Abstract
Background: Improved nutrition early in life is associated with better pulmonary function for patients with cystic fibrosis (CF). However, nutritional status is poorly correlated with the CFTR genotype.
Objective: We investigated the extent to which modifier genes influence nutrition in children with CF.
Design: BMI data were longitudinally collected from the CF Twin-Sibling Study and Cystic Fibrosis Foundation Patient Registry for twins and siblings from 2000 to 2010. A nutritional phenotype was derived for 1124 subjects by calculating the average BMI z score from 5–10 y of age (BMI-z5to10). The genetic contribution to the variation in BMI-z5to10 (ie, heritability) was estimated by comparing the similarity of the phenotype in monozygous twins to that in dizygous twins and siblings. Linkage analysis identified potential modifier-gene loci.
Results: The median BMI-z5to10 was −0.07 (range: −3.89 to 2.30), which corresponded to the 47th CDC percentile. BMI-z5to10 was negatively correlated with pancreatic insufficiency, history of meconium ileus, and female sex but positively correlated with later birth cohorts and lung function. Monozygous twins showed greater concordance for BMI-z5to10 than did dizygous twins and siblings; heritability estimates from same-sex twin-only analyses ranged from 0.54 to 0.82. For 1010 subjects with pancreatic insufficiency, genome-wide significant linkage was identified on chromosomes 1p36.1 [log of odds (LOD): 5.3] and 5q14 (LOD: 5.1). These loci explained ≥16% and ≥15%, respectively, of the BMI variance.
Conclusions: The analysis of twins and siblings with CF indicates a prominent role for genes other than CFTR to BMI variation. Specifically, regions on chromosomes 1 and 5 appear to harbor genetic modifiers of substantial effect.
INTRODUCTION
Cystic fibrosis (CF)5 is an autosomal recessive disease that affects ∼70,000 individuals worldwide and is caused by mutations in the CFTR (cystic fibrosis transmembrane conductance regulator) gene (1). It is marked by a progressive decline in lung function and malnutrition, although improved nutrition early in life has been associated with better pulmonary function later in life (2–5). Nutritional status is not well correlated with the CFTR genotype (6, 7), which suggests the additional influence of environmental, genetic, or stochastic factors (8). Identified nongenetic influences include the diagnosis of CF via a newborn screen (NBS) and placement of a gastrostomy, which are both associated with improved nutritional status (9, 10). Alternately, pancreatic insufficiency (PI) and meconium ileus (MI) negatively influence nutrition and growth in CF patients (11, 12). We also have previously shown that genes independent of CFTR contribute to the average lifetime nutritional status in CF (13). The identification of the extent to which these modifier genes modulate nutritional status in children with CF is important to elucidate the cause of poor nutrition in these patients when they have otherwise been nutritionally optimized.
Twin studies allow for the estimation of the relative contributions of genes and environment to an observed phenotype. With the use of data from the CF Twin-Sibling Study (www.clinicaltrials.gov; NCT00037778) (14), we investigated the influence of genetic and nongenetic factors on nutrition in young CF patients who experience the greatest changes in growth rates. In accordance with the recommendation of the Cystic Fibrosis Foundation (CFF), BMI (in kg/m2) was used as the marker of nutritional status (15). BMI percentiles more accurately predict nutritional failure in CF patients than do conventional anthropometric measures, including the height-for-age percentile, weight-for-age percentile, and percentage of ideal body weight (16, 17). Because BMI varies during childhood and adolescence, we derived measures from a specific age range. The age range of 5–10 y was selected as a stable period in the disease process after the majority of patients have been diagnosed with CF but before the onset of disease-related comorbidities, such as CF-related diabetes (18, 19). We hypothesized that this BMI phenotype would be heritable (ie, within-pair similarity would be greater for monozygous twins than for dizygous twins and siblings) and could be used to identify modifier genes that contribute to the variability in nutritional status of children with CF.
SUBJECTS AND METHODS
Study population
All subjects were part of the CF Twin-Sibling Study and recruited on the basis of having a twin or sibling with CF. Family members all shared the same CFTR genotype. Written consent or assent was obtained from all subjects or guardians, and the study was approved by the Johns Hopkins University Institutional Review Board. The zygosity status of twins was confirmed by short tandem repeat analysis with use of an AmpFLSTR Profiler (Applied Biosystems). Clinical data were supplemented by using data provided by the US CFF Data Registry.
Phenotype generation
Longitudinal height and weight data were collected from 2000 to 2010. The raw data were subjected to a cleaning process whereby biologically implausible dates, ages, heights, and weights were corrected or excluded. For each clinic visit, BMI was calculated if the age of the subject age at the time of the visit was ≥2 y and if both height and weight measurements were available. z scores for height, weight, and BMI were generated by using CDC reference equations (20). To reduce the possible bias associated with more-frequent measurements taken during times of sickness, average-per-quarter z scores were calculated for each subject with a quarter being defined as a 3-mo period beginning with the subject's month of birth. For generation of the longitudinal BMI phenotype used in our analysis, BMI z scores were dropped if the subject's age was <5 or ≥11 y at the time BMI was obtained. Exclusion criteria included the following: 1) having BMI data during <2 quarters between 5 and 10 y of age, 2) being diagnosed with CF at >5 y of age, and 3) half-siblings and twins of indeterminate zygosity. In addition, individuals without a twin or sibling who remained in the study sample after the exclusion criteria were applied were also excluded. For the remaining individuals, a phenotype was derived by calculating the average of average-per-quarter BMI z scores from 5 to 10 y of age (BMI-z5to10).
Covariates
The following covariates have been previously shown to confound nutritional status in CF so were evaluated for their contributions to the variability in BMI-z5to10 phenotype: sex (4), birth cohort (19), age at CF diagnosis (y) (21), diagnosis by NBS (9), homozygosity for the F508del CFTR mutation (21), severe exocrine PI (12), history of MI (11), presence of a gastrostomy (10), pulmonary function (forced expiratory volume in 1 s) (19), and socioeconomic status (22). Birth cohorts of subjects were defined according to the year in which subjects were born by using the following intervals: <1980 (1), 1980–1984 (2), 1985–1989 (3), 1990–1994 (4), 1995–1999 (5), and >1999 (6). The CFTR genotype is known to be associated with the severity of pancreatic exocrine disease in CF patients, which, in turn, is correlated with nutritional status (23–26). Thus, pancreatic sufficiency was defined as having at least one CFTR mutation associated with pancreatic sufficiency, as described previously (27, 28). In the 7% of cases in which the CFTR genotype was missing or indeterminate, clinical data, including fecal elastase, fecal fat, serum trypsin, and the use of pancreatic enzymes, respectively, were used. To further control for the influence of CFTR on nutritional status, we analyzed patients who were homozygous for the F508del mutation (∼55% of the study population). Other CFTR genotypes were too infrequent to perform statistically robust comparisons. Diagnostic criteria for MI were previously defined (29); in addition, pancreatic-sufficient patients with an unknown diagnosis of MI were considered to have no history of MI. The presence of a gastrostomy was defined at the time of enrollment in the study. Lung function was quantified by calculating CF-specific percentiles for the best FEV measurement during each quarter (14) between age 6 and age 10; each best-per-quarter percentile was averaged to obtain a single value per subject (FEVq6to10). FEV obtained at <6 y of age were excluded owing to lack of reference values (30). Insurance type, which was defined as public or private on the basis of the most recent CFF data, was selected as the marker of socioeconomic status because it was previously shown to be associated with lower lung function for patients in the CF Twin-Sibling Study (31).
Statistical analysis
Fisher's exact test and 2-tailed t tests were used to compare subjects stratified by zygosity. The association of covariates with BMI-z5to10 was tested by using univariate linear regression. Covariates that were significant (P < 0.05) in the univariate analyses were included in multivariate regressions. Residuals from multivariate models were used in heritability estimations and linkage analyses. These analyses were also performed with the inclusion of only subjects with PI and only F508del homozygotes to control for the effect of CFTR on nutritional status. To account for the possibility of changes in the mean BMI z score across the 6-y window used to construct the BMI-z5to10 phenotype, a regression analysis was performed by using age centered on 8 y as a covariate; a residual phenotype was generated from this regression and used in separate heritability and linkage analyses. All linear regressions accounted for within-family correlations by using a generalized estimating equations methodology. Stata IC 11 software (StataCorp) was used for all analyses.
Estimation of heritability
Pearson's correlation coefficient for BMI-z5to10 and each residual phenotype was determined for monozygous twins, dizygous twins, same-sex dizygous twins, and a dizygous twin and sibling group. The dizygous twin and sibling group included same-sex dizygous twins and siblings born ≤3 y of each other; this group was used as a proxy for dizygous twins because of the limited number of pairs of dizygous twins. Heritability was estimated by subtracting the correlation coefficient for dizygous twins from the correlation coefficient for monozygous twins and multiplying the difference by 2; estimates were also generated from the group of monozygous twins and same-sex dizygous twins and the group of monozygous twins, dizygous twins, and siblings (32). Heritability estimates >1.0 were reported as 1. CIs on heritability were estimated by using a bootstrapping technique (33). From the set of M monozygous twins and N dizygous twins and siblings, a set of M monozygous twins and N dizygous twins and siblings was sampled (with replacement) in each iteration with random assignment of the order of individuals within the pair. Heritability was calculated by using this randomly selected set of pairs. After 106 iterations, means and 95% CIs were calculated.
Linkage analysis
Linkage marker selection was completed as previously described (28). Linkage analysis was performed with each residual phenotype by using both the variance-components algorithm as implemented in MERLIN 1.1.2 (34) and SOLAR 4.3.1 (35) software packages. Because essentially identical results were obtained, only the results from the MERLIN software package are reported. The log of odds (LOD) score was defined as genome-wide significant at a LOD score ≥3.6 and genome-wide suggestive at a LOD score ≥2.2 (36).
RESULTS
Phenotype description
Longitudinal height and weight data were collected for 1618 twins and siblings with CF from 800 families, which totaled 120,245 clinic visits and 17,140 patient-years (average: 10.6 y per patient). After the application of inclusion and exclusion criteria, 1124 subjects remained, including 130 monozygous twins, 42 dizygous twins, and 952 siblings (Figure 1). Demographic and clinical characteristics that have been shown to influence BMI are summarized for all included subjects in Table 1. Compared with dizygous twins, siblings, and the dizygous twin and sibling group, monozygous twins had a greater representation in the earlier birth cohorts and a higher percentage of F508del homozygotes (P < 0.05). Compared with monozygous twins, dizygous twins had a greater number of diagnoses by newborn screening, individuals with PI, and presence of gastrostomies (P < 0.05).
FIGURE 1.
Flowchart for the application of the inclusion and exclusion criteria to the study population.
TABLE 1.
Demographic and clinical characteristics of the study sample1
| MZ twins | DZ twins | Siblings | DZ twins and siblings2 | Entire study sample | |
| Individuals (n) | 1303 | 424 | 9525 | 2966 | 1124 |
| Female sex [n (%)] | 62 (48) | 14 (33) | 468 (49) | 134 (45) | 544 (48) |
| White ethnicity [n (%)] | 118 (91) | 34 (81) | 868 (91) | 258 (87) | 1020 (91) |
| Birth cohort [n (%)] | |||||
| 1 (<1980) | 20 (15) | 0 | 35 (4) | 14 (5) | 55 (5) |
| 2 (1980–1984) | 18 (14) | 2 (5) | 81 (9) | 23 (8) | 101 (9) |
| 3 (1985–1989) | 20 (15) | 6 (14) | 167 (18) | 59 (20) | 193 (17) |
| 4 (1990–1994) | 24 (18) | 8 (19) | 267 (28) | 81 (27) | 299 (27) |
| 5 (1995–1999) | 40 (31) | 8 (19) | 273 (29) | 70 (24) | 321 (29) |
| 6 (>1999) | 8 (6) | 18 (43)7 | 129 (14)7 | 49 (17)7 | 155 (14) |
| Age at CF diagnosis (y) | 0.34 (0–4.44)8 | 0.18 (0.01–2.93) | 0.17 (0–4.96) | 0.21 (0–4.61) | 0.20 (0–4.96) |
| n | 115 | 42 | 738 | 239 | 895 |
| Diagnosis by NBS [n (%)] | |||||
| Yes | 5 (4) | 8 (19) | 56 (6) | 22 (7) | 69 (6) |
| No | 122 (94) | 34 (81) | 750 (79) | 274 (93) | 906 (81) |
| Unknown | 3 (2) | 07 | 146 (15) | 0 | 149 (13) |
| F508del homozygotes [n (%)] | 88 (68) | 18 (43)7 | 513 (54)7 | 154 (52)7 | 619 (55) |
| Pancreatic insufficient [n (%)] | 118 (91) | 42 (100)7 | 850 (89) | 277 (94) | 1010 (90) |
| History of meconium ileus [n (%)] | |||||
| Yes | 30 (23) | 9 (21) | 153 (16) | 51 (17) | 192 (17) |
| No | 97 (75) | 33 (79) | 643 (68) | 245 (83) | 773 (69) |
| Unknown | 3 (2) | 0 | 156 (16) | 0 | 159 (14) |
| Presence of a gastrostomy [n (%)] | |||||
| Yes | 14 (11) | 10 (24) | 120 (13) | 42 (14) | 144 (13) |
| No | 108 (83) | 30 (71) | 636 (67) | 254 (86) | 774 (69) |
| Unknown | 8 (6) | 2 (5)7 | 196 (21) | 0 | 206 (18) |
| FEVq6to10 | 0.61 (0.08–0.98) | 0.66 (0.07–0.93) | 0.60 (0.01–1) | 0.59 (0.04–1) | 0.60 (0.01–1) |
| n | 122 | 42 | 932 | 282 | 1096 |
| Insurance status [n (%)] | |||||
| Private insurance | 61 (47) | 24 (57) | 501 (53) | 158 (53) | 586 (52) |
| Public insurance | 61 (47) | 15 (36) | 393 (41) | 128 (43) | 469 (42) |
| Unknown | 8 (6) | 3 (7) | 58 (6) | 10 (3) | 69 (6) |
| Height-z5to10 | −0.79 (−4.61 to 1.12) | −0.80 (−2.86 to 1.76) | −0.51 (−4.40 to 2.46) | −0.60 (−4.40 to 2.12) | −0.58 (−4.61 to 2.46) |
| Weight-z5to10 | −0.54 (−6.23 to 1.56) | −0.72 (−2.41 to 1.80) | −0.40 (−4.71 to 2.38) | −0.39 (−4.71 to 2.34) | −0.42 (−6.23 to 2.38) |
| BMI-z5to10 | −0.10 (−3.89 to 1.89) | −0.32 (−1.74 to 1.58) | −0.06 (−3.60 to 2.30) | −0.07 (−2.83 to 2.11) | −0.07 (−3.89 to 2.30) |
BMI-z5to10, average BMI z score from 5 to 10 y of age; CF, cystic fibrosis; DZ, dizygous; FEVq6to10, average cystic fibrosis–specific forced expiratory volume in 1 s percentile for all measures obtained from 6 to 10 y of age; Height-z5to10, average height z score from 5 to 10 y of age; MZ, monozygous; NBS, newborn screen; Weight-z5to10, average weight z score from 5 to 10 y of age.
Same-sex DZ twins and siblings with <3 y of difference in age.
Included 61 pairs of MZ twins and 4 families with MZ twins and a sibling.
Included 21 pairs of DZ twins.
Included 431 pairs of siblings, 27 families with 3 siblings, 1 family with 5 siblings, and 4 families with MZ twins and a sibling.
Included 148 pairs.
Compared with MZ twins, P < 0.05 (Fisher's exact test or 2-tailed t test).
Median; range in parentheses (all such values).
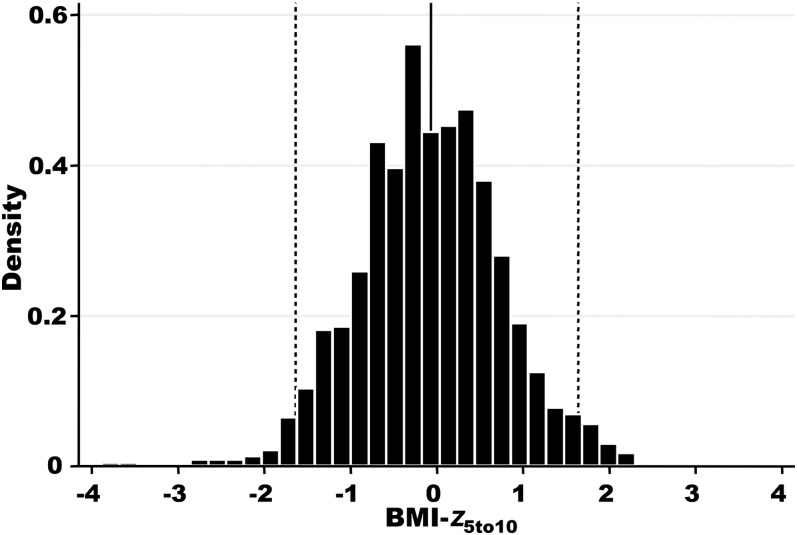
Average height z score from 5 to 10 y of age (height-z5to10) and average weight z score from 5 to 10 y of age (weight-z5to10) scores were derived for each patient. The median height-z5to10 for all included subjects was −0.58 (range: −4.61 to 2.46), which corresponded to the 28th CDC percentile; the median weight-z5to10 was −0.42 (range: −6.23 to 2.38), which corresponded to the 34th CDC percentile (Table 1). Height and weight z scores were highly correlated (r = 0.82), and both measures showed skewing toward lower z scores (see Figure 1 under “Supplemental data” in the online issue). With the use of height and weight measures, a BMI-z5to10 phenotype was derived for each included subject. The median BMI-z5to10 was −0.07 (range: −3.89 to 2.30), which corresponded to the 47th CDC percentile, as shown in Figure 2. The phenotype encompassed the full range of severity, and as seen in the height and weight z-score distributions, there were more individuals with z scores indicative of poor nutritional status (BMI-z5to10 less than or equal to −1.64; 5th CDC percentile) than with z scores indicative of obesity (BMI-z5to10 ≥1.64; 95th CDC percentile).
FIGURE 2.
Distribution of the BMI-z5to10 phenotype. The solid vertical line represents the median BMI-z5to10 (−0.07; 47th CDC percentile); the dashed vertical lines represent the 5th and 95th CDC percentiles (−1.64 and 1.64, respectively). BMI-z5to10, average BMI z score from 5 to 10 y of age.
There were no significant differences in the average BMI-z5to10 between monozygous twins and siblings and dizygous twins and siblings (P = 0.07 and 0.54, respectively). With the use of linear regression, the mean BMI z score was shown to decrease an average of 0.06 units/y from 5 to 10 y of age (P < 0.001), ant thus, an adjusted BMI-z5to10 phenotype was generated by using the residual from this regression. However, there were no appreciable differences in results generated by using this phenotype, and thus, only analyses that used the original BMI-z5to10 phenotype are described in the following sections. The BMI-z5to10 phenotype generated for the 1010 subjects with PI had a median (range) of −0.12 (−3.89 to 2.30). The BMI-z5to10 in the 619 subjects who were homozygous for F508del had a similar distribution (median: −0.11; range: −3.89 to 2.09).
Covariate analysis
Sex, birth cohort, age at CF diagnosis, diagnosis by NBS, F508del homozygosity, PI, history of MI, presence of a gastrostomy, FEVq6to10, and insurance type were evaluated for their contributions to variability in BMI-z5to10. The results for the univariate linear regression analyses are shown in Table 2. When considered independently, birth cohort, age at CF diagnosis, PI, history of MI, presence of a gastrostomy, and FEVq6to10 covaried with BMI-z5to10 (P < 0.05). Female sex was also included in subsequent analyses because there was weak evidence of a correlation (P = 0.08). In the multivariate analyses, age at CF diagnosis and presence of a gastrostomy were no longer significant, likely because of confounding with other covariates, notably PI and history of MI.
TABLE 2.
Contributions of covariates to variability in the BMI-z5to10 phenotype as determined by using univariate linear regression analysis with the subsequent generation of residual phenotypes (BMI-zadj and BMI-zadjFEV) by using multivariate regression analyses1
| Univariate regression analysis |
Multivariate regression analyses |
|||
| BMI-z5to10 |
BMI-zadj (n = 965) | BMI-zadjFEV (n = 943) | ||
| Coefficient (95% CI) | No. of subjects in the analysis | Coefficient (95% CI) | Coefficient (95% CI) | |
| Female sex | −0.09 (−0.19 to 0.01) | 1124 | −0.12 (−0.23 to −0.02) | −0.15 (−0.25 to −0.05) |
| P | 0.08 | 0.02 | 0.004 | |
| Birth cohort | 0.10 (0.06–0.14) | 1124 | 0.10 (0.06–0.14) | 0.08 (0.03–0.12) |
| P | <0.001 | <0.001 | <0.001 | |
| Age at CF diagnosis | 0.06 (0.01–0.11) | 895 | — | — |
| P | 0.01 | — | — | |
| Diagnosis by NBS | 0.09 (−0.15 to 0.33) | 975 | — | — |
| P | 0.45 | — | — | |
| F508del homozygosity | −0.06 (−0.18 to 0.06) | 1124 | — | — |
| P | 0.33 | — | — | |
| Pancreatic insufficiency | −0.35 (−0.55 to −0.14) | 1124 | −0.33 (−0.53 to −0.13) | −0.32 (−0.52 to −0.12) |
| P | 0.001 | 0.001 | 0.002 | |
| History of MI | −0.19 (−0.33 to −0.05) | 965 | −0.16 (−0.29 to −0.02) | −0.13 (−0.25 to 0.00) |
| P | 0.01 | 0.03 | 0.04 | |
| Presence of a gastrostomy | −0.45 (−0.61 to −0.29) | 918 | — | — |
| P | <0.001 | — | — | |
| FEVq6to10 (/10 percentile points) | 0.10 (0.08–0.13) | 1096 | — | 0.10 (0.08–0.13) |
| P | <0.001 | — | <0.001 | |
| Public insurance | −0.03 (−0.15 to 0.08) | 1055 | — | — |
| P | 0.57 | — | — | |
Coefficient (95% CI) denotes the magnitude of the effect of each covariate with the 95% CI of each coefficient in parentheses. P values are for the significance of the coefficient derived by using linear regression (Stata IC 11 software; StataCorp). BMI-zadj, average BMI z score from 5 to 10 y of age adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-z5to10, average BMI z score from 5 to 10 y of age; CF, cystic fibrosis; FEVq6to10, average cystic fibrosis–specific forced expiratory volume in 1 s percentile for all measures obtained from 6 to 10 y of age; MI, meconium ileus; NBS, newborn screen.
Two multivariate regression analyses were performed (Table 2). All covariates in the model remained independent predictors of BMI-z5to10 in the multivariate models. With the use of these models, adjusted BMI z-score phenotypes were generated for subsequent heritability and linkage analyses. The first analysis generated BMI-z5to10 values adjusted for female sex, birth cohort, PI, and history of MI (BMI-zadj); there was a strong correlation between nutritional status and lung function (37), and thus, FEVq6to10 was not included as a covariate in this model to separate the effects of BMI and lung function. After the exclusion of subjects with missing covariate data, BMI-zadj included 965 subjects with a median (range) BMI z score of 0.01 (−3.67 to 2.37). The second analysis generated BMI-z5to10 values adjusted as for BMI-zadj plus for FEVq6to10 (BMI-zadjFEV), which considered the influence of lung function on BMI. The resulting adjusted trait (ie, BMI-zadjFEV) was generated for 943 subjects with a median (range) BMI z score of 0.02 (−2.93 to 2.45). Residual phenotypes that used the same covariates, except for the omission of PI, were generated for pancreatic-insufficient subjects [BMI-zadj-PI (n = 851); BMI-zadjFEV-PI (n = 832)] and F508del homozygotes [BMI-zadj values including only subjects homozygous for F508del (BMI-zadj-F508del; n = 532) and BMI-zadj values including only subjects homozygous for F508del (BMI-zadjFEV-F508del; n = 518)].
Heritability
Intrapair correlations for monozygous twins, dizygous twins, same-sex dizygous twins, and the dizygous twin and sibling group are reported in Table 3 for BMI-z5to10, BMI-zadj, and BMI-zadjFEV. For each phenotype, the correlation coefficient was consistently high for monozygous twins (0.80–0.85 for 58–65 pairs), which indicated a high degree of concordance for BMI in individuals who shared 100% of genes. The dizygous twin-only groups had lower correlation coefficients (0.58–0.66 for 21 dizygous twin pairs; 0.41–0.57 for 13 same-sex dizygous twin pairs), which indicated a lower concordance in this group of twins who shared, on average, 50% of genes. Correlation coefficients for the dizygous twin and sibling group were <0.5 (0.25–0.31 for 122–148 pairs).
TABLE 3.
Pearson's correlation coefficients for BMI-z5to10 and residual phenotypes1
| MZ twins | DZ twins | Same-sex DZ twins | DZ twins and siblings2 | |
| BMI-z5to10 | 0.80 (65) | 0.59 (21) | 0.47 (13) | 0.31 (148) |
| BMI-z5to103 | 0.84 (55) | 0.62 (17) | 0.35 (9) | 0.34 (123) |
| BMI-zadj | 0.81 (63) | 0.58 (21) | 0.41 (13) | 0.27 (127) |
| BMI-zadjFEV | 0.85 (58) | 0.66 (21) | 0.57 (13) | 0.25 (122) |
All values are Pearson's correlation coefficients; no. of pairs in parentheses. BMI-zadj, average BMI z score from 5 to 10 y of age adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-z5to10, average BMI z score from 5 to 10 y of age; DZ, dizygous; MZ, monozygous.
Same-sex DZ twins and siblings with <3 y of difference in age.
After exclusion of pairs in whom one or both members of the pair had an average height z score from 5 to 10 y of age less than −1.96.
Heritability estimates and 95% CIs are reported in Table 4. Heritability was significantly different from zero except for the estimate generated for BMI-zadjFEV in monozygous and dizygous twins only. The same-sex twin-only analyses generated high heritability estimates (0.54–0.82) for unadjusted and adjusted BMI phenotypes. The addition of same-sex siblings as a proxy for dizygous twins resulted in increased heritability estimates (0.99–1). Heritability was generally greater in adjusted BMI than unadjusted BMI traits. Because all pairs in the analysis had identical CFTR mutations, these results indicated that a substantial proportion of the variability in BMI-z5to10 could be attributed to genes other than CFTR.
TABLE 4.
Heritability estimates for BMI-z5to10 and the residual phenotypes1
| MZ and DZ twins | MZ and same-sex DZ twins | MZ twins and DZ twins and siblings2 | |
| BMI-z5to10 | 0.44 (0.04–0.94) | 0.67 (0.09–1.43) | 0.99 (0.61–1.40) |
| BMI-z5to103 | 0.48 (0.04–1.06) | 0.97 (0.20–1.86) | 1 (0.60–1.44) |
| BMI-zadj | 0.47 (0.06–0.98) | 0.82 (0.19–1.61) | 1 (0.68–1.50) |
| BMI-zadjFEV | 0.38 (−0.01–0.83) | 0.54 (0.04–1.17) | 1 (0.76–1.62) |
All values are means; 95% CIs in parentheses. Confidence intervals on heritability were estimated by using a bootstrapping technique (33). BMI-zadj, average BMI z score from 5 to 10 y of age adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-z5to10, average BMI z score from 5 to 10 y of age; DZ, dizygous; MZ, monozygous.
Same-sex DZ twins and siblings with <3 y of difference in age.
After exclusion of pairs in whom one or both members of the pair had an average height z score from 5 to 10 y of age less than −1.96.
The intrapair correlations and heritability estimates for each of the phenotypes were essentially unchanged when the analyses were limited to subjects with height-z5to10 of at least −1.96 (to exclude patients with short stature defined as <3rd CDC percentile) (Tables 3 and 4), PI (see Tables 1 and 2 under “Supplemental data” in the online issue), or F508del homozygosity (see Table 3 under “Supplemental data” in the online issue). However, only monozygous twins and dizygous twins and siblings were included in the F508del analysis because of small numbers of dizygous twins and same-sex dizygous twins (9 and 5 pairs, respectively).
Linkage
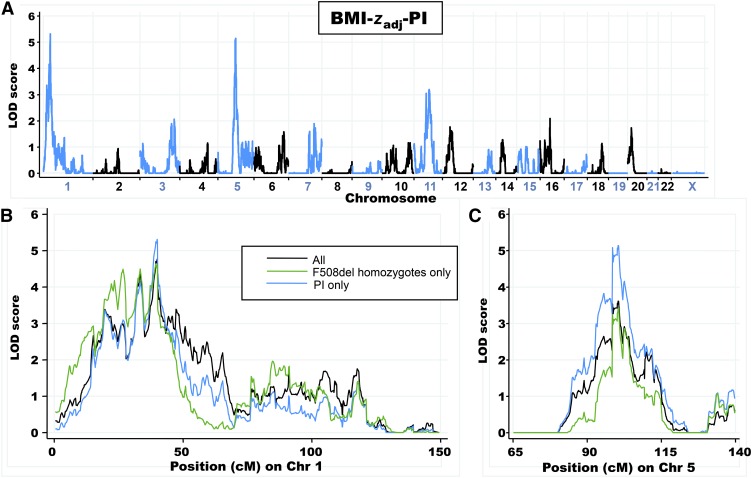
Linkage results for individuals with PI (BMI-zadj-PI; 358 sibling pairs) revealed 2 prominent genome-wide significant peaks on chromosomes 1p36.1 (LOD: 5.3) and 5q14 (LOD: 5.1) (Figure 3A; blue line in Figure 3, B and C). The exclusion of 91 subjects with short stature (height-z5to10 less than −1.96) resulted in minor changes in peak scores on chromosome 1p36.1 (LOD: 4.3) and chromosome 5q14 (LOD: 4.8), which were consistent with the reduction in the number of sibling pairs included in the analysis. Restriction of the analysis to the 219 sibling pairs who were homozygous for the F508del mutation (BMI-zadj-F508del) preserved the peak on chromosome 1p36.1 (LOD: 4.6) but decreased the evidence for linkage on chromosome 5q14 (LOD: 3.4) (green line in Figure 3, B and C; see Figure 2 under “Supplemental data” in the online issue). Similarly, the inclusion of the additional 10% of subjects with pancreatic sufficiency in BMI-zadj (402 sibling pairs) resulted in no change to the linkage peak on chromosome 1p36.1 (LOD: 4.8) with a decrease in the peak on chromosome 5q14 (LOD: 3.6) (black line in Figure 3, B and C; see Figure 3 under “Supplemental data” in the online issue). Additional loci that achieved suggestive evidence of linkage are shown in Table 5.
FIGURE 3.
A: Linkage-analysis results for BMI-zadj-PI (358 sibling pairs). Alternating blue and black lines represent Chrs labeled on the x axis, and the strength of linkage is indicated by the LOD score on the y axis. B: Comparison of Chr 1 linkage results for BMI-zadj-PI, BMI-zadj-F508del (219 sibling pairs), and BMI-zadj (402 sibling pairs). C: Comparison of Chr 5 linkage results for each phenotype. Linkage analysis was performed with each residual phenotype by using the variance-components algorithm as implemented in the MERLIN 1.1.2 software program (34). BMI-zadj, BMI-z5to10 values adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadj-F508del, BMI-zadj values with the inclusion of only subjects homozygous for F508del; BMI-zadj-PI, BMI-zadj values with the inclusion of only pancreatic-insufficient subjects; BMI-z5to10, average BMI z score from 5 to 10 y of age; Chr, chromosome; LOD, log of odds; PI, pancreatic insufficiency.
TABLE 5.
Chromosomal regions that exhibited significant (LOD ≥3.6) and suggestive (LOD ≥2.2) linkage to BMI1
| LOD scores |
||||||
| Region | BMI-zadj (402 pairs) | BMI-zadj-PI (358 pairs) | BMI-zadj-F508del (219 pairs) | BMI-zadjFEV (393 pairs) | BMI-zadjFEV-PI (350 pairs) | BMI-zadjFEV-F508del (213 pairs) |
| 1p36.1 | 4.82 | 5.32 | 4.62 | 3.2 | 3.4 | 3.82 |
| 1p31-22 | — | — | — | 3.0 | 2.4 | 2.9 |
| 2q14.3 | — | — | — | — | — | 2.3 |
| 5q14 | 3.62 | 5.12 | 3.4 | 3.2 | 5.22 | 3.3 |
| 6q25 | — | — | — | 2.5 | 2.3 | — |
| 7q33 | 2.8 | — | — | 2.9 | — | 2.4 |
| 9q34 | — | — | 2.5 | — | — | 2.7 |
| 10q25-26 | — | — | 2.5 | — | — | 2.6 |
| 11q12 | 2.7 | — | — | 2.3 | — | — |
| 11q14 | — | 3.2 | 3.4 | — | 2.7 | 3.82 |
| 12p12 | — | — | — | — | 2.5 | — |
| 13q33 | — | — | 2.5 | — | — | — |
| 14q21 | — | — | 2.5 | — | — | 3.62 |
| 16p11.2 | 2.7 | — | — | — | — | — |
BMI-zadj, average BMI z score from 5 to 10 y of age adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-zadjFEV-F508del, BMI-zadjFEV values with the inclusion of only subjects homozygous for F508del; BMI-zadjFEV-PI, BMI-zadjFEV values with the inclusion of only pancreatic-insufficient subjects; BMI-zadj-F508del, BMI-zadj values with the inclusion of only subjects homozygous for F508del; BMI-zadj-PI, BMI-zadj values with the inclusion of only pancreatic-insufficient subjects; LOD, log of odds.
LOD score met or exceeded genome-wide significance (36).
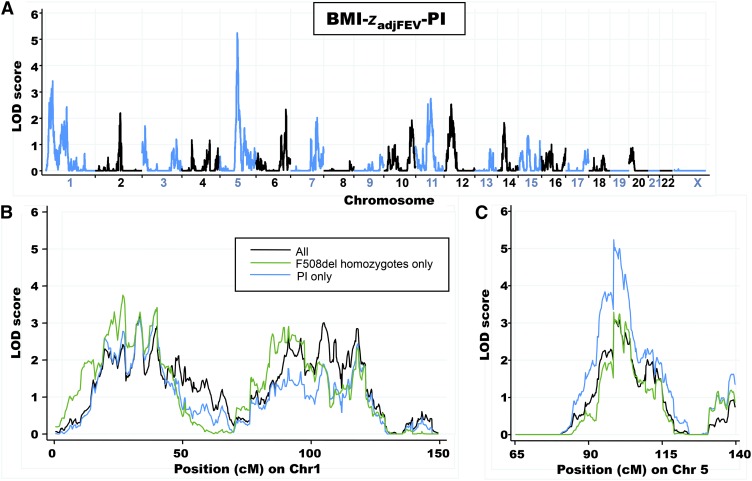
Adjustment of BMI for lung function (BMI-zadjFEV-PI; 350 sibling pairs) decreased the LOD score for the locus on chromosome 1p36.1 (LOD: 3.4) but increased the evidence for linkage at an adjacent region from chromosome 1p31-22 (LOD: 2.4); linkage on chromosome 5q14 (LOD: 5.2) was unaffected (Figure 4A; blue line in Figure 4, B and C). Linkage results for chromosome 1p36.1 were similarly decreased when adjusted for lung function in all subjects (BMI-zadjFEV; LOD: 3.2 in 393 sibling pairs) or only F508del homozygous subjects (BMI-zadjFEV-F508del; LOD: 3.8 in 213 sibling pairs) (Figure 4B). Evidence for suggestive linkage for the region from chromosome 1p31-22 (LOD: 2.9) was retained when sibling pairs who were homozygous for F508del were analyzed. Adjustment for lung function had little effect on LOD scores at chromosome 5q14 when any of the 3 subgroups were analyzed (PI subjects, F508del homozygotes, or all subjects; Figure 4C compared with Figure 3C). As in the unadjusted analyses, LOD scores were greater when PI subjects were analyzed (LOD: 5.2) and lower when all subjects (LOD: 3.2) or F508del homozygotes (LOD: 3.3) were included (Figure 4C). Significant and suggestive LOD scores for these phenotypes are summarized in Table 5.
FIGURE 4.
A. Linkage-analysis results for BMI-zadjFEV-PI (350 sibling pairs). Alternating blue and black lines represent Chrs labeled on the x axis, and the strength of linkage is indicated by the LOD score on the y axis. B: Comparison of Chr 1 linkage results for BMI-zadjFEV-PI, BMI-zadjFEV-F508del, (213 sibling pairs), and BMI-zadjFEV (393 sibling pairs). C: Comparison of Chr 5 linkage results for each phenotype. Linkage analysis was performed with each residual phenotype by using the variance-components algorithm as implemented in the MERLIN 1.1.2 software program (34). BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-zadjFEV-F508del, BMI-zadjFEV values with the inclusion of only subjects homozygous for F508del; BMI-zadjFEV-PI, BMI-zadjFEV values with the inclusion of only pancreatic-insufficient subjects; Chr, chromosome; LOD, log of odds; PI, pancreatic insufficiency.
Quantitative trait locus (QTL) heritability at 1p36.1 and 5q14 as estimated with the MERLIN program for PI subjects suggested that these loci respectively contributed to 54% and 53% of the variation in BMI. However, these estimates were highly likely to be biased upward because of the winner's curse (38); after potential bias was corrected for, ≥16% and ≥15% of the BMI variance was estimated to be explained by the respective loci.
DISCUSSION
To our knowledge, this was the first study to quantify the relative contribution of genetic modifiers to the nutritional status of young children with CF. Heritability estimates suggested strong genetic control of variation in our nutritional phenotype BMI-z5to10. In addition, linkage analyses pointed to genes on chromosomes 1 and 5 as having substantial influence on this phenotype.
Studies of heritability and linkage first require reliable phenotype measures. The BMI-z5to10 phenotype was roughly normally distributed across the full range of BMI z scores represented in the CDC reference population. In particular, there were children at both extremes of BMI-z5to10 of −1.64 or less or 5th CDC percentile (2.7% of subjects) and ≥1.64 or 95th CDC percentile (2.2% of subjects). The mean was slightly less than zero, which indicated that, on average, children in this study had lower BMI than the CDC reference population did. This result was similarly described in a study of school-age children with CF who consumed more calories per day than their healthy peers did but still had significantly lower height and weight measurements (39). The median height-z5to10 of −0.58 and median weight-z5to10 of −0.42 corresponded to published literature that cited height z scores of −0.5 to −0.8 and weight z scores of −0.4 to −0.7 for children aged 3–10 y with CF in the United States (40). Subjects with height-z5to10 less than −1.96 (or 3rd CDC percentile) had BMI-z5to10 scores distributed equally from low to high. Thus, some of the subjects may have had poor nutritional status that manifested primarily as short stature but preserved body weight that resulted in a BMI z score that did not reflect their nutritional status. However, the exclusion of individuals with height-z5to10 less than −1.96 did not appreciably change heritability estimates or linkage results. The nutritional phenotype used in this study encompassed the age range from 5 to 10 y because growth status has been reported to be fairly stable in childhood for patients with CF (18), whereas there is an increased prevalence of abnormal nutritional variables, such as weight for height, as patients mature into adulthood (41). The evaluation of our phenotype revealed a small but significant decline in the mean BMI z score of 0.06 units/y, which was supported by another study of growth in CF that suggested a decline in weight z scores during the prepubertal period (4). Despite this annual decrease in the mean BMI z score, adjustment for the decrease did not lead to appreciable differences in the remainder of the analyses.
BMI-z5to10 was significantly positively correlated with later birth cohorts and lung function. These results are supported by the 2009 CFF Patient Registry Report that showed both the increasing median BMI percentile for children with CF and the relation between higher BMI and improved lung function (19). Alternately, BMI-z5to10 was negatively correlated with PI and history of MI. A previous study on pancreatic status in CF suggested that children with residual pancreatic function had improved growth and nutritional status (12), whereas associations have been made between MI and malnutrition (11). The phenotype was also negatively correlated with female sex, although just below the level of significance. However, sex was maintained as a covariate because there have been reported sex-related differences in growth during childhood in CF such that girls have a greater rate of decrease in weight z score than do boys (4).
For BMI-z5to10 and each of the residual phenotypes, correlation coefficients were consistently highest for monozygous twins, which suggested a genetic contribution to the variation in nutritional status. However, monozygous twins share 100% of genes, and thus, correlation coefficients <1 indicate that variation is also influenced by environmental or stochastic factors. On average, dizygous twins share 50% of genes, and their correlation coefficients were close to 0.5 as expected for a trait under strong genetic control. Consequently, heritability estimates from the same-sex twin-only analyses were high for each of the BMI-z5to10 phenotypes (0.54–0.82), and similar estimates have been reported for heritability of BMI variation in the general population (42, 43). The analysis of monozygous and dizygous twins may produce the most accurate heritability estimates because twin pairs have greater similarity in environmental exposures than siblings or unrelated patients (44). Heritability estimates were higher when the subgroup of dizygous twins and siblings was used in the calculations (0.99–1), but this analysis was limited by the higher chance for nongenetic influences in siblings. Overall, heritability estimates for the twin-only analyses were higher for BMI-zadj than for BMI-zadjFEV; one possible explanation for this result is that genetic factors that influence lung function may be different from those that are important to nutritional status.
Although all patients in this study had classic CF, ∼10% retained exocrine pancreatic function (pancreatic sufficiency), which is strongly correlated with specific mutations in CFTR that confer a slightly milder disease course (24). Therefore, analyses were also performed after exclusion of patients with pancreatic sufficiency and in the subset of subjects who were homozygous for F508del, which is the most common CF-causing mutation in CFTR, an approach that traded sample size for increased homogeneity. The heritability of BMI-z5to10 remained high for each of these subgroups, which indicated that linkage analysis with these subgroups could control for allelic variation in CF while preserving the power to uncover modifier loci.
The linkage analysis by using BMI-zadj-PI showed significant linkage on chromosomes 1p36.1 (LOD: 5.3) and 5q14 (LOD: 5.1) with a combined contribution to variation in BMI ≤31%. Adjustment for lung function in BMI-zadjFEV-PI caused the linkage evidence to decrease on chromosome 1p36.1 but did not affect the locus on chromosome 5q14. These results suggested that modifier genes located at the former locus influence both nutritional status and lung function, whereas chromosome 5q14 encompasses a gene that modifies nutritional status that is independent of lung-disease severity. Additional support for a non-CFTR influence on nutritional status is provided by the observation that the linkage at chromosome 1p36.1 in subjects with identical CFTR genotypes (F508del homozygotes) was of similar magnitude as in the pancreatic-insufficient group, even though fewer sibling pairs were included (219 compared with 358). Therefore, a modifier gene at this locus may be specific to CF, such as a gene involved in protein processing. In contrast, genes that influence BMI in the general population have been mapped to the region on chromosome 5 linked to BMI in CF patients (45–47). Intriguingly, the critical region of linkage (1 LOD drop from maximum linkage) on chromosome 5q14 contains the arrestin domain-containing 3 (ARRCD3) gene that has recently been implicated as a regulator of body mass and energy expenditure in males (47). We observed higher linkage scores in male than in female patients, which suggested that ARRCD3 may be viable candidate for the modification of BMI in CF. Another BMI-susceptibility locus in the general population is located on chromosome 16p11.2, which is a region that, in our study, had a suggestive linkage with an LOD score of 2.7 (48). Because patients with CF are under nutritional stress, they may be uniquely sensitive to genetic modifiers that are also important to weight homeostasis in the general population.
In conclusion, our study of the nutritional status of young twins and siblings with CF shows that genes other than CFTR influence the variation in BMI. Specifically, genetic modifiers located at loci on chromosomes 1 and 5 contribute to a considerable percentage of the BMI variance. These results support additional investigation of CF-modifier genes, which can provide greater insight into the CF disease process and targets for future nutritional therapies.
Acknowledgments
We acknowledge the numerous patients with CF, their families, research coordinators, and clinicians who participated in the CF Twin-Sibling Study. We thank J Michael Collaco for his thoughtful discussions of this material and Patricia Cornwall for her administrative assistance.
The authors’ responsibilities were as follows—GMB, SMB, and GRC: designed the research and wrote the manuscript; GMB and SMB: conducted the research; GMB, CPW, VKD, and SMB: analyzed data; GRC: had primary responsibility for the final content of the manuscript; and all authors: read and approved the final manuscript. None of the authors had a conflict of interest.
Footnotes
Abbreviations used: BMI-zadj, average BMI z score from 5 to 10 y of age adjusted for female sex, birth cohort, pancreatic insufficiency, and history of meconium ileus; BMI-zadjFEV, average BMI z score from 5 to 10 y of age adjusted as for BMI-zadj plus for FEVq6to10; BMI-zadjFEV-F508del, BMI-zadjFEV values with the inclusion of only subjects homozygous for F508del; BMI-zadjFEV-PI, BMI-zadjFEV values with the inclusion of only pancreatic-insufficient subjects; BMI-zadj-F508del, BMI-zadj values with the inclusion of only subjects homozygous for F508del; BMI-zadj-PI, BMI-zadj values with the inclusion of only pancreatic-insufficient subjects; BMI-z5to10, average BMI z score from 5 to 10 y of age; CF, cystic fibrosis; CFF, Cystic Fibrosis Foundation; FEVq6to10, average cystic fibrosis–specific forced expiratory volume in 1 s percentile for all measures obtained from 6 to 10 y of age; height-z5to10, average height z score from 5 to 10 y of age; LOD, log of odds; MI, meconium ileus; NBS, newborn screen; PI, pancreatic insufficiency; weight-z5to10, average weight z score from 5 to 10 y of age.
REFERENCES
- 1.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science 1989;245:1073–80 [DOI] [PubMed] [Google Scholar]
- 2.Lai HJ, Shoff SM, Farrell PM. Recovery of birth weight z score within 2 years of diagnosis is positively associated with pulmonary status at 6 years of age in children with cystic fibrosis. Pediatrics 2009;123:714–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matel JL. Nutritional management of cystic fibrosis. JPEN J Parenter Enteral Nutr 2012;36:60S–7S [DOI] [PubMed] [Google Scholar]
- 4.Zemel BS, Jawad AF, Fitzsimmons S, Stallings VA. Longitudinal relationship among growth, nutritional status, and pulmonary function in children with cystic fibrosis: analysis of the cystic fibrosis foundation national CF patient registry. J Pediatr 2000;137:374–80 [DOI] [PubMed] [Google Scholar]
- 5.Konstan MW, Butler SM, Wohl ME, Stoddard M, Matousek R, Wagener JS, Johnson CA, Morgan WJ. Growth and nutritional indexes in early life predict pulmonary function in cystic fibrosis. J Pediatr 2003;142:624–30 [DOI] [PubMed] [Google Scholar]
- 6.Salvatore F, Scudiero O, Castaldo G. Genotype-phenotype correlation in cystic fibrosis: the role of modifier genes. Am J Med Genet 2002;111:88–95 [DOI] [PubMed] [Google Scholar]
- 7.McKone EF, Goss CH, Aitken ML. CFTR genotype as a predictor of prognosis in cystic fibrosis. Chest 2006;130:1441–7 [DOI] [PubMed] [Google Scholar]
- 8.Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci 2010;1214:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southern KW, Merelle MM, Dankert-Roelse JE, Nagelkerke AD. Newborn screening for cystic fibrosis. Cochrane Database Syst Rev 2009;CD001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley GM, Carson KA, Leonard AR, Mogayzel PJ, Jr, Oliva-Hemker M. Nutritional outcomes following gastrostomy in children with cystic fibrosis. Pediatr Pulmonol 47:743–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai HC, Kosorok MR, Laxova A, Davis LA, FitzSimmon SC, Farrell PM. Nutritional status of patients with cystic fibrosis with meconium ileus: a comparison with patients without meconium ileus and diagnosed early through neonatal screening. Pediatrics 2000;105:53–61 [DOI] [PubMed] [Google Scholar]
- 12.Cohen JR, Schall JI, Ittenbach RF, Zemel BS, Stallings VA. Fecal elastase: pancreatic status verification and influence on nutritional status in children with cystic fibrosis. J Pediatr Gastroenterol Nutr 2005;40:438–44 [DOI] [PubMed] [Google Scholar]
- 13.Blackman SM, Vanscoy LL, Collaco JM, Naughton KM, Cutting GR. Variability in body mass index in cystic fibrosis is determined partly by a genetic locus on chromosome 5. Pediatric Pulmonology 2008;271(suppl 31):10-23-0008. [Google Scholar]
- 14.Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, Algire M, McWilliams R, Beck S, Hoover-Fong J, et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med 2007;175:1036–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stallings VA, Stark LJ, Robinson KA, Feranchak AP, Quinton H. Evidence-based practice recommendations for nutrition-related management of children and adults with cystic fibrosis and pancreatic insufficiency: results of a systematic review. J Am Diet Assoc 2008;108:832–9 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Lai HJ. Comparison of the use of body mass index percentiles and percentage of ideal body weight to screen for malnutrition in children with cystic fibrosis. Am J Clin Nutr 2004;80:982–91 [DOI] [PubMed] [Google Scholar]
- 17.Wiedemann B, Paul KD, Stern M, Wagner TO, Hirche TO. Evaluation of body mass index percentiles for assessment of malnutrition in children with cystic fibrosis. Eur J Clin Nutr 2007;61:759–68 [DOI] [PubMed] [Google Scholar]
- 18.Haeusler G, Frisch H, Waldhor T, Gotz M. Perspectives of longitudinal growth in cystic fibrosis from birth to adult age. Eur J Pediatr 1994;153:158–63 [DOI] [PubMed] [Google Scholar]
- 19.Cystic Fibrosis Foundation Patient Registry 2009 annual data report. Bethesda, MD: Cystic Fibrosis Foundation, 2009 [Google Scholar]
- 20.Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR, Roche AF, Johnson CL. 2000 CDC Growth Charts for the United States: methods and development. Vital Health Stat 11 2002:1–190 [PubMed] [Google Scholar]
- 21.Umławska W, Susanne C. Growth and nutritional status in children and adolescents with cystic fibrosis. Ann Hum Biol 2008;35:145–53 [DOI] [PubMed] [Google Scholar]
- 22.Schechter MS. Nongenetic influences on cystic fibrosis outcomes. Curr Opin Pulm Med 2011;17:448–54 [DOI] [PubMed] [Google Scholar]
- 23.Kerem E, Corey M, Kerem B-S, Rommens J, Markiewicz D, Levison H, Tsui LC, Durie P. The relation between genotype and phenotype in cystic fibrosis–analysis of the most common mutation (deltaF508). N Engl J Med 1990;323:1517–22 [DOI] [PubMed] [Google Scholar]
- 24.Kristidis P, Bozon D, Corey M, Markiewicz D, Rommens J, Tsui LC, Durie P. Genetic determination of exocrine pancreatic function in cystic fibrosis. Am J Hum Genet 1992;50:1178–84 [PMC free article] [PubMed] [Google Scholar]
- 25.The Cystic Fibrosis Genotype-Phenotype Consortium Correlation between genotype and phenotype in patients with cystic fibrosis. N Engl J Med 1993;329:1308–13 [DOI] [PubMed] [Google Scholar]
- 26.Ahmed N, Corey M, Forstner G, Zielenski J, Tsui LC, Ellis L, Tullis E, Durie P. Molecular consequences of cystic fibrosis transmembrane regulator (CFTR) gene mutations in the exocrine pancreas. Gut 2003;52:1159–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green DM, McDougal KE, Blackman SM, Sosnay PR, Henderson LB, Naughton KM, Collaco JM, Cutting GR. Mutations that permit residual CFTR function delay acquisition of multiple respiratory pathogens in CF patients. Respir Res 2010;11:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, Berthiaume Y, Cutler D, Cojocaru A, Collaco JM, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet 2011;43:539–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackman SM, Deering-Brose R, McWilliams R, Naughton K, Coleman B, Lai T, Algire M, Beck S, Hoover-Fong J, Hamosh A, et al. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology 2006;131:1030–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulich M, Rosenfeld M, Campbell J, Kronmal R, Gibson RL, Goss CH, Ramsey B. Disease-specific reference equations for lung function in patients with cystic fibrosis. Am J Respir Crit Care Med 2005;172:885–91 [DOI] [PubMed] [Google Scholar]
- 31.Collaco JM, Blackman SM, McGready J, Naughton KM, Cutting G. Quantification of the contribution of environmental and genetic modifiers to cystic fibrosis lung disease. Ped Pulm 2009;(suppl 32):268 [Google Scholar]
- 32.Falconer DS, Mackay TFC, eds. Introduction to quantitative genetics. Essex, United Kingdom: Longman, 1996 [Google Scholar]
- 33.Efron B. Better bootstrap confidence intervals. J Am Stat Assoc 1987;82:171–85 [Google Scholar]
- 34.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101 [DOI] [PubMed] [Google Scholar]
- 35.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998;62:1198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 1995;11:241–7 [DOI] [PubMed] [Google Scholar]
- 37.Matel JL, Milla CE. Nutrition in cystic fibrosis. Semin Respir Crit Care Med 2009;30:579–86 [DOI] [PubMed] [Google Scholar]
- 38.Göring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet 2001;69:1357–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark LJ, Mulvihill MM, Jelalian E, Bowen AM, Powers SW, Tao S, Creveling S, Passero MA, Harwood I, Light M, et al. Descriptive analysis of eating behavior in school-age children with cystic fibrosis and healthy control children. Pediatrics 1997;99:665–71 [PubMed] [Google Scholar]
- 40.Lai HC, Corey M, Fitzsimmons S, Kosorok MR, Farrell PM. Comparison of growth status of patients with cystic fibrosis between the United States and Canada. Am J Clin Nutr 1999;69:531–8 [DOI] [PubMed] [Google Scholar]
- 41.Steinkamp G, Wiedemann B. Relationship between nutritional status and lung function in cystic fibrosis: cross sectional and longitudinal analyses from the German CF quality assurance (CFQA) project. Thorax 2002;57:596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rokholm B, Silventoinen K, Tynelius P, Gamborg M, Sorensen TI, Rasmussen F. Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLoS ONE 2011;6:e27135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, Davis CJ, Fagnani C, Hjelmborg J, Kaprio J, et al. Sex differences in heritability of BMI: a comparative study of results from twin studies in eight countries. Twin Res 2003;6:409–21 [DOI] [PubMed] [Google Scholar]
- 44.Segal NL, Feng R, McGuire SA, Allison DB, Miller S. Genetic and environmental contributions to body mass index: comparative analysis of monozygotic twins, dizygotic twins and same-age unrelated siblings. Int J Obes (Lond) 2009;33:37–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell CG, Benzinou M, Siddiq A, Lecoeur C, Dina C, Lemainque A, Clement K, Basdevant A, Guy-Grand B, Mein CA, et al. Genome-wide linkage analysis for severe obesity in french caucasians finds significant susceptibility locus on chromosome 19q. Diabetes 2004;53:1857–65 [DOI] [PubMed] [Google Scholar]
- 46.Cheng CY, Kao WH, Patterson N, Tandon A, Haiman CA, Harris TB, Xing C, John EM, Ambrosone CB, Brancati FL, et al. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet 2009;5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, et al. The arrestin domain-containing 3 protein regulates body mass and energy expenditure. Cell Metab 2011;14:671–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Magi R, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–48 [DOI] [PMC free article] [PubMed] [Google Scholar]