The crystal structure of full-length HypF from T. kodakarenesis has been determined.
Keywords: HypF, [NiFe]-hydrogenase maturation proteins, Thermococcus kodakarenesis
Abstract
HypF is involved in the biosynthesis of the CN ligand of the NiFe(CN)2CO centre of [NiFe]-hydrogenases. Here, the full-length structure of HypF from Thermococcus kodakarenesis is reported at 4.5 Å resolution. The N-terminal acylphosphatase-like (ACP) domain interacts with the zinc-finger domain with some flexibility in its relative position. Molecular-surface analysis shows that a deep pocket formed between the ACP and zinc-finger domains is highly conserved and has positive potential. These results suggest that the positively charged pocket identified is involved in the hydrolysis of carbamoyl phosphate and the formation of a carbamoyl intermediate.
1. Introduction
Hydrogenases catalyze the reversible oxidation of hydrogen and play an important role in energy metabolism and regulation of proton concentration. Hydrogenases are classified into three types according to the metals in the reaction centre: [NiFe]-hydrogenases, [FeFe]-hydrogenases and [Fe]-hydrogenases (Vignais & Billoud, 2007 ▶; Thauer et al., 2010 ▶). Studies of hydrogenases are expected to have applications in the biological production of hydrogen and in the development of fuel cells (Kruse & Hankamer, 2010 ▶; Winkler et al., 2011 ▶).
The core component of [NiFe]-hydrogenases consists of a large subunit and a small subunit. The reaction centre in the large subunit contains an [NiFe]-cluster composed of an Ni atom and an Fe atom, which are bound to the conserved cysteine residues of the large subunit. The Fe atom is coordinated by two CN ligands and one CO ligand (Volbeda & Fontecilla-Camps, 2003 ▶; Fontecilla-Camps et al., 2007 ▶). The biosynthesis of the [NiFe]-cluster is catalyzed by six Hyp proteins: HypA, HypB, HypC, HypD, HypE and HypF (Böck et al., 2006 ▶). HypA and HypB are involved in insertion of the Ni atom (Kaluarachchi et al., 2010 ▶). HypC and HypD are involved in insertion of the Fe atom. HypE and HypF are involved in the synthesis of the CN ligand. Firstly, HypF catalyzes the transfer of the carbamoyl moiety of carbamoylphosphate to the C-terminal cysteine residue of HypE. HypE then catalyzes an ATP-dependent dehydration to produce a thiocyanate group (Blokesch, Paschos et al., 2004 ▶; Reissmann et al., 2003 ▶). The cyano group is transferred to the Fe atom, and the Fe ligand is inserted into the hydrogenase large subunit by HypC and HypD (Blokesch, Albracht et al., 2004 ▶). It is thought that the CO ligand is also inserted, but the biological process for the CO ligand is still unclear (Bürstel et al., 2011 ▶). HypA and HypB then insert the Ni atom into the hydrogenase large subunit. Finally, the C-terminus of the hydrogenase large subunit is cleaved by a specific protease and maturation is completed (Theodoratou et al., 2005 ▶).
The largest Hyp protein, HypF (∼86 kDa), consists of an N-terminal acylphosphatase domain (ACP), a zinc-finger domain, a middle domain and a C-terminal domain (Paschos et al., 2002 ▶). The N-terminal ACP domain contains the acylphosphatase motif. The ACP domain is thought to be involved in the hydrolysis of carbamoylphosphate and the formation of the intermediate. The zinc-finger domain contains two zinc-finger motifs (CXXCX 18CXXC) carrying zinc ions. Its function remains unclear. Both the middle and the C-terminal domains contain nucleotide-binding sites. The two nucleotide-binding sites are assumed to catalyze the carbamoylation of the C-terminal cysteine of HypE (Paschos et al., 2002 ▶; Petkun et al., 2011 ▶).
While structures of all of the Hyp proteins except HypF have been determined from several organisms (Watanabe et al., 2007 ▶, 2009 ▶; Xia et al., 2009 ▶; Gasper et al., 2006 ▶, Wang et al., 2007 ▶; Rangarajan et al., 2008 ▶; Shomura et al., 2007 ▶), only the crystal structure of the ACP domain in HypF was initially reported (Rosano et al., 2002 ▶). Crystal structures of Escherichia coli HypF lacking the ACP domain [EcHypF(92–750)] with or without nucleotides have been reported, revealing the formation of a carbamoyl adenylate intermediate (Petkun et al., 2011 ▶). Moreover, crystal structures of the Caldanaerobacter subterranus full-length HypF (CsHypF) and HypE–HypF complex have been reported very recently (Shomura & Higuchi, 2012 ▶). The structure of the HypE–HypF complex suggests that the S-carbamoylation of the C-terminal cysteine of HypE is catalyzed by the C-terminal domain of HypF.
Here, we report the crystal structure of full-length HypF from Thermococcus kodakarenesis KOD1 (TkHypF) at 4.5 Å resolution, which clarifies the relative position of each domain. Molecular-surface analysis combined with structural comparison provides insights into the molecular mechanism of HypF.
2. Materials and methods
2.1. Cloning, expression and purification
T. kodakarensis KOD1 was the source of the genomic DNA. E. coli DH5α and plasmid pUC18 were used for gene cloning. After confirming the sequences of the DNA fragments, they were inserted into the NdeI/EcoRI sites of pET21a(+). The HypF gene was amplified from the genomic DNA of T. kodakarensis. The primer set used was HypF5 (5′-GGGTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGAAGGCCTATCAATTCACG-3′) and HypF3 (5′-GCGGAATTCACTAGTTTACAGCATCAGATCCTCC-3′). After introduction into E. coli Rosetta2 (DE3) pLysS cells, gene expression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside at the late exponential growth phase, with further incubation for 24 h at 293 K. After induction of gene expression, the cells were washed with 50 mM Tris–HCl pH 8.5, 1 mM DTT and resuspended in the same buffer. Cells were sonicated on ice and the supernatant after centrifugation (34 600g, 30 min at 277 K) was subjected to heat treatment at 348 K for 15 min. After centrifugation (34 600g, 30 min at 277 K), the supernatant was applied onto an anion-exchange column (HiTrap Q HP, GE Healthcare, Little Chalfont, England) equilibrated with 50 mM HEPES–NaOH pH 7.0, 1 mM DTT. Protein was eluted with a linear gradient of NaCl from 0.3 to 1.0 M. Ammonium sulfate was then added to the sample to 0.5 M. The sample was applied onto a hydrophobic interaction column (Resource Phe, GE Healthcare). Proteins were eluted with a linear gradient of ammonium sulfate from 0.5 to 0 M. The sample was then applied onto a gel-filtration column (Superdex 200 10/300 GL, GE Healthcare) equilibrated with 50 mM Tris–HCl pH 8.0, 150 mM NaCl and 1 mM DTT or TCEP, where TCEP was used as a matter of convenience.
2.2. Crystallization
An initial search for crystallization conditions was performed using the following screening kits: Crystal Screen, Crystal Screen 2, Index, Crystal Screen Cryo, PEG/Ion, Natrix, MembFac, Grid Screen (Ammonium Sulfate, Sodium Chloride), SaltRx (Hampton Research, Aliso Viejo, California, USA), Wizard I and II, Cryo I and II and Precipitant Synergy (Primary 64, 33% of Primary 64, 67% of Primary 64) (Emerald BioSystems, Bedford, Massachusetts, USA). Crystallization was performed by the sitting-drop vapour-diffusion method at 293 K using 96-well Intelli-Plates (Hampton Research) and a Hydra II Plus One system (Matrix Technologies Corporation, Hudson, New Hampshire, USA). For the Grid Screen kits, crystallization was performed manually using CrystalClear D Strips (Hampton Research). The protein sample for crystallization was concentrated to 15–27 mg ml−1 in 20 mM Tris–HCl pH 8.0, 1 mM DTT. Initial crystals were obtained using Grid Screen Sodium Chloride condition D6 (0.1 M Bicine pH 9.0, 4.0 M NaCl). The crystallization condition was optimized by varying the pH and the concentration of NaCl.
2.3. Incorporation of selenomethionine
Selenomethionine-labelled HypF was expressed in M9 medium (Nakamura & Ishikawa, 2008 ▶). Cells were cutivated at 310 K until the optical density at 600 nm reached 0.3. Lysine, phenylalanine and threonine at 100 mg l−1, leucine, isoleucine and valine at 50 mg l−1 and selenomethionine (SeMet) at 60 mg l−1 were then added. Subsequent expression, purification and crystallization were carried out in the same way as for native HypF.
2.4. Data collection and structure determination
X-ray diffraction experiments were performed on beamlines BL41XU at SPring-8 and AR-NE3A at Photon Factory. The drops were substituted by cryoprotectant solution [0.1 M Tris–HCl pH 8.6, 3.7 M NaCl, 10%(v/v) ethylene glycol]. Crystals were then mounted in a nylon loop (Hampton Research) and flash-cooled in a nitrogen stream at 100 K. In order to irradiate with a beam perpendicular to the c axis, we used an extended-arc goniometer. Diffraction data were collected with an MX225HE detector (Rayonix, Evanston, Illinois, USA) using an oscillation angle of 0.5°, an exposure time of 0.5 s per frame and a wavelength of 0.97600 Å. The data sets were processed using the HKL-2000 package (Otwinowski & Minor, 1997 ▶). Data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection and refinement statistics for HypF.
Values in parentheses are for the highest resolution shell.
| Native | SeMet-labelled | |
|---|---|---|
| Data collection | ||
| Space group | P6522 | P6522 |
| Unit-cell parameters | a = 265.8, b = 265.8, c = 693.7 | a = 265.8, b = 265.8, c = 693.7 |
| Resolution (Å) | 50–4.50 (4.66–4.50) | 50–4.70 (4.87–4.70) |
| R merge † (%) | 17.4 (68.9) | 19.6 (75.0) |
| 〈I/σ(I)〉 | 12.6 (3.4) | 21.7 (5.9) |
| Completeness (%) | 99.9 (99.8) | 99.9 (99.9) |
| Multiplicity | 9.3 (9.5) | 27.6 (28.3) |
| Refinement | ||
| Resolution (Å) | 50–4.50 | |
| No. of reflections | 86501 | |
| R work ‡/R free § (%) | 28.2/30.3 | |
| Ramachandran plot (%) | ||
| Residues in preferred regions | 77.8 | |
| Residues in allowed regions | 19.4 | |
| Outliers | 2.8 | |
R
merge = 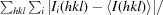
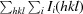 , where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
, where I
i(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
R
work = 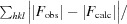
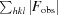 , where F
calc and F
obs are the calculated and the observed structure factors, respectively.
, where F
calc and F
obs are the calculated and the observed structure factors, respectively.
R free is the same as R work but calculated for a randomly chosen 5% of reflections that were omitted from the refinement.
The structure was determined by a combined molecular-replacement (MR) and single-wavelength anomalous dispersion (SAD) method. The initial phases were determined by MR using MOLREP (Vagin & Teplyakov, 2010 ▶). Search models were created using CHAINSAW in CCP4 (Winn et al., 2011 ▶) using the structure of EcHypF(92–750) (PDB entry 3tsp; Petkun et al., 2011 ▶) as a model. Se sites were found using an anomalous difference Fourier map calculated from the MR solution. Experimental phases were determined by the SAD method using Phaser (Read & McCoy, 2011 ▶; McCoy et al., 2007 ▶) and were further improved by DM (Cowtan, 1994 ▶). The structure model was automatically built by Buccaneer (Cowtan, 2006 ▶) using the structures of EcHypF(92–750) and EcHypF acylphosphatase-like domain (PDB entry 1gxu; Rosano et al., 2002 ▶) as initial templates. Manual model building was performed using Coot (Emsley & Cowtan, 2004 ▶). The structure was refined with CNS (Brünger et al., 1998 ▶) and was validated with MolProbity (Chen et al., 2010 ▶). Coordinates and structure factors have been deposited in the Protein Data Bank with accession code 4g9i.
3. Results and discussion
3.1. Crystallization, data collection and structure determination
Recombinant HypF from T. kodakarensis (772 amino acids, 86.7 kDa) was overexpressed in E. coli and purified by anion-exchange, hydrophobic-interaction and size-exclusion column chromatography. Crystals of HypF grew in two weeks using a reservoir solution consisting of 0.1 M Tris–HCl pH 8.6, 3.6–3.7 M NaCl. The crystals grew to typical dimensions of 0.2 × 0.2 × 0.02 mm. SeMet-labelled HypF was purified and crystallized using the same conditions. The largest crystal of SeMet-labelled HypF grew to dimensions of 0.7 × 0.7 × 0.03 mm.
The hexagonal crystal of HypF belonged to space group P6522, with unit-cell parameters a = b = 265.8, c = 693.7 Å. These crystals diffracted X-rays to beyond 4.5 Å resolution, but the long c axis caused significant overlapping of diffraction spots. In order to avoid overlapping spots, we used an extended-arc goniometer and rotated the crystals along the long c axis during data collection. Complete X-ray diffraction data for native and SAD phasing were successfully collected to 4.5 Å resolution from the largest crystal of SeMet-labelled HypF.
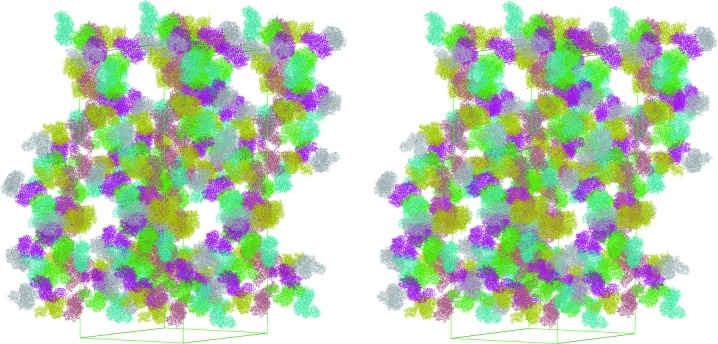
The structure of full-length HypF was determined by a combined MR and SAD method at 4.5 Å resolution. Despite the low resolution, an electron-density map calculated using the density-modified SAD phases was interpretable for most of the protein. The estimated Matthews coefficient (V M) indicated that the asymmetric unit contained more than 12 HypF molecules (V M of 3.4 Å3 Da−1, with a solvent content of 63.8%). However, the MR solution and electron-density map revealed that there were only six molecules in the asymmetric unit. Although the corresponding calculated Matthews coefficient (V M of 6.8 Å3 Da−1; 81.9% solvent content) is unusual, the tight crystal packing supports the presence of only six HypF molecules in the crystallographic asymmetric unit (Fig. 1 ▶).
Figure 1.
Stereoview of the crystal packing of HypF. Independent molecules in the asymmetric units are shown in different colours.
3.2. Description of the structure
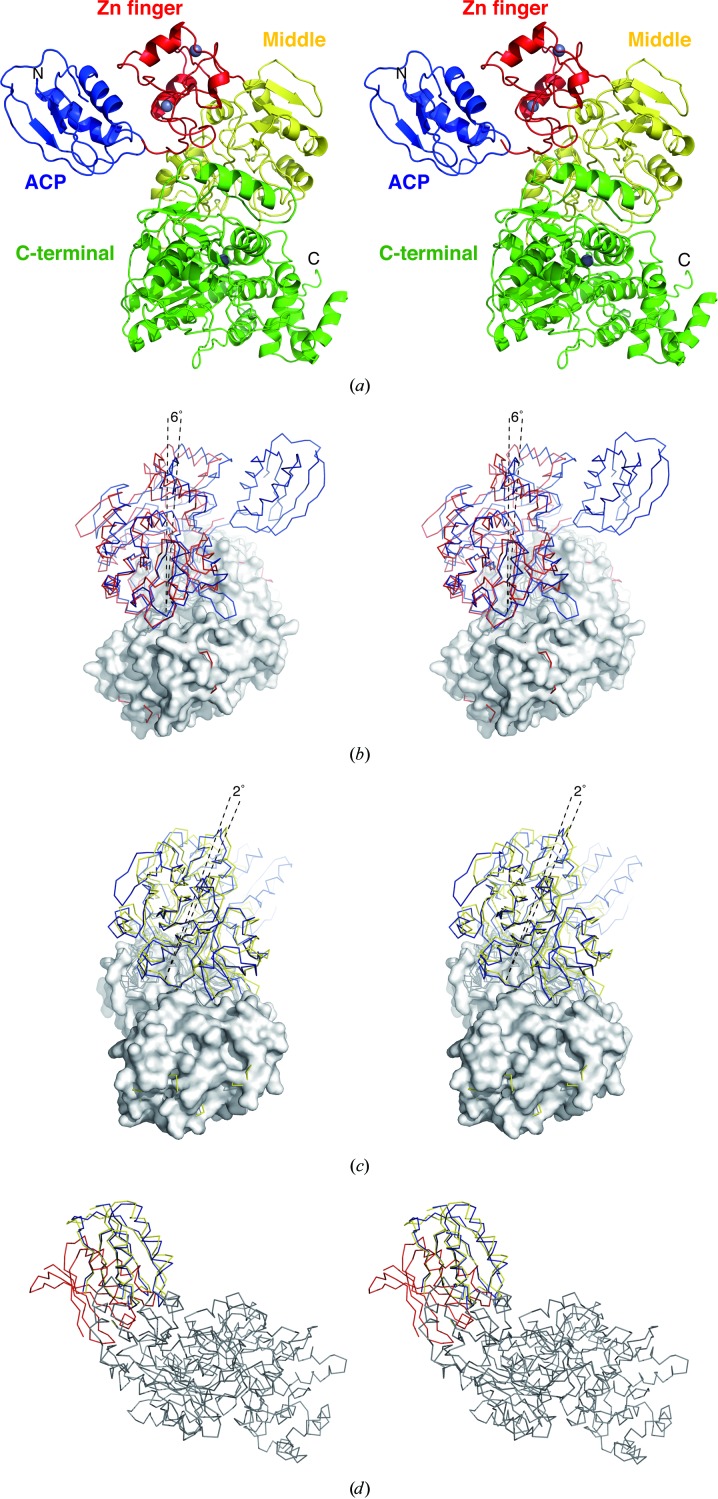
The density-modified SAD electron-density map clearly showed the overall architecture of full-length HypF, which consists of ACP, zinc-finger, middle and C-terminal domains. The ACP domain is located adjacent to the zinc-finger domain (Fig. 2 ▶ a). The two α-helices of the ACP domain face the zinc-finger domain. Compared with the structure of EcHypF, the three N-terminal domains of TkHypF are rotated by about 6° and closed with respect to the C-terminal domain (Fig. 2 ▶ b). Those of CsHypF are further closed, with a rotation of about 2° compared with TkHypF (Fig. 2 ▶ c). These observations suggest that HypF undergoes an open–closed motion during ATP hydrolysis.
Figure 2.
Structure of full-length HypF. (a) Stereoview of the overall structure of full-length HypF shown as a ribbon representation. The ACP, zinc-finger, middle and C-terminal domains are coloured blue, red, yellow and green, respectively. Zn atoms are shown as grey spheres. (b) Stereoview of a comparison of TkHypF (blue) with EcHypF(92–750) (red). The C-terminal domains of the two structures are superimposed on each other and are shown in surface representation. (c) Stereoview of a comparison of TkHypF (blue) with CsHypF (yellow). The C-terminal domains of the two structures are superimposed on each other. (d) Stereoview of a superposition of three of the six TkHypF molecules in the asymmetric unit. The ACP domains are coloured blue, red and yellow, respectively.
A comparison of the six independent HypF molecules in the asymmetric unit shows that relative positions of the ACP domains are different (Fig. 2 ▶ d). In the three of six HypF molecules in the asymmetric unit the electron density for the ACP domain is not well ordered. The linker between the ACP and zinc-finger domains is also disordered. These observations indicate that the ACP domain of HypF is flexible.
An anomalous difference Fourier map revealed three strong anomalous peaks apart from those from the Se atoms in the HypF monomer structure. Two of the three peaks are found in the two zinc-finger motifs and one is in the C-terminal domain (Fig. 2 ▶ a). The position of the third peak is at the same position as the Zn ion in the C-terminal domain of EcHypF. Therefore, these peaks probably correspond to anomalous signals from Zn atoms. The third Zn ion seems to be a common feature of HypF proteins.
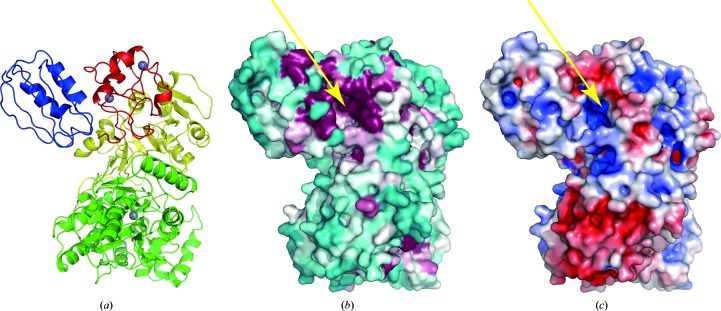
3.3. Conserved positively-charged pocket and mechanistic implications
Mapping of conserved residues on the molecular surface revealed three highly conserved regions in HypF (Fig. 3 ▶). As expected, the residues around the two nucleotide-binding sites in the middle and the C-terminal domains are well conserved. The other significant conserved region is found in a deep pocket formed between the ACP and the zinc-finger domains (Fig. 3 ▶ b). This pocket is highly positively charged (Fig. 3 ▶ c). In the CsHypF structure a phosphate ion is bound in the pocket (Shomura & Higuchi, 2012 ▶). The residues located in the conserved pocket include motifs that are conserved in the acylphosphatase family (Rosano et al., 2002 ▶). HypF mutants that have impaired ATP/ADP-binding regions in the middle and C-terminal domains showed carbamoylphosphate phosphatase activity (Petkun et al., 2011 ▶). These results suggested that the conserved positively-charged pocket catalyzes the hydrolysis of carbamoylphosphate to carbamate. The expected carbamate substrate can be delivered to the nucleotide-binding site in the middle domain through a tunnel from the zinc-finger domain to the middle domain (Petkun et al., 2011 ▶; Shomura & Higuchi, 2012 ▶). On the other hand, the fact that the size of the cavity is sufficient to accommodate ATP/ADP raises the possibility that the ACP domain is also involved in the formation of a carbamoyl intermediate such as carbamoyl adenylate. The flexibility of the ACP domain may regulate substrate recognition of HypF.
Figure 3.
Molecular-surface properties of HypF. (a) Ribbon representations of the molecule as a reference. (b) Conserved regions of HypF are coloured from burgundy (highly conserved) to cyan (variable). The sequence-conservation scores were calculated with ConSurf (Landau et al., 2005 ▶). (c) Electrostatic surface-potential representation of TkHypF calculated using APBS (Baker et al., 2001 ▶). Orange arrows indicate the conserved positively-charged pocket of HypF.
Supplementary Material
PDB reference: HypF, 4g9i
Acknowledgments
We thank the beamline scientists at SPring-8 and the Photon Factory for their help with X-ray data collection. This work was supported by Grants-in-Aid for Scientific Research (A) (20247009 and 23247014 to KM) and for Creative Scientific Research (18GS0421 to TI) from the Japan Society for the Promotion of Science and by a grant from the National Project on Protein Structural and Functional Analysis (to KM and TI) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J. & McCammon, J. A. (2001). Proc. Natl Acad. Sci. USA, 98, 10037–10041. [DOI] [PMC free article] [PubMed]
- Blokesch, M., Albracht, S. P., Matzanke, B. F., Drapal, N. M., Jacobi, A. & Böck, A. (2004). J. Mol. Biol. 344, 155–167. [DOI] [PubMed]
- Blokesch, M., Paschos, A., Bauer, A., Reissmann, S., Drapal, N. & Böck, A. (2004). Eur. J. Biochem. 271, 3428–3436. [DOI] [PubMed]
- Böck, A., King, P. W., Blokesch, M. & Posewitz, M. C. (2006). Adv. Microb. Physiol. 51, 1–71. [DOI] [PubMed]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed]
- Bürstel, I., Hummel, P., Siebert, E., Wisitruangsakul, N., Zebger, I., Friedrich, B. & Lenz, O. (2011). J. Biol. Chem. 286, 44937–44944. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cowtan, K. (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 31, 34–38.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fontecilla-Camps, J. C., Volbeda, A., Cavazza, C. & Nicolet, Y. (2007). Chem. Rev. 107, 4273–4303. [DOI] [PubMed]
- Gasper, R., Scrima, A. & Wittinghofer, A. (2006). J. Biol. Chem. 281, 27492–27502. [DOI] [PubMed]
- Kaluarachchi, H., Chan Chung, K. C. & Zamble, D. B. (2010). Nat. Prod. Rep. 27, 681–694. [DOI] [PubMed]
- Kruse, O. & Hankamer, B. (2010). Curr. Opin. Biotechnol. 21, 238–243. [DOI] [PubMed]
- Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T. & Ben-Tal, N. (2005). Nucleic Acids Res. 33, W299–W302. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Nakamura, T. & Ishikawa, K. (2008). PSSJ Arch. 1, e45.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Paschos, A., Bauer, A., Zimmermann, A., Zehelein, E. & Böck, A. (2002). J. Biol. Chem. 277, 49945–49951. [DOI] [PubMed]
- Petkun, S., Shi, R., Li, Y., Asinas, A., Munger, C., Zhang, L., Waclawek, M., Soboh, B., Sawers, R. G. & Cygler, M. (2011). Structure, 19, 1773–1783. [DOI] [PubMed]
- Rangarajan, E. S., Asinas, A., Proteau, A., Munger, C., Baardsnes, J., Iannuzzi, P., Matte, A. & Cygler, M. (2008). J. Bacteriol. 190, 1447–1458. [DOI] [PMC free article] [PubMed]
- Read, R. J. & McCoy, A. J. (2011). Acta Cryst. D67, 338–344. [DOI] [PMC free article] [PubMed]
- Reissmann, S., Hochleitner, E., Wang, H., Paschos, A., Lottspeich, F., Glass, R. S. & Böck, A. (2003). Science, 299, 1067–1070. [DOI] [PubMed]
- Rosano, C., Zuccotti, S., Bucciantini, M., Stefani, M., Ramponi, G. & Bolognesi, M. (2002). J. Mol. Biol. 321, 785–796. [DOI] [PubMed]
- Shomura, Y. & Higuchi, Y. (2012). J. Biol. Chem. 287, 28409–28419. [DOI] [PMC free article] [PubMed]
- Shomura, Y., Komori, H., Miyabe, N., Tomiyama, M., Shibata, N. & Higuchi, Y. (2007). J. Mol. Biol. 372, 1045–1054. [DOI] [PubMed]
- Thauer, R. K., Kaster, A. K., Goenrich, M., Schick, M., Hiromoto, T. & Shima, S. (2010). Annu. Rev. Biochem. 79, 507–536. [DOI] [PubMed]
- Theodoratou, E., Huber, R. & Böck, A. (2005). Biochem. Soc. Trans. 33, 108–111. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vignais, P. M. & Billoud, B. (2007). Chem. Rev. 107, 4206–4272. [DOI] [PubMed]
- Volbeda, A. & Fontecilla-Camps, J. C. (2003). Dalton Trans., pp. 4030–4038. [DOI] [PubMed]
- Wang, L., Xia, B. & Jin, C. (2007). Biochem. Biophys. Res. Commun. 361, 665–669. [DOI] [PubMed]
- Watanabe, S., Arai, T., Matsumi, R., Atomi, H., Imanaka, T. & Miki, K. (2009). J. Mol. Biol. 394, 448–459. [DOI] [PubMed]
- Watanabe, S., Matsumi, R., Arai, T., Atomi, H., Imanaka, T. & Miki, K. (2007). Mol. Cell, 27, 29–40. [DOI] [PubMed]
- Winkler, M., Kawelke, S. & Happe, T. (2011). Bioresour. Technol. 102, 8493–8500. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xia, W., Li, H., Sze, K.-H. & Sun, H. (2009). J. Am. Chem. Soc. 131, 10031–10040. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: HypF, 4g9i