Abstract
Background: Carotenoids have been hypothesized to reduce the risk of many diseases, but associations with intakes or blood concentrations may arise from other constituents of fruit and vegetables. Use of genetic variation in β-carotene 15,15′-monooxygenase 1 (BCMO1), a key enzyme in provitamin A carotenoid metabolism, as a surrogate for carotenoid exposure may aid in determining the role of carotenoids unconfounded by other carotenoid-containing food constituents, but important variants must be identified.
Objective: Our goal was to select BCMO1 single nucleotide polymorphisms (SNPs) that predict plasma carotenoid concentrations for use in future epidemiologic studies.
Design: We assessed the associations between 224 SNPs in BCMO1 ± 20 kb imputed from the 1000 Genomes Project EUR reference panel with plasma carotenoid and retinol concentrations by using 7 case-control data sets (n = 2344) within the Nurses’ Health Study, randomly divided into training (n = 1563) and testing (n = 781) data sets. SNPs were chosen in the training data set through stepwise selection in multivariate linear regression models; β-coefficients were used as weights in weighted gene scores.
Results: Two or 3 SNPs were selected as predictors of β-carotene, α-carotene, β-cryptoxanthin, and lutein/zeaxanthin. In the testing data set, the weighted gene scores were significantly associated with plasma concentrations of the corresponding carotenoid (P = 6.4 × 10−12, 3.3 × 10−3, 0.02, and 1.8 × 10−17, respectively), and concentrations differed by 48%, 15%, 15%, and 36%, respectively, across extreme score quintiles.
Conclusions: SNPs in BCMO1 are associated with plasma carotenoid concentrations. Given adequate sample size, the gene scores may be useful surrogates for carotenoid exposure in future studies.
INTRODUCTION
Carotenoids are pigments obtained in the diet primarily from fruit and vegetables. Certain carotenoids can be converted to vitamin A, but the health effects of carotenoids may instead or additionally result from their antioxidant activity (1) or other mechanisms, such as selective induction of apoptosis (2). β-Carotene 15,15′-monooxygenase 1 (BCMO1)4 centrally cleaves dietary provitamin A carotenoids, including β-carotene, α-carotene, and β-cryptoxanthin, to form retinal, which is the first step in converting provitamin A carotenoids to vitamin A (3–6). Leung et al (7) identified 2 nonsynonymous single nucleotide polymorphisms (SNPs; A379V, rs7501331 and R267S, rs12934922) in the coding region of BCMO1 that were associated with higher fasting plasma β-carotene and reduced conversion of a pharmacologic dose of β-carotene to retinyl palmitate. Additionally, through a genome-wide association study (GWAS), Ferrucci et al (8) identified 4 other SNPs in modest (r2 = 0.22–0.55) linkage disequilibrium (LD) with each other near BCMO1 that were related to circulating carotenoid concentrations. Specifically, rs6564851 was associated with increased β- and α-carotene and decreased lutein, zeaxanthin, and lycopene (8); rs6564851 was recently reported to reduce BCMO1 activity by 48% in females (9).
Other research suggests that carotenoids may reduce the risk of several diseases, including breast cancer. A meta-analysis suggests that dietary intake of α- and β-carotene is associated with a 9% and 6% lower breast cancer risk, respectively, comparing highest versus lowest intakes (10). These modest associations may result from attenuation of a stronger causal association due to misclassification of an individual's internal carotenoid dose as measured by his or her dietary intake. Alternatively, the apparent inverse associations may be due to residual confounding by personal characteristics and/or other beneficial components of fruit and vegetables. In accordance with the principles of Mendelian randomization (11, 12), genetic variation in carotenoid metabolism is likely to be uncorrelated with dietary and lifestyle factors, thus providing an unconfounded surrogate for carotenoid exposure that may also improve prediction of internal carotenoid dose when used in combination with dietary intake. Thus, the aim of this analysis was to identify important genetic variants related to plasma carotenoid concentrations and develop gene scores for use in future studies as unconfounded measures of carotenoid exposure. Retinol was included in these analyses because of its relation with the provitamin A carotenoids. BCMO1 was selected as the candidate locus of interest given its known role in provitamin A carotenoid metabolism (3–6) and previous identification of polymorphisms in the region associated with circulating carotenoid concentrations (7, 8).
SUBJECTS AND METHODS
Subjects and data sources
The Nurses’ Health Study (NHS) was established in 1976 when 121,700 female registered nurses in the United States aged 30–55 y completed a mailed questionnaire regarding medical history and other health-related exposures. The women have since been followed biennially by mailed self-administered questionnaires. In 1989–1990, 32,826 NHS participants provided blood samples. Details of the blood collection were published previously (13). In brief, women arranged to have their blood drawn and shipped, via overnight courier with an icepack, to our laboratory where samples were immediately centrifuged, portioned into aliquots, and stored in liquid nitrogen freezers. Ninety-seven percent of samples were received within 26 h of collection.
This study includes participants from case-control studies, nested within the blood subcohort, of breast cancer, cataract, colorectal cancer, colorectal adenoma, and myocardial infarction for whom plasma carotenoid and retinol concentrations were assayed. Because the subclinical presence of a major chronic disease could alter plasma biomarker concentrations, women with breast cancer, colon cancer, myocardial infarction, or diabetes diagnosed within 2 y after blood collection were excluded (n = 592).
Some individuals in the biomarker-assayed groups were included in GWASs of breast cancer [National Cancer Institute's Cancer Genetic Markers of Susceptibility (14)], ischemic heart disease, type 2 diabetes, or kidney stones. SNPs were imputed for these 2344 women, hereafter referred to as women with genome-wide scans. rs12934922, rs7501331, and rs6564851, which were previously identified (7, 8), and rs28380273, rs4889286, and rs4448930, which were identified during our analysis, were genotyped by the TaqMan assay in 3492, 3317, 872, 2205, 2495, and 2535 women, respectively (see Supplemental Table 1 under “Supplemental data” in the online issue). rs7501331 and rs6564851 were genotyped by GWAS in 661 and 2340 women, respectively (see Supplemental Table 1 under “Supplemental data” in the online issue). When both genotyped and imputed data were available, genotyped data were used; TaqMan genotyped data were prioritized over GWAS genotyped data when both were available. To eliminate concern over population stratification, the analyses were limited to women of European descent. The final sample consisted of 4135 women, of whom 2149 were controls. Of the 2344 women with genome-wide scans, 1197 were controls. The study protocol was approved by the Institutional Review Board of Brigham and Women's Hospital.
All participants in this analysis completed a semiquantitative food-frequency questionnaire (FFQ) in 1986 and/or 1990 (3632 completed both FFQs, 383 completed only the 1990 FFQ, and 120 completed only the 1986 FFQ). FFQs were considered complete if a woman reported a plausible total energy intake (600–3500 kcal/d) and left ≤70 food items blank. To reduce within-person variation, average nutrient intakes from the 1986 and 1990 FFQs were used when possible. To determine nutrient intakes, reported consumption frequencies of commonly used units or portion sizes of food items were multiplied by the nutrient contents of the specified unit and summed over all foods. Food nutrient values were derived primarily from USDA sources. Use of multivitamins and other supplements, as well as dose and duration of use, also were assessed and incorporated into retinol and β-carotene intake calculations. The reproducibility and validity of the FFQs were reported previously (15–17). The correlation between vitamin A intake from food and supplements estimated from the FFQ compared with four 1-wk dietary records was 0.49; the validity for specific carotenoids was not calculated (16). In another analysis, the correlations between 2 averaged FFQs and plasma carotenoid concentrations in nonsmoking women were 0.48 for α-carotene, 0.27 for β-carotene, 0.32 for β-cryptoxanthin, 0.21 for lycopene, and 0.27 for lutein; retinol was not calculated (18). Except for alcohol, nutrient intakes in this analysis were adjusted for total energy intake by the residual method (19).
Nondietary variables were obtained from questionnaires completed by participants. Menopausal status and postmenopausal hormone (PMH) use were determined from a questionnaire completed at the time of blood collection. Date of birth and height were determined from the baseline NHS questionnaire. BMI was calculated by using height reported in 1976 and weight reported on the blood collection questionnaire. If weight was missing, weight reported on the 1990 NHS questionnaire was used (n = 109). For the 3 women with both weights missing, weight reported on the 1988 NHS questionnaire was used. For the 9 women (4 with genome-wide scans) missing all weights or height, the median BMI value for all women was used. Smoking status was determined from the 1990 NHS questionnaire. Given the low percentage of smokers among women with available data (13.7%), women missing smoking status were assumed to be never/former smokers (n = 10 for all women, 4 for women with genome-wide scans).
Genotyping and imputation
Genotypes for rs12934922, rs7501331, rs28380273, rs4889286, and rs4448930 in breast cancer cases and controls were determined by measuring endpoint fluorescence by using the 5′ nuclease assay (TaqMan) on the ABI PRISM 7900HT Sequence Detection System (20). Genotypes for rs12934922, rs7501331, and rs6564851 in the colorectal cancer and adenoma cases and controls were determined according to the manufacturer's instructions by using the TaqMan OpenArray SNPGenotyping Platform (Applied Biosystems).
For women with genome-wide scans, SNP genotypes were imputed by GWAS data set in 2 steps. First, genotypes were phased by using MaCH (v1.0.16) (21). Then, Minimac (v.beta.2010) was used to impute (22) ∼11 million SNPs by using 1000 Genomes Project EUR data [release August 2010/National Center for Biotechnology Information (NCBI) 37] as the reference panel. Imputation results for each SNP were expressed as “allele dosages” (fractional values from 0 to 2) and were extracted for the BCMO1 gene region plus 20 kilobases up- and downstream (NCBI 37 chr16:81252296–81344747). For women in the Cancer Genetic Markers of Susceptibility and kidney stones GWAS groups (genotyped by using Illumina 550k and 610Q, respectively), 17 SNPs in the region were GWAS genotyped. Thirty SNPs were GWAS genotyped for women in the ischemic heart disease and type 2 diabetes GWAS groups (genotyped by using Affymetrix 6.0). SNPs with imputation r2 ≥ 0.50 in all 4 GWAS data sets were initially considered for further analysis (n = 225 of 548). Later, rs28380273 was removed because of poor genotyping results, which left 224 SNPs. Reported minor allele frequencies and imputation r2 are weighted averages based on sample size across the 4 individual GWAS data sets.
Plasma biomarker assays data set
Plasma carotenoid and retinol concentrations were measured by the Micronutrient Analysis Laboratory in the Department of Nutrition at the Harvard School of Public Health by using reversed-phase HPLC by the methods described by El-Sohemy et al (23). Lycopene was assayed as total lycopene. Blinded quality-control samples (10%) were randomly placed throughout sample boxes. Laboratory technicians also were blinded to case-control status. CVs were calculated for each biomarker within each laboratory batch. Across the 12 batches for each biomarker, CVs were generally <15%, except for 1 batch for β-carotene (CV = 20.7%), 2 batches for β-cryptoxanthin (CV = 19.4% and 20.6%), 1 batch for lycopene (CV = 17.5%), and 3 batches for retinol (CV = 16.2%, 17.8%, and 25.8%). Values were missing for some biomarkers because of laboratory technical difficulties (9 α-carotene, 10 β-carotene, 10 β-cryptoxanthin, 9 lutein/zeaxanthin, 9 lycopene, and 13 retinol).
To account for laboratory variation that was evident across batches, plasma concentrations were standardized to an average batch. β-Coefficients from linear regression models adjusted for the covariates listed below with batch indicators as the independent variables and natural log–transformed biomarker concentrations as the dependent variable were averaged. The differences between coefficients for each biomarker measurement's batch and the average across batches were subtracted from each biomarker value. A similar method was used previously to account for study effects among 11 blood pressure studies (24).
Total plasma cholesterol was assayed in 15 batches by using the enzymatic methods described by Allain et al (25). CVs were 2.1–15.6%. Plasma cholesterol data were not available for 482 women (355 with genome-wide scans), and 1 outlier identified by the generalized extreme Studentized deviate many-outlier detection method (26) was set to missing.
Statistical analysis
All plasma biomarker values were natural log transformed to improve normality, and all SNP dosages or genotypes were coded as additive variables (ie, allele counts). Outlying biomarker concentrations among all women were identified within batch by a generalized extreme Studentized deviate many-outlier detection method (26) and set to missing (6 of 4126 α-carotene, 9 of 4125 β-carotene, 6 of 4125 β-cryptoxanthin, 3 of 4126 lutein/zeaxanthin, 24 of 4126 lycopene, and 7 of 4122 retinol).
Biomarker residuals were derived from multivariate linear regression of each plasma biomarker on covariates. Correlations bewteen back-transformed plasma biomarker residuals were evaluated by Spearman correlation coefficients. For reduction of extraneous variation in biomarker concentrations, multivariate linear regression models included the following covariates: age at blood collection (y), case-control status (as 7 sets of indicator variables coded as not in data set, control, or case for breast cancer, colon cancer, colorectal adenoma, cataract, myocardial infarction, kidney stones, and diabetes), menopausal status and PMH (premenopausal; postmenopausal, no PMH; postmenopausal, current PMH; or dubious/unknown), BMI (kg/m2), smoking status (never/former, current <15 cigarettes/d, or current ≥15 cigarettes/d or an unknown amount/d), plasma cholesterol (mg/dL or missing indicator), alcohol intake (g/d), energy-adjusted fat intake (g/d), and energy-adjusted intake of the nutrient of interest (μg/d for carotenoids; IU/d for retinol). Plasma β-carotene, α-carotene, and β-cryptoxanthin models also were adjusted for dietary retinol intake (IU/d) to account for possible reduced conversion of provitamin A carotenoids in the presence of high preformed retinol intake. Results from multivariate linear regressions of each plasma biomarker on each SNP among women with genome-wide scans were used to create regional association plots based on NCBI Build 36 positions by using SNAP version 2.2 (27).
To select SNPs associated with the plasma biomarkers and provide unbiased estimates of model performance after SNP selection, women with genome-wide scans were randomly divided into 2 groups: a two-thirds training subset (n = 1563) and a one-third testing subset (n = 781). In the training subset, SNPs were chosen by stepwise selection by using SAS PROC GLMSELECT and the Schwartz Bayesian information selection criterion with the previously listed covariates forced into the model. In the testing subset, unweighted gene scores were calculated by adding dosages of biomarker-increasing alleles across selected SNPs. Weighted gene scores were calculated by multiplying each selected SNP's biomarker-increasing allele dosage by its β-coefficient in the final selected model and adding across SNPs.
Composite LD among SNPs was evaluated by Pearson correlation coefficients. Percentage variation explained by the genetic terms (gene scores or SNPs) was determined by a multiple partial R2, which was calculated by subtracting the sum of squared errors (SSE) for the model including both the genetic term and covariates from the SSE for the covariate-only model and then dividing by the SSE for the covariate-only model. This method provides the percentage of variation in the residual biomarker concentration explained by the genetic term after accounting for variation explained by the covariates. Covariate-adjusted geometric mean plasma biomarker concentrations were determined by back-transforming the least-squares means from SAS PROC GLM. Gene-diet interactions were assessed among all women with genome-wide scans by including cross-product terms for dietary intake (represented as quintile medians of the nutrient of interest) and the continuous gene score in multivariate models as well as the main effects of dietary intake (quintile medians) and gene score. All reported P values are 2-sided and unadjusted for multiple testing. All statistical analyses were performed by using SAS version 9 (SAS Institute).
RESULTS
Participant characteristics are shown in Table 1. Spearman correlations among back-transformed plasma biomarker concentration residuals after regressing out covariates are shown elsewhere (see Supplemental Table 2 under “Supplemental data” in the online issue). The β-carotene and α-carotene residuals were the most correlated (r = 0.74), and the β-carotene and retinol residuals were the least correlated (r = 0.02).
TABLE 1.
Participant characteristics at the time of blood collection
| All women |
Women with genome-wide scans |
Women without genome-wide scans 1 |
||||
| Characteristic | n | Value | n | Value | n | Value |
| Age (y) | 4135 | 57.8 ± 6.82 | 2344 | 58.8 ± 6.4 | 1791 | 56.5 ± 7.2 |
| BMI (kg/m2) | 4135 | 25.3 ± 4.4 | 2344 | 25.4 ± 4.6 | 1791 | 25.1 ± 4.2 |
| Plasma cholesterol (mg/dL) | 3653 | 219 ± 40 | 1989 | 222 ± 40 | 1664 | 217 ± 40 |
| Postmenopausal or unknown status (%) | 3411 | 82.5 | 2059 | 87.8 | 1352 | 75.5 |
| Current postmenopausal hormone use (%)3 | 1482 | 48.8 | 951 | 50.7 | 531 | 45.6 |
| Current smoking <15 cigarettes/d (%) | 230 | 5.6 | 148 | 6.3 | 82 | 4.6 |
| Current smoking ≥15 cigarettes/d (%) | 336 | 8.1 | 213 | 9.1 | 123 | 6.9 |
| Daily dietary intake4 | ||||||
| β-Carotene (μg) | 4135 | 4306 ± 2209 | 2344 | 4394 ± 2322 | 1791 | 4191 ± 2046 |
| α-Carotene (μg) | 4135 | 762 ± 505 | 2344 | 775 ± 520 | 1791 | 745 ± 485 |
| β-Cryptoxanthin (μg) | 4135 | 177 ± 94 | 2344 | 179 ± 94 | 1791 | 174 ± 94 |
| Lutein/zeaxanthin (μg) | 4135 | 2881 ± 1608 | 2344 | 2914 ± 1673 | 1791 | 2836 ± 1518 |
| Lycopene (μg) | 4135 | 6357 ± 3301 | 2344 | 6413 ± 3382 | 1791 | 6285 ± 3192 |
| Retinol (IU) | 4135 | 4184 ± 3957 | 2344 | 4256 ± 4137 | 1791 | 4090 ± 3707 |
| Total fat (g) | 4135 | 57 ± 9 | 2344 | 56 ± 9 | 1791 | 57 ± 9 |
| Alcohol (g) | 4135 | 6 ± 10 | 2344 | 6 ± 10 | 1791 | 6 ± 9 |
| Plasma biomarker concentration (μg/L) | ||||||
| β-Carotene | 4116 | 301 ± 250 | 2331 | 303 ± 258 | 1785 | 300 ± 239 |
| α-Carotene | 4120 | 74 ± 54 | 2330 | 74 ± 55 | 1790 | 75 ± 53 |
| β-Cryptoxanthin | 4119 | 84 ± 50 | 2332 | 85 ± 50 | 1787 | 83 ± 51 |
| Lutein/zeaxanthin | 4123 | 187 ± 78 | 2333 | 187 ± 78 | 1790 | 187 ± 78 |
| Lycopene | 4102 | 432 ± 194 | 2323 | 429 ± 195 | 1779 | 436 ± 193 |
| Retinol | 4115 | 568 ± 138 | 2340 | 578 ± 136 | 1775 | 554 ± 139 |
Differences between women with and without genome-wide scans for the variables presented in the table were tested by Wilcoxon's 2-sample and chi-square tests by using a Bonferroni-corrected α = 0.0025. Age (P < 0.0001), plasma cholesterol concentration (P = 0.0002), menopausal status (P < 0.0001), smoking status (P = 0.0007), and plasma retinol concentration (P < 0.0001) were significantly different between groups.
Mean ± SD (all such values).
Among postmenopausal women not missing data.
Energy-adjusted, except for alcohol.
Associations between the 3 previously identified SNPs (7, 8) and each plasma biomarker among all women, as determined by multivariate linear regression β-coefficients, are shown elsewhere (see Supplemental Table 3 under “Supplemental data” in the online issue). The rs6564851 G allele was significantly associated with higher β-carotene, α-carotene, and β-cryptoxanthin and with lower lutein/zeaxanthin (P = 5.2 × 10−26, 1.5 × 10−4, 2.2 × 10−4, and < 1.0 × 10−30, respectively); no association was observed with lycopene or retinol. The rs12934922 T allele was significantly associated with higher β-carotene, α-carotene, β-cryptoxanthin, and lutein/zeaxanthin (P = 1.1 × 10−12, 3.1 × 10−7, 2.4 × 10−3, and 5.9 × 10−6, respectively) and was selected for the β-carotene score. The SNP was not associated with lycopene or retinol. The rs7501331 T allele was significantly associated with lower β-carotene and α-carotene (P = 1.6 × 10−5 and 0.03, respectively); no associations were observed with the other biomarkers. Results for rs12934922 and rs7501331 were similar when restricted to genotyped data (see Supplemental Table 3 under “Supplemental data” in the online issue); results are shown only for rs6564851 genotyped data because the SNP was only imputed in 4 women with available GWAS data. When we limited the rs7501331 analysis to genotyped data among currently nonsmoking fasting controls with a BMI (in kg/m2) <30 who did not report use of multivitamins, β-carotene supplements, or vitamins A, C, or E to best approximate the inclusion criteria of the previous study (7), the T allele was nonsignificantly inversely associated with plasma β-carotene (n = 427, P = 0. 26; data not shown). A gene score composed of counts of T alleles in rs12934922 and rs7501331 was borderline-significantly positively associated with plasma β-carotene concentration in this group (n = 369, P = 0.07; data not shown).
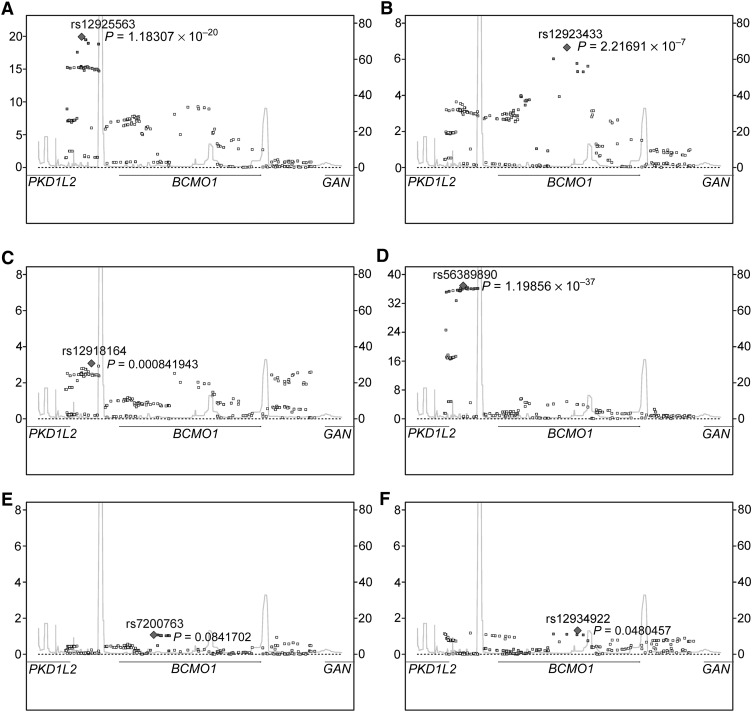
Regional association plots for each biomarker and 224 SNPs for women with genome-wide scans indicate that the most significant SNPs for β-carotene (rs12925563), β-cryptoxanthin (rs12918164), and lutein/zeaxanthin (rs56389890) were slightly upstream of BCMO1 near PKD1L2 (polycystic kidney disease 1-like 2) (Figure 1). The most significant SNPs for α-carotene (rs12923433) and retinol (rs12934922) were in the center of BCMO1.
FIGURE 1.
Regional (NCBI 36 chr16:79829797–79882248) association plots between SNPs in BCMO1 ± 20 kb and plasma carotenoid and retinol concentrations among women with genome-wide scans (n = 2323–2340). Plotted P values are from multivariate linear regression of each natural log–transformed plasma biomarker concentration on additively modeled SNPs adjusted for the following covariates: age, case-control status, cholesterol (mg/dL or missing indicator), BMI (kg/m2), smoking (never/former, current <15 cigarettes/d, current ≥15 cigarettes/d), menopausal status and PMH use (premenopausal; postmenopausal, no PMH; postmenopausal, PMH; unknown), alcohol intake (g/d), total fat intake (g/d), and dietary intake of the outcome of interest (μg/d for carotenoids or IU/d for retinol). Dietary retinol intake (IU/d) was also included in the models for α-carotene, β-carotene, and β-cryptoxanthin. A: β-carotene; B, α-carotene; C, β-cryptoxanthin; D, lutein/zeaxanthin; E, lycopene; F, retinol. Left y axis = observed (-logP); right y axis = recombination rate (centiMorgans/megabase); x axis = chromosome 16 position (human genome 18) (kb). chr, chromosome; NCBI, National Center for Biotechnology Information; PMH, postmenopausal hormone; SNPs, single nucleotide polymorphisms.
In the original stepwise selection from 225 SNPs for an association with plasma biomarker concentrations in the training data set, zero to 3 SNPs were selected for each biomarker (3 for β-carotene, α-carotene, and β-cryptoxanthin; 1 for lutein/zeaxanthin; and 0 for lycopene and retinol). rs28380273 was selected for lutein/zeaxanthin and also was the individual SNP most significantly associated with β-carotene (P = 6.3 × 10−21), despite not being selected by stepwise selection for the β-carotene gene score. We thus sought confirmation of these associations via genotyping. The SNP was subsequently removed from further analyses because of the 85% genotyping success rate and lack of Hardy-Weinberg equilibrium (P < 1 × 10−30). After repeating stepwise selection from the 224 remaining SNPs, SNP selection changed only for lutein/zeaxanthin, for which 2 other SNPs were selected.
Associations between the selected SNPs and respective biomarkers are shown in Table 2 for the training and testing subsets. Because no SNPs were selected for lycopene or retinol, these biomarkers are not included in Table 2. rs4889293 was selected for both α-carotene and β-cryptoxanthin with the G allele being associated with higher concentrations of both carotenoids. rs4889286 was selected for both β- and α-carotene, with the T allele being associated with higher concentrations of both carotenoids. rs4448930 was selected for β-carotene, α-carotene, and β-cryptoxanthin, with the C allele being associated with higher concentrations of all 3 carotenoids. Because of their selection in multiple gene scores, we genotyped rs4448930 and rs4889286 for confirmation. After the gene scores derived in the training subset from imputed data were applied to genotyped data in the testing subset, associations for rs4448930 were substantially attenuated. (Table 2). Because of this attenuation, and the moderate-to-high composite LD between the remaining selected SNPs and previously identified SNPs (see Supplemental Table 4 under “Supplemental data” in the online issue), we elected to exclude rs4448930 from the 3 gene scores for which it was selected.
TABLE 2.
Associations between selected SNPs in BCMO1 and plasma carotenoid concentrations among women with genome-wide scans by subset1
| Training subset (n = 1554–1556) |
Testing subset (n = 775–778) |
|||||||||
| Biomarker and SNP | MAF (allele)2 | Imputation3 r2 | Imputation4 | Multiple partial5 R2 | β (SE)6 | P value | Imputation | Multiple partial R2 | β (SE) | P value |
| % | % | |||||||||
| β-Carotene | ||||||||||
| rs4889286 | 0.50 (T) | 0.98 | 100 | 0.061 | 0.1762 (0.022) | 8.5 × 10−16 | 29 | 0.061 | 0.1748 (0.030) | 9.0 × 10−9 |
| rs12934922 | 0.44 (T) | 0.90 | 25 | 0.1377 (0.022) | 5.9 × 10−10 | 26 | 0.1412 (0.031) | 5.9 × 10−6 | ||
| rs4448930 | 0.15 (C) | 0.74 | 100 | 0.09747 (0.034) | 3.8 × 10−3 | 28 | 0.01753 (0.042) | 0.68 | ||
| α-Carotene | ||||||||||
| rs4889293 | 0.42 (G) | 0.85 | 73 | 0.027 | 0.1183 (0.022) | 8.7 × 10−8 | 71 | 0.012 | 0.06480 (0.032) | 0.04 |
| rs4448930 | 0.15 (C) | 0.74 | 100 | 0.09750 (0.032) | 2.3 × 10−3 | 28 | −0.006095 (0.041) | 0.88 | ||
| rs4889286 | 0.50 (T) | 0.98 | 100 | 0.06581 (0.020) | 1.3 × 10−3 | 29 | 0.06844 (0.029) | 0.02 | ||
| β-Cryptoxanthin | ||||||||||
| rs4448930 | 0.15 (C) | 0.74 | 100 | 0.019 | 0.09872 (0.027) | 2.2 × 10−4 | 28 | 0.008 | 0.02774 (0.035) | 0.43 |
| rs12918164 | 0.17 (A) | 0.83 | 100 | 0.07583 (0.024) | 1.9 × 10−3 | 100 | 0.05948 (0.035) | 0.09 | ||
| rs4889293 | 0.42 (G) | 0.85 | 73 | 0.06043 (0.018) | 1.0 × 10−3 | 71 | 0.04819 (0.027) | 0.07 | ||
| Lutein/zeaxanthin | ||||||||||
| rs56389940 | 0.36 (A) | 0.94 | 100 | 0.074 | 0.1636 (0.015) | 2.6 × 10−27 | 100 | 0.097 | 0.1792 (0.021) | 2.0 × 10−17 |
| rs10048138 | 0.14 (A) | 0.92 | 73 | 0.06580 (0.020) | 1.3 × 10−3 | 71 | 0.1334 (0.030) | 1.1 × 10−5 | ||
Multivariate linear regression of each natural log–transformed plasma biomarker concentration (μg/L) on additively modeled SNPs selected by stepwise selection by using the Schwartz Bayesian information criterion and the following covariates were forced into the model: age, case-control status, cholesterol (mg/dL, or missing indicator), BMI (kg/m2), smoking (never/former, current <15 cigarettes/d, or current ≥15 cigarettes/d), menopausal status and PMH use (premenopausal; postmenopausal, no PMH; postmenopausal, PMH; or unknown), alcohol intake (g/d), total fat intake (g/d), and dietary intake of the outcome of interest (μg/d for carotenoids or IU/d for retinol). Dietary retinol intake (IU/d) was also forced into the models for β-carotene, α-carotene, and β-cryptoxanthin. Results are shown for rs4889286 and rs4448930 original imputed values in the training subset on which selection was performed. Results in the testing subset include later genotyped values when available. MAF, minor allele frequency; PMH, postmenopausal hormone; SNP, single nucleotide polymorphism; SSE, sum of squared errors.
Weighted average MAF among all women with genome-wide scans.
Minimac imputation r2 among all women with genome-wide scans.
Percentage of women with imputed data.
Multiple partial R2: SSE for the covariate-only model − SSE for the model with SNPs and covariates divided by SSE for the covariate-only model.
β-Coefficient from the final selected model described above.
After finalizing the gene scores, we determined geometric mean plasma concentrations for each weighted gene score quintile among the testing subset (Table 3). The β-carotene–weighted gene score represented the largest difference across extreme quintiles (48%), and the α-carotene- and β-cryptoxanthin-weighted gene scores represented the smallest difference (15%). Results for the unweighted gene scores were similar to the results for the weighted gene scores (data not shown).
TABLE 3.
BCMO1-weighted gene score associations with plasma carotenoid concentrations among the testing subset of women with genome-wide scans (n = 775–778)1
| Geometric mean concentration by gene score quintile (95% CI) 2 |
||||||||
| Biomarker | 1 | 2 | 3 | 4 | 5 | β (SE)3 | P value | Multiple partial4R2 |
| μg/L | ||||||||
| β-Carotene5 | 194 (176, 214) | 197 (178, 218) | 218 (190, 250) | 244 (227, 262) | 288 (264, 314) | 1.076 (0.154) | 6.4 × 10−12 | 0.061 |
| α-Carotene6 | 57 (52, 63) | 57 (52, 62) | 54 (49, 60) | 61 (56, 66) | 66 (60, 71) | 0.7362 (0.250) | 3.3 × 10−3 | 0.011 |
| β-Cryptoxanthin7 | 68 (63, 73) | 69 (64, 75) | 74 (69, 80) | 70 (64, 75) | 78 (72, 84) | 0.8191 (0.354) | 0.02 | 0.007 |
| Lutein/zeaxanthin8 | 140 (133, 147) | 167 (157, 178) | 176 (166, 186) | 190 (180, 202) | 191 (181, 203) | 1.099 (0.126) | 1.8 × 10−17 | 0.092 |
PMH, postmenopausal hormone; SSE, sum of squared errors.
Covariate-adjusted geometric mean plasma biomarker concentrations determined by back-transforming the least-squares means from the linear regression model described below with gene scores modeled as quintile indicator variables.
β-Coefficient and SE from multivariate linear regression of each natural log–transformed plasma biomarker concentration (μg/L) on corresponding continuous weighted gene score were adjusted for age, case-control status, cholesterol (mg/dL, or missing indicator), BMI (kg/m2), smoking (never/former, current <15 cigarettes/d, or current ≥15 cigarettes/d), menopausal status and PMH use (premenopausal; postmenopausal, no PMH; postmenopausal, PMH; or unknown), alcohol intake (g/d), total fat intake (g/d), and dietary intake of the outcome of interest (μg/d for carotenoids or IU/d for retinol). β-Carotene, α-carotene, and β-cryptoxanthin were also adjusted for dietary retinol intake (IU/d).
Multiple partial R2: SSE for the covariate-only model − SSE for the model with gene score and covariates divided by SSE for the covariate-only model.
Weighted gene score = (0.1637 × rs4889286 T) + (0.1274 × rs12934922 T); weights derived from testing data set were 0.1744 and 0.1397, respectively.
Weighted gene score = (0.06530 × rs4889286 T) + (0.1075 × rs4889293 G); weights derived from testing data set were 0.06860 and 0.06538, respectively.
Weighted gene score = (0.07709 × rs12918164 A) + (0.04916 × rs4889293 G); weights derived from testing data set were 0.05722 and 0.04560, respectively.
Weighted gene score = (0.1636 × rs56389940 A) + (0.06580 × rs10048138 A); weights derived from testing data set were 0.1792 and 0.1334, respectively.
Spearman correlations between each weighted gene score and dietary intake of the corresponding carotenoid among women with genome-wide scans (n = 2344) ranged from −0.03 to 0.03 (P ≥ 0.17). We may thus assume the weighted gene scores are unassociated with dietary intake of other constituents of carotenoid-containing foods as well.
To compare the ability of dietary carotenoid intake compared with BCMO1 SNPs to explain variation in plasma carotenoid concentrations, we determined the multiple partial R2 for the dietary carotenoid variables adjusted for all covariates, including the relevant weighted gene score. The percentages of residual variation explained by dietary intake of the specific carotenoids in the testing data set were 9% for β-carotene, 8% for α-carotene, 8% for β-cryptoxanthin, and 6% for lutein/zeaxanthin.
Each weighted gene score for one carotenoid was significantly associated with plasma concentrations of other carotenoids among the testing subset (n = 775–780, data not shown). Whereas the β-carotene–weighted gene score was associated with 48% higher β-carotene across extreme quintiles (Table 3), it was also associated with 24% higher α-carotene (P-continuous score = 1.1 × 10−3) and 18% lower lutein/zeaxanthin (P = 6.2 × 10−7), explaining 1.4% and 3.3% of the residual variation, respectively. The score was borderline-significantly associated with 10% higher β-cryptoxanthin (P = 0.07) and was not associated with lycopene or retinol. The α-carotene–weighted gene score, associated with 15% higher α-carotene across extreme quintiles (Table 3), was also associated with 40% higher β-carotene (P = 8.2 × 10−9), 10% higher β-cryptoxanthin (P = 0.04), and 10% lower lutein/zeaxanthin (P = 0.01), explaining 4.3%, 0.6%, and 0.9% of the residual variation, respectively. The score was not associated with lycopene or retinol. Whereas the β-cryptoxanthin–weighted gene score was associated with 15% higher β-cryptoxanthin across extreme quintiles (Table 3), the score also was associated with 25% higher β-carotene (P = 3.3 × 10−5) and 6% lower retinol (P = 4.1 × 10−3), explaining 2.0% and 1.1% of the residual variation, respectively. The score was borderline-significantly associated with 3% higher α-carotene (P = 0.09) and was not associated with lutein/zeaxanthin or lycopene. The lutein/zeaxanthin-weighted gene score, associated with 36% higher lutein/zeaxanthin across extreme quintiles (Table 3), was also associated with 14% lower α-carotene (P = 7.7 × 10−3) and 28% lower β-carotene (P = 1.1 × 10−8), explaining 0.9% and 4.2% of the residual variation, respectively. The score was borderline-significantly associated with 7% lower β-cryptoxanthin (P = 0.06) and not associated with lycopene or retinol. Because the SNPs in the other gene scores included or were in composite LD with at least one of the SNPs in the β-carotene gene score, we examined the relation between rs4889286 and rs12934922 and plasma biomarker concentrations among all women with available genotyped data (see Supplemental Table 5 under “Supplemental data” in the online issue). Gene scores composed of these SNPs were significantly associated with all biomarkers other than lycopene (P = 0.20 and 0.18 for unweighted and weighted scores, respectively; data not shown). An increasing number of T alleles was associated with higher β-carotene, α-carotene, and β-cryptoxanthin and lower retinol. In comparison with rs12934922, lutein/zeaxanthin was strongly influenced by rs4889286, in which an increasing number of T alleles was associated with lower lutein/zeaxanthin.
The β-carotene–, α-carotene–, and β-cryptoxanthin–weighted gene scores did not appear to interact with dietary intake of their respective nutrients (Table 4). A significant (P = 0.04) weighted gene score × diet interaction was found for lutein/zeaxanthin, wherein the association between the weighted gene score and plasma lutein/zeaxanthin was weakest in women with the lowest dietary lutein/zeaxanthin intake. P values for the unweighted gene score × diet cross-product terms were 0.69 for β-carotene, 0.05 for α-carotene, 0.68 for β-cryptoxanthin, and 0.13 for lutein/zeaxanthin (data not shown).
TABLE 4.
BCMO1-weighted gene score associations with plasma carotenoid concentration stratified by quintile of dietary intake of the nutrient of interest among women with genome-wide scans (n = 465–468 per quintile)1
| Diet quintile 1 |
Diet quintile 2 |
Diet quintile 3 |
Diet quintile 4 |
Diet quintile 5 |
|||||||
| Biomarker | β (SE)2 | P value | β (SE)2 | P value | β (SE)2 | P value | β (SE)2 | P value | β (SE)2 | P value | P-interaction3 |
| β-Carotene4 | 1.275 (0.193) | 1.2 × 10−10 | 0.9422 (0.186) | 6.2 × 10−7 | 0.9096 (0.208) | 1.5 × 10−5 | 0.8386 (0.212) | 8.7 × 10−5 | 1.283 (0.219) | 8.8 × 10−9 | 0.77 |
| α-Carotene5 | 1.617 (0.316) | 4.7 × 10−7 | 0.7327 (0.299) | 0.01 | 0.8791 (0.316) | 5.7 × 10−3 | 0.1871 (0.324) | 0.56 | 0.9488 (0.360) | 8.7 × 10−3 | 0.26 |
| β-Cryptoxanthin6 | 1.327 (0.510) | 9.6 × 10−3 | 1.115 (0.477) | 0.02 | 0.8857 (0.443) | 0.05 | 0.0724 (0.451) | 0.87 | 1.324 (0.446) | 3.2 × 10−3 | 0.62 |
| Lutein/zeaxanthin7 | 0.6766 (0.156) | 1.7 × 10−5 | 1.287 (0.161) | 7.4 × 10−14 | 0.9160 (0.166) | 5.9 × 10−8 | 1.106 (0.166) | 8.9 × 10−11 | 1.188 (0.177) | 6.1 × 10−11 | 0.04 |
PMH, postmenopausal hormone.
β-Coefficient and SE from multivariate linear regression of each natural log–transformed plasma biomarker concentration (μg/L) on corresponding continuous weighted gene score adjusted for age, case-control status, cholesterol (mg/dL, or missing indicator), BMI (kg/m2), smoking (never/former, current <15 cigarettes/d, or current ≥15 cigarettes/d), menopausal status and PMH use (premenopausal; postmenopausal, no PMH; postmenopausal, PMH; or unknown), alcohol intake (g/d), total fat intake (g/d), and dietary intake of the outcome of interest (μg/d for carotenoids or IU/d for retinol). β-Carotene, α-carotene, and β-cryptoxanthin were also adjusted for dietary retinol intake (IU/d).
value for gene score × diet interaction cross-product term included in the multivariate linear regression models described above by using diet quintile medians for the cross-product and diet main effect.
Weighted gene score = (0.1637 × rs4889286 T) + (0.1274 × rs12934922 T).
Weighted gene score = (0.06530 × rs4889286 T) + (0.1075 × rs4889293 G).
Weighted gene score = (0.07709 × rs12918164 A) + (0.04916 × rs4889293 G).
Weighted gene score = (0.1636 × rs56389940 A) + (0.06580 × rs10048138 A).
To assess the influence on our results of subsequent disease diagnosed after blood collection, we ran multivariate linear regression models with weighted gene scores in the testing subset, excluding cases (n = 404–406; data not shown). The associations between the weighted gene scores and the respective biomarker concentrations were not materially different for β-carotene, β-cryptoxanthin, and lutein/zeaxanthin, although the association was no longer significant for β-cryptoxanthin. The association between the α-carotene–weighted gene score and plasma α-carotene was both attenuated and no longer significant.
DISCUSSION
Here, BCMO1 SNPs were associated with plasma carotenoid concentrations. Similar to the previous GWAS (8), the rs6564851 G allele was associated with higher β- and α-carotene and lower lutein/zeaxanthin. Rs4889286 or rs56389940, in composite LD with rs6564851 and in the same upstream area, was selected for the β-carotene, α-carotene, and lutein/zeaxanthin gene scores. SNPs upstream of BCMO1, rs7501331 + rs12934922, and rs7501331 alone were previously reported to reduce BCMO1 activity and were associated with fasting plasma β-carotene (7, 9). Here, the rs12934922 T allele was positively associated with plasma β-carotene, and either rs12934922 or rs4889293, in composite LD with rs12934922, was selected for the provitamin A carotene gene scores. The inverse association between the rs7501331 T allele and plasma β-carotene, however, was opposite from that expected based on the positive association reported previously (7). The magnitude of our inverse association was similar, although nonsignificant, when we limited our data to an approximation of the previous study's data, but other differences between study groups, such as age, remained.
The provitamin A carotenoid-weighted gene scores were significantly associated with their corresponding plasma concentrations in the testing subset. Human or chicken BCMO1 catalyzes central cleavage of carotenoids with ≥1 unsubstituted β-ionone ring, explaining its activity toward β-carotene, α-carotene, and β-cryptoxanthin (4, 6). Chicken BCMO1, the gene for which has 67% sequence identity with human BCMO1 (4), is catalytically more efficient with a higher retinoid conversion yield with β-carotene as substrate compared with α-carotene and β-cryptoxanthin (6). Thus, as observed, we expected a stronger association between BCMO1 variants and plasma β-carotene than α-carotene or β-cryptoxanthin. The lack of a major association with plasma retinol and the low correlation between the plasma carotenoids and retinol also was expected. A recent GWAS of circulating retinol did not detect any genome-wide significantly associated SNPs in or near BCMO1 (28). Circulating retinol concentrations are maintained homeostatically in the absence of critically low liver stores (29). All plasma retinol concentrations were >200 μg/L, implying sufficient vitamin A status (30) and thus limited interindividual variation. Regardless, the β-cryptoxanthin gene score was significantly inversely associated with plasma retinol in the testing subset, and the β-carotene gene score, when weighted for retinol, was significantly associated with plasma retinol in all women with genotyped data for the relevant SNPs. Liver retinol stores may be more affected by BCMO1 variation, as suggested by the reduced liver vitamin A concentrations in β-carotene–fed BCMO1 knockout mice (31, 32). Overall, our results support the hypothesis that BCMO1 variants affect provitamin A carotenoid conversion, presumably because of altered BCMO1 function (7, 9).
BCMO1 is not thought to catalyze lutein/zeaxanthin or lycopene cleavage because of the lack of unsubstituted β-ionone rings (4, 6). Our null lycopene results thus agree with biological expectation and provide further evidence that BCMO1 does not cleave lycopene. Our observed lutein/zeaxanthin associations, while confirming previous findings (8), remain biologically unsubstantiated. The associations may be due to an indirect effect through provitamin A carotenoids (8), because carotenoids interact at many stages of absorption and metabolism (33). BCMO1 SNPs resulting in higher absorption of intact β-carotene may lead to reduced plasma lutein/zeaxanthin. The area under the serum lutein-versus-time curve with oral lutein plus β-carotene is smaller than the area with lutein alone, which suggests that β-carotene can reduce lutein absorption (34). β-Carotene supplementation in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study in male smokers reduced serum lutein concentrations by 11% (35), but plasma lutein concentrations were not significantly different after β-carotene supplementation in mostly nonsmoking men in the Physicians Health Study (36). Alternatively, β,β-carotene-9′,10′-oxygenase (BCDO2), which catalyzes asymmetric cleavage of provitamin A and nonprovitamin A carotenoids, may be involved (37–39). Both BCMO1 and BCDO2 are expressed in several human tissues, including the small intestine epithelium and liver hepatocytes (40). BCMO1-deficient mice have elevated liver BCDO2 mRNA (41), so SNPs reducing BCMO1 function may indirectly alter BCDO2 expression. BCDO2’s apparent preferential cleavage of lutein and zeaxanthin over β-cryptoxanthin (37) may explain the stronger effect of this indirect pathway on lutein/zeaxanthin compared with the other carotenoids. Finally, BCMO1 may have as yet unknown direct activity on lutein/zeaxanthin.
The gene scores may be useful markers of carotenoid exposure in future studies. In an analysis of plasma carotenoids and breast cancer risk in the NHS, the multivariate-adjusted ORs for the comparison of the top with the bottom quintiles were 0.64 (0.47–0.88) for a 113-μg/L difference in α-carotene, 0.73 (0.53–1.02) for a 521-μg/L difference in β-carotene, and 0.74 (0.55–1.01) for a 148-μg/L difference in lutein/zeaxanthin (42). From these ORs and our observed genetic associations with the plasma carotenoids, the expected ORs of breast cancer for comparisons of extreme weighted gene score quintiles were 0.97 (α-carotene), 0.94 (β-carotene), and 0.90 (lutein/zeaxanthin). Although the expected ORs might be larger for the weighted gene scores because they presumably represent lifetime exposure rather than a single blood measurement, a large sample may be required when these scores are used in future studies. Some SNPs were selected as predictors of multiple carotenoids and/or were in composite LD with other selected SNPs, causing significant associations between gene scores for one carotenoid with concentrations of other carotenoids. Gene scores composed of rs4889286 and rs12934922 predicted plasma concentrations of all biomarkers other than lycopene. This pleiotropy may not only limit attribution of genetic associations to specific carotenoids but may also attenuate associations if both provitamin A carotenoids and lutein/zeaxanthin are similarly associated with disease risk, given their opposite associations with some BCMO1 SNPs. Whereas no SNPs were selected for retinol, pleiotropy also may exist for gene scores modeling differential converter phenotypes because they may be associated with site-specific retinol concentrations that are less regulated than plasma concentrations.
Exclusion of other genes was a limitation. We also did not adjust P values for multiple testing. Some of our nominally statistically significant results may be false positive; thus, results from the more exploratory analyses (eg, gene-diet interaction tests) should be interpreted with special care. However, our main findings (replication of the β-carotene and lutein/zeaxanthin gene scores) are quite robust, passing a conservative Bonferroni correction for the number of marker-biomarker tests conducted and surpassing a widely used genome-wide significance threshold (P < 5 × 10−8). The failure of the TaqMan genotyping assay for rs28380273 exemplifies a limitation of our reliance on mostly imputed genetic data. This failure may have been due to the apparently triallelic nature of rs28380273 that the other selected SNPs do not seem to share (43). The high LD between particular SNPs in this region combined with random variation also means that SNP selection is sensitive to the data set; other analysts may select different SNPs. Differences also may occur in non-European study populations given the variation in allele frequencies noted for some BCMO1 SNPs across ethnicities (7, 9), which limits this study's generalizability. Despite limitations, the gene scores may still be useful in epidemiologic analyses because they should be largely unconfounded and may be more readily available in large data sets than dietary intakes or plasma concentrations. The SNPs were selected by using a large data set with thorough covariates and were comprehensive across the entire BCMO1 region ± 20 kb.
Our results support an important role for BCMO1 variation on plasma carotenoid concentrations, particularly β-carotene and lutein/zeaxanthin. Genetic surrogates for 4 plasma carotenoids are now available for future studies of disease risk, which provides an additional avenue for research into the health effects of carotenoids.
Acknowledgments
We thank Hannia Campos, Hardeep Ranu, and Patrice Soule for laboratory assistance and Carolyn Guo for programming assistance. We also thank the participants and staff of the NHS for their valuable contributions and the cancer registries from following states for their help: Alabama, Arizona, Arkansas, California, Colorado, Connecticut, Delaware, Florida, Georgia, Idaho, Illinois, Indiana, Iowa, Kentucky, Louisiana, Maine, Maryland, Massachusetts, Michigan, Nebraska, New Hampshire, New Jersey, New York, North Carolina, North Dakota, Ohio, Oklahoma, Oregon, Pennsylvania, Rhode Island, South Carolina, Tennessee, Texas, Virginia, Washington, and Wyoming.
The authors’ responsibilities were as follows—SJH, AH, PK, BAR, and WCW: designed the research; AH, AHE, and PK: provided the essential materials; CC: performed the imputations; and SJH: analyzed the data, wrote the manuscript, and had primary responsibility for the final content. All authors read and approved the final manuscript and had no conflicts of interest to declare. Merck played no role in the design, implementation, analysis, or interpretation of the data for this study.
Footnotes
Abbreviations used: BCDO2, β,β-carotene-9′,10′-oxygenase; BCMO1, β-carotene 15,15′-monooxygenase 1; FFQ, food-frequency questionnaire; GWAS, genome-wide association study; LD, linkage disequilibrium; NCBI,National Center for Biotechnology Information; NHS, Nurses' Health Study; PMH, postmenopausal hormone; SNP, single nucleotide polymorphism; SSE, sum of squared errors.
REFERENCES
- 1.Krinsky NI. Antioxidant functions of carotenoids. Free Radic Biol Med 1989;7:617–35 [DOI] [PubMed] [Google Scholar]
- 2.Sumantran VN, Zhang R, Lee DS, Wicha MS. Differential regulation of apoptosis in normal versus transformed mammary epithelium by lutein and retinoic acid. Cancer Epidemiol Biomarkers Prev 2000;9:257–63 [PubMed] [Google Scholar]
- 3.Olson JA, Hayaishi O. The enzymatic cleavage of beta-carotene into vitamin A by soluble enzymes of rat liver and intestine. Proc Natl Acad Sci USA 1965;54:1364–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindqvist A, Andersson S. Biochemical properties of purified recombinant human β-carotene 15,15′-monooxygenase. J Biol Chem 2002;277:23942–8 [DOI] [PubMed] [Google Scholar]
- 5.Goodman DS, Huang H. Biosynthesis of vitamin A with rat intestinal enzymes. Science 1965;149:879–80 [DOI] [PubMed] [Google Scholar]
- 6.Kim Y-S, Oh D-K. Substrate specificity of a recombinant chicken beta-carotene 15,15′-monooxygenase that converts beta-carotene into retinal. Biotechnol Lett 2009;31:403–8 [DOI] [PubMed] [Google Scholar]
- 7.Leung WC, Hessel S, Meplan C, Flint J, Oberhauser V, Tourniaire F, Hesketh JE, von Lintig J, Lietz G. Two common single nucleotide polymorphisms in the gene encoding {beta}-carotene 15,15'-monoxygenase alter {beta}-carotene metabolism in female volunteers. FASEB J 2009;23:1041–53 [DOI] [PubMed] [Google Scholar]
- 8.Ferrucci L, Perry JRB, Matteini A, Perola M, Tanaka T, Silander K, Rice N, Melzer D, Murray A, Cluett C, et al. Common variation in the [beta]-carotene 15,15'-monooxygenase 1 gene affects circulating levels of carotenoids: a genome-wide association study. Am J Hum Genet 2009;84:123–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lietz G, Oxley A, Leung W, Hesketh J. Single nucleotide polymorphisms upstream from the β-carotene 15,15’-monoxygenase gene influence provitamin a conversion efficiency in female volunteers. J Nutr 2012;142:161S–5S [DOI] [PubMed] [Google Scholar]
- 10.Hu F, Wang Yi B, Zhang W, Liang J, Lin C, Li D, Wang F, Pang D, Zhao Y. Carotenoids and breast cancer risk: a meta-analysis and meta-regression. Breast Cancer Res Treat 2012;131:239–53 [DOI] [PubMed] [Google Scholar]
- 11.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22 [DOI] [PubMed] [Google Scholar]
- 12.Schatzkin A, Abnet CC, Cross AJ, Gunter M, Pfeiffer R, Gail M, Lim U, Davey-Smith G. Mendelian randomization: how it can—and cannot—help confirm causal relations between nutrition and cancer. Cancer Prev Res (Phila) 2009;2:104–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hankinson SE, Willett WC, Manson JE, Colditz GA, Hunter DJ, Spiegelman D, Barbieri RL, Speizer FE. Plasma sex steroid hormone levels and risk of breast cancer in postmenopausal women. J Natl Cancer Inst 1998;90:1292–9 [DOI] [PubMed] [Google Scholar]
- 14.Hunter DJ, Kraft P, Jacobs K, Cox D, Yeager M, Hankinson S, Wacholder S, Zhaoming W, Welch R, Hutchinson A, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 2007;39:870–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvini S, Hunter DJ, Sampson L, Stampfer MJ, Colditz GA, Rosner B, Willett WC. Food-based validation of a dietary questionnaire: the effects of week-to-week variation in food consumption. Int J Epidemiol 1989;18:858–67 [DOI] [PubMed] [Google Scholar]
- 16.Willett WC, Sampson L, Stampfer M, Rosner B, Bain C, Witschi J, Hennekens C, Speizer F. Reproducibility and validity of a semiquantitative food frequency questionnaire. Am J Epidemiol 1985;122:51–65 [DOI] [PubMed] [Google Scholar]
- 17.Willett WC, Sampson L, Browne M, Stampfer M, Rosner B, Hennekens C, Speizer F. The use of a self-administered questionnaire to assess diet four years in the past. Am J Epidemiol 1988;127:188–99 [DOI] [PubMed] [Google Scholar]
- 18.Michaud DS, Giovannucci EL, Ascherio A, Rimm EB, Forman MR, Sampson L, Willett WC. Associations of plasma carotenoid concentrations and dietary intake of specific carotenoids in samples of two prospective cohort studies using a new carotenoid database. Cancer Epidemiol Biomarkers Prev 1998;7:283–90 [PubMed] [Google Scholar]
- 19.Willett W. Nutritional epidemiology. 2nd ed. New York, NY: Oxford University Press, Inc, 1998 [Google Scholar]
- 20.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 1999;14:143–9 [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010;34:816–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet 2009;10:387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Sohemy A, Baylin A, Kabagambe E, Ascherio A, Spiegelman D, Campos H. Individual carotenoid concentrations in adipose tissue and plasma as biomarkers of dietary intake. Am J Clin Nutr 2002;76:172–9 [DOI] [PubMed] [Google Scholar]
- 24.Rosner B, Cook N, Portman R, Daniels S, Falkner B. Determination of blood pressure percentiles in normal-weight children: Some methodologic issues. Am J Epidemiol 2008;167:653–66 [DOI] [PubMed] [Google Scholar]
- 25.Allain CC, Poon L, Chan C, Richmon W, Fu P. Enzymatic determination of total serum cholesterol. Clin Chem 1974;20:470–5 [PubMed] [Google Scholar]
- 26.Rosner B. Percentage points for a generalized ESD many-outlier procedure. Technometrics 1983;25:165–72 [Google Scholar]
- 27.Johnson AD, Handsaker R, Pulit S, Nizzari M, O'Donnell C, de Bakker P. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondul AM, Yu K, Wheeler W, Zhang H, Weinstein SJ, Major JM, Cornelis MC, Männistö S, Hazra A, Hsing AW, et al. Genome-wide association study of circulating retinol levels. Hum Mol Genet 2011;20:4724–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanumihardjo SA. Assessing vitamin a status: past, present and future. J Nutr 2004;134:290S–3S [DOI] [PubMed] [Google Scholar]
- 30.Sommer A, Davidson FR. Assessment and control of vitamin a deficiency: the Annecy Accords. J Nutr 2002;132:2845S–50S [DOI] [PubMed] [Google Scholar]
- 31.Lindshield BL, King JL, Wyss A, Goralczyk R, Lu C-H, Ford NA, Erdman JW. Lycopene biodistribution is altered in 15,15'-carotenoid monooxygenase knockout mice. J Nutr 2008;138:2367–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hessel S, Eichinger A, Isken A, Amengual J, Hunzelmann S, Hoeller U, Elste V, Hunziker W, Goralczyk R, Oberhauser V, et al. CMO1 deficiency abolishes vitamin a production from β-carotene and alters lipid metabolism in mice. J Biol Chem 2007;282:33553–61 [DOI] [PubMed] [Google Scholar]
- 33.van den Berg H. Carotenoid interactions. Nutr Rev 1999;57:1–10 [DOI] [PubMed] [Google Scholar]
- 34.Kostic D, White W, Olson J. Intestinal absorption, serum clearance, and interactions between lutein and beta-carotene when administered to human adults in separate or combined oral doses. Am J Clin Nutr 1995;62:604–10 [DOI] [PubMed] [Google Scholar]
- 35.Albanes D, Virtamo J, Taylor P, Rautalahti M, Pietinen P, Heinonen O. Effects of supplemental beta-carotene, cigarette smoking, and alcohol consumption on serum carotenoids in the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study. Am J Clin Nutr 1997;66:366–72 [Published erratum appears in Am J Clin Nutr 1997 Dec;66(6):1491.] [DOI] [PubMed] [Google Scholar]
- 36.Fotouhi N, Meydani M, Santos M, Meydani S, Hennekens C, Gaziano J. Carotenoid and tocopherol concentrations in plasma, peripheral blood mononuclear cells, and red blood cells after long-term beta-carotene supplementation in men. Am J Clin Nutr 1996;63:553–8 [DOI] [PubMed] [Google Scholar]
- 37.Mein JR, Dolnikowski GG, Ernst H, Russell RM, Wang X-D. Enzymatic formation of apo-carotenoids from the xanthophyll carotenoids lutein, zeaxanthin and [beta]-cryptoxanthin by ferret carotene-9',10'-monooxygenase. Arch Biochem Biophys 2011;506:109–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem 2001;276:14110–6 [DOI] [PubMed] [Google Scholar]
- 39.Amengual J, Lobo GP, Golczak M, Li HNM, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J 2011;25:948–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindqvist A, He Y-G, Andersson S. Cell type-specific expression of β-carotene 9',10'-monooxygenase in human tissues. J Histochem Cytochem 2005;53:1403–12 [DOI] [PubMed] [Google Scholar]
- 41.Shmarakov I, Fleshman MK, D'Ambrosio DN, Piantedosi R, Riedl KM, Schwartz SJ, Curley RW, Jr, von Lintig J, Rubin LP, Harrison EH, et al. Hepatic stellate cells are an important cellular site for [beta]-carotene conversion to retinoid. Arch Biochem Biophys 2010;504:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamimi RM, Hankinson SE, Campos H, Spiegelman D, Zhang S, Colditz GA, Willett WC, Hunter DJ. Plasma carotenoids, retinol, and tocopherols and risk of breast cancer. Am J Epidemiol 2005;161:153–60 [DOI] [PubMed] [Google Scholar]
- 43. Database of single nucleotide polymorphisms. Bethesda, MD: National Center for Biotechnology Information, National Library of Medicine. dbSNP accession:{rs10048138, rs12918164, rs12934922, rs28380273, rs34500060, rs4448930, rs4889286, rs4889293, rs56389940}. Available from: http://www.ncbi.nlm.nih.gov/SNP/ (cited 22 October 2011)