vaccenate
Footnotes
This work was supported by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (grant no. 97-35301-4426 to J.B.), the Office of Basic Energy Sciences of the U.S. Department of Energy (E.C., J.S.), and the Agricultural Research Center, Washington State University.
LITERATURE CITED
- Cahoon EB, Coughlan SJ, Shanklin J. Characterization of a structurally and functionally diverged acyl-acyl carrier protein desaturase from milkweed seed. Plant Mol Biol. 1997a;33:1106–1110. doi: 10.1023/a:1005821007291. [DOI] [PubMed] [Google Scholar]
- Cahoon EB, Cranmer AM, Shanklin J, Ohlrogge JB. Δ6 Hexadecenoic acid is synthesized by the activity of a soluble Δ6 palmitoyl-acyl carrier protein desaturase in Thunbergia alata endosperm. J Biol Chem. 1994a;269:27519–27526. [PubMed] [Google Scholar]
- Cahoon EB, Lindqvist Y, Schneider G, Shanklin J. Redesign of soluble fatty acid desaturases from plants for altered substrate specificity and double bond position. Proc Natl Acad Sci USA. 1997b;94:4872–4877. doi: 10.1073/pnas.94.10.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Ohlrogge JB. Metabolic evidence for the involvement of a Δ4-palmitoyl-acyl carrier protein desaturase in petroselinic acid synthesis in coriander endosperm and transgenic tobacco cells. Plant Physiol. 1994b;104:827–837. doi: 10.1104/pp.104.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB. Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA. 1992;89:11184–11188. doi: 10.1073/pnas.89.23.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Westler WM, Xia B, Oh BH, Markley JL. Protein expression, selective isotopic labeling, and analysis of hyperfine-shifted NMR signals of Anabaena 7120 vegetative [2Fe-2S] ferredoxin. Arch Biochem Biophys. 1995;316:619–634. doi: 10.1006/abbi.1995.1082. [DOI] [PubMed] [Google Scholar]
- Chisholm MJ, Hopkins CY. Fatty acids of Doxantha seed oil. J Am Oil Chem Soc. 1965;42:49–50. doi: 10.1007/BF02915346. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Joshi CP. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987;15:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. Manipulation of lipid metabolism in transgenic plants: biotechnological goals and biochemical realities. Biochem Soc Trans. 1994;22:926–931. doi: 10.1042/bst0220926. [DOI] [PubMed] [Google Scholar]
- Nagai J, Bloch K. Enzymatic desaturation of stearyl acyl carrier protein. J Biol Chem. 1968;243:4626–4633. [PubMed] [Google Scholar]
- Ohlrogge JB. Design of new plant products: engineering of fatty acid metabolism. Plant Physiol. 1994;104:821–826. doi: 10.1104/pp.104.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SW, Redinbaugh MG, Shiraishi N, Vrba JM, Campbell WH. Identification of a maize root transcript expressed in the primary response to nitrate: characterization of a cDNA with homology to ferredoxin-NADP+ oxidoreductase. Plant Mol Biol. 1994;26:679–690. doi: 10.1007/BF00013753. [DOI] [PubMed] [Google Scholar]
- Rock CO, Garwin JL. Preparative enzymatic synthesis and hydrophobic chromatography of acyl-acyl carrier protein. J Biol Chem. 1979;254:7123–7128. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schultz EJ, Cahoon DB, Shanklin J, Craig R, Cox-Foster DL, Mumma RO, Medford JI. Expression of a Δ9 14,0-acyl carrier protein fatty acid desaturase gene is necessary for the production of ω5 anacardic acids found in pest-resistant geranium (Pelargonium xhortorum) Proc Natl Acad Sci USA. 1996;93:8771–8775. doi: 10.1073/pnas.93.16.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Xin Z, Browse J. Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol. 1997;114:1533–1539. doi: 10.1104/pp.114.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, Somerville C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA. 1991;88:2510–2514. doi: 10.1073/pnas.88.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA, Scherer DE, Foxall-VanAken S, Kenny JW, Young HL, Shintani DK, Kridl JC, Knauf VC. Primary structures of the precursor and mature forms of stearoyl-acyl carrier protein desaturase from safflower embryos and requirement of ferredoxin for enzyme activity. Proc Natl Acad Sci. 1991;88:2578–2582. doi: 10.1073/pnas.88.6.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]