The structure of the HECT C-lobe of UBR5 shows a unique tertiary conformation. The sub-domain is also able to form a thioester bond with ubiquitin and may thus confer E2 binding specificity to UBR5.
Keywords: UBR5, HECT domains, E3 ubiquitin ligases, C-lobe
Abstract
UBR5 ubiquitin ligase (also known as EDD, Rat100 or hHYD) is a member of the E3 protein family of HECT (homologous to E6-AP C-terminus) ligases as it contains a C-terminal HECT domain. In ubiquitination cascades involving E3s of the HECT class, ubiquitin is transferred from an associated E2 ubiquitin-conjugating enzyme to the acceptor cysteine of the HECT domain, which consists of structurally distinct N- and C-lobes connected by a flexible linker. Here, the high-resolution crystal structure of the C-lobe of the HECT domain of human UBR5 is presented. The structure reveals important features that are unique compared with other HECT domains. In particular, a distinct four-residue insert in the second helix elongates this helix, resulting in a strikingly different orientation of the preceding loop. This protruding loop is likely to contribute to specificity towards the E2 ubiquitin-conjugating enzyme UBCH4, which is an important functional partner of UBR5. Ubiquitination assays showed that the C-lobe of UBR5 is able to form a thioester-linked E3–ubiquitin complex, although it does not physically interact with UBCH4 in NMR experiments. This study contributes to a better understanding of UBR5 ubiquitination activity.
1. Introduction
Ubiquitination is an important regulatory mechanism involved in diverse cellular processes such as cell-cycle control and signal transduction (Hershko & Ciechanover, 1986 ▶, 1998 ▶), and deregulation of targeted proteolysis has been implicated in several human diseases (Ardley et al., 2005 ▶; Arnold et al., 1998 ▶). Ubiquitination is a three-step enzymatic reaction carried out by three distinct classes of enzymes, E1, E2 and E3, that not only catalyze the covalent conjugation of ubiquitin to specific protein substrates but also promote the formation of long polyubiquitin chains through one of seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) of ubiquitin. UBR5, also known as EDD, Rat100 and hHYD, is the 300 kDa human orthologue of Drosophila melanogaster tumour suppressor hyperplastic discs protein (HYD). UBR5 belongs to the family of HECT (homologous to E6-AP C-terminus) E3 ubiquitin ligases and is frequently overexpressed in breast and ovarian cancer, suggesting a role in cancer development (Clancy et al., 2003 ▶; Fuja et al., 2004 ▶). Previous studies using a yeast two-hybrid screen identified an interaction between UBR5 and DNA topoisomerase II-binding protein 1 (TopBP1). Additionally, in vitro ubiquitination assays demonstrated that UBR5 was able to ubiquitinate TopBP1 in the presence of the E2 enzyme UBCH4 (Honda et al., 2002 ▶). UBR5 is involved in DNA-damage signalling; TopBP1, a target for ubiquitination by UBR5, co-localizes with BRCA1 at stalled DNA-replication forks (Honda et al., 2002 ▶; Mäkiniemi et al., 2001 ▶). UBR5 was shown to activate the DNA-damage checkpoint kinase CHK2 (Henderson et al., 2006 ▶). UBR5 also interacts with the calcium and integrin-binding protein (CIB) in a DNA-damage-dependent manner (Henderson et al., 2002 ▶). Additionally, UBR5 is an in vivo substrate of the extracellular signal-regulated kinases (ERKs) 1 and 2 (Eblen et al., 2003 ▶). As a ubiquitin ligase, UBR5 has been shown to control the levels of poly(A)-binding protein-interacting protein 2 (Paip2; Yoshida et al., 2006 ▶). UBR5 has recently been reported to ubiquitinate and up-regulate β-catenin (Hay-Koren et al., 2010 ▶), regulate transcription (Cojocaru et al., 2010 ▶) and activate smooth-muscle differentiation through its ability to stabilize myocardin (Hu et al., 2010 ▶).
The extreme C-terminus of UBR5 harbours a catalytic HECT domain. UBR5 also contains two nuclear localization signals and three protein–protein interaction domains: a ubiquitin-associated (UBA) domain at its N-terminus (Kozlov et al., 2007 ▶), a zinc-finger-like domain termed the ubiquitin-recognin (UBR) box near the middle of the protein (Tasaki et al., 2005 ▶) and an approximately 70-residue MLLE (mademoiselle) domain named after its conserved and essential signature motif KITGMLLE that is immediately adjacent to the HECT domain (Deo et al., 2001 ▶; Kozlov et al., 2001 ▶). Structures of the UBR domains from two related ubiquitin ligases have recently been solved and shown to be involved in recognition of protein N-terminal arginine and lysine residues (Matta-Camacho et al., 2010 ▶; Choi et al., 2010 ▶).
Previous structural studies have shown that HECT domains have two distinct lobes joined by a flexible linker (Huang et al., 1999 ▶; Verdecia et al., 2003 ▶). The N-terminal lobe interacts with the E2 ubiquitin-conjugating enzyme and the C-terminal lobe (C-lobe) contains the catalytic cysteine that receives ubiquitin from the E2 enzyme to form a thioester-linked E3–ubiquitin complex. The C-lobe of the HECT domain is the site of functional specialization that defines both the topology of the chain product and the mode of its assembly (Wang & Pickart, 2005 ▶; Kim & Huibregtse, 2009 ▶).
In the present study, we determined the high-resolution crystal structure of the C-lobe of human UBR5. The structure reveals unique features of UBR5 that could affect its specificity for E2 enzymes and linkage types.
2. Materials and methods
2.1. Protein expression and purification
The HECT C-lobe from human UBR5 ubiquitin ligase (residues Val2687–Val2799) was PCR-amplified, cloned into a pGEX-6P-1 vector (Amersham Pharmacia) and expressed in Escherichia coli BL21 (DE3) in rich (LB) medium as a fusion with an N-terminal GST tag. The GST-fusion protein was purified by affinity chromatography on glutathione Sepharose resin and the tag was removed by cleavage with PreScission Protease (GE Healthcare), leaving a Gly-Pro-Leu-Gly-Ser N-terminal extension. The HECT C-lobe was further purified by gel filtration (Superdex 75; GE Healthcare) in buffer consisting of 50 mM MES pH 6.5, 100 mM NaCl, 1 mM dithiothreitol (DTT).
Bovine ubiquitin was purchased from Sigma–Aldrich and used without further purification. To obtain 15N-labelled ubiquitin, the K48R mutant of yeast ubiquitin was overexpressed in E. coli BL21 (DE3) and purified as described previously (Kozlov et al., 2007 ▶). The E2 conjugating enzyme pRSET-UBCH4 was kindly donated by Dr Yoshiomi Honda (Kumamoto University School of Medicine) and expressed as a His6-tagged fusion protein. Both proteins were further purified by anion-exchange and gel-filtration chromatography. For NMR experiments, proteins were labelled by growth in M9 minimal medium with 15N-labelled ammonium chloride as the unique source of nitrogen.
2.2. Crystallization and structure determination
Crystallization conditions were screened by the hanging-drop vapour-diffusion method using The Classics II Suite (Qiagen). The best crystals were obtained by equilibrating a drop consisting of 1 µl HECT C-lobe solution (6 mg ml−1 in 50 mM MES pH 6.5, 100 mM NaCl, 1 mM DTT) mixed with 1 µl reservoir solution consisting of 0.2 M NaCl, 0.1 M bis-Tris pH 6.5, 25%(w/v) PEG 3350 against 1 ml reservoir solution. Crystals grew in 3 d at 293 K. For data collection, crystals were cryoprotected by the addition of 15%(v/v) glycerol and flash-cooled in an N2 cold stream. The crystals belonged to space group P1 with two protein molecules per asymmetric unit, corresponding to a solvent content of 33.1%.
Diffraction data were collected from a single crystal of HECT C-lobe using an ADSC Quantum 210 CCD detector (Area Detector Systems Corp.) on beamline A1 of Cornell High-Energy Synchrotron Source (CHESS), Ithaca, New York, USA. Data processing and scaling were performed with HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure was determined by molecular replacement with Phaser (Read, 2001 ▶) using the coordinates of the corresponding fragment of the E6AP E3 ligase (PDB entry 1c4z; Huang et al., 1999 ▶) as a search model. The initial model obtained from Phaser was improved by several cycles of refinement using REFMAC (Murshudov et al., 2011 ▶) and model refitting with Coot (Emsley & Cowtan, 2004 ▶). The refinement statistics are given in Table 1 ▶. The coordinates and structure factors have been deposited in the RCSB Protein Data Bank as entry 3pt3. Structure figures were produced using PyMOL (http://www.pymol.org).
Table 1. Data-collection and refinement statistics for the UBR5 C-lobe.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 29.24, b = 39.07, c = 44.17, α = 90.04, β = 84.59, γ = 81.77 |
| Resolution (Å) | 50.0–1.98 (2.01–1.98) |
| R merge | 0.103 (0.152) |
| 〈I/σ(I)〉 | 10.3 (6.4) |
| Completeness (%) | 93.0 (93.6) |
| Multiplicity | 2.1 (1.8) |
| Refinement | |
| Resolution (Å) | 23.1–1.97 |
| No. of reflections for refinement | 11665 |
| R work/R free | 0.219/0.276 |
| No. of atoms | |
| Protein | 1508 |
| Water | 77 |
| B factors (Å2) | |
| Protein | 14.2 |
| Water | 28.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.30 |
| Ramachandran statistics (%) | |
| Residues in favoured regions | 98.3 |
| Residues in additional allowed regions | 1.7 |
2.3. In vitro ubiquitination assays
Reactions were carried out in a total volume of 25 µl PBDM8 ubiquitination buffer (250 mM Tris–HCl pH 8.0, 25 mM MgCl2, 50 mM creatine phosphate, 3 U ml−1 inorganic pyrophosphatase, 3 U ml−1 creatine phosphokinase) containing 100 nM human recombinant His6-ubiquitin activating enzyme UBE1 (Boston Biochem), 4 mM ATP, 0.5 mM DTT, 0.4 mM bovine ubiquitin (Sigma–Aldrich), 0.04 mg ml−1 UBCH4, 0.04 mg ml−1 HECT C-lobe. The reactions were incubated for 1 h at 298 K and stopped by the addition of loading buffer for analysis by SDS–PAGE. To assess the type of C-lobe–ubiquitin complex formation (thioester-linked or amide-linked), 50 mM DTT was added to the ubiquitin reaction 10 min before the reaction was stopped.
2.4. NMR spectroscopy
NMR samples were prepared in 90% NMR buffer (50 mM MES pH 6.6, 100 mM NaCl, 1 mM DTT) and 10% D2O. For NMR titrations, unlabelled UBR5 HECT C-lobe was added stepwise to 0.15 mM 15N-labelled protein (ubiquitin or UBCH4) until reaching saturation. All NMR experiments were performed at 301 K on a Bruker 600 MHz spectrometer. NMR spectra were processed using NMRPipe (Delaglio et al., 1995 ▶) and were analyzed with XEASY (Bartels et al., 1995 ▶).
3. Results and discussion
3.1. Crystallization and structure determination
We performed structural studies of the HECT domain of UBR5 in order to obtain insight into the functional aspects of its activity as a ubiquitin ligase. The full-length HECT domain could be cloned, expressed and purified from bacteria, but showed a tendency to aggregate and did not yield crystals despite extensive screening. The absence of crystals may reflect intrinsic intradomain mobility between the N- and C-lobes, as has been previously observed for other HECT domains. To overcome this problem, we cloned the N- and C-lobes individually and subjected both fragments to crystallization trials. We obtained small but well diffracting crystals of the C-terminal lobe. The 2.0 Å resolution data set acquired using synchrotron radiation was phased by molecular replacement using the corresponding fragment of the E6AP HECT domain (Huang et al., 1999 ▶) as the model. While the C-lobes of UBR5 and E6AP have only 30% sequence identity, an 80-residue stretch of the two proteins shows 37% identity. This allowed a weak molecular-replacement solution, which was subsequently improved via multiple rounds of model building and refinement.
The asymmetric unit contains two molecules that are very similar, with an r.m.s.d. value of 0.17 Å over 84 Cα atoms. The final structure does not include 19 of the 113 residues, including the seven last C-terminal residues, as these regions were disordered in the crystal.
3.2. Structure of the UBR5 HECT C-lobe
The structure of the C-lobe closely resembles those of other structurally characterized HECT domains. The lobe fold is an independent unit composed of four α-helices (α1–α4) and four β-strands (β1–β4) (Fig. 1 ▶ a). The β-strands form a mixed parallel/antiparallel sheet which caps a bundle of four α-helices. Helices α2 and α3 form the centre of the bundle, surrounded by helices α1 and α4. The structure also features three loops, L1 (seven residues), L2 (17 residues) and L3 (seven residues); the longest loop L2 is disordered and is absent from the density map. In the crystal of a homologous HECT domain from E6AP (PDB entry 1c4z), the conformation of the L2 loop is stabilized by hydrophobic (Ala792, Ile605, Val793 and Asn603) interactions with the N-lobe (Fig. 1 ▶ c).
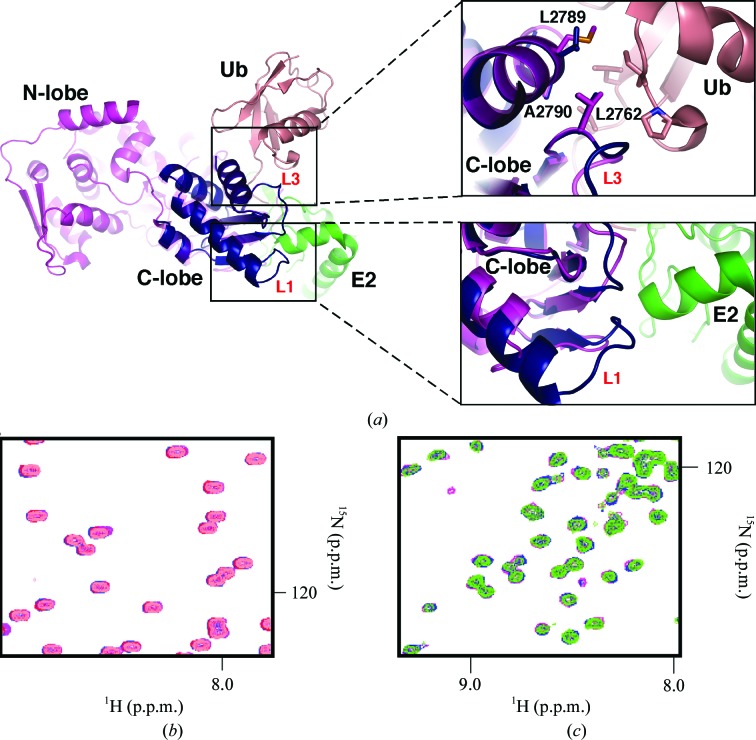
Figure 1.
Structure of the HECT C-lobe of the ubiquitin ligase UBR5. (a) Structural comparison of the C-lobes of UBR5 (light blue) and E6AP (orange). The top panel shows the extended helix α2, the strikingly different conformation of loop L1 and the disordered loop L2 of UBR5. The star indicates the catalytic cysteine residue that forms the thioester linkage to ubiquitin. The bottom panel is a 90° rotation around the y axis to highlight the four β-strands present in the C-lobe. (b) Sequence alignment of human HECT domains performed using ClustalW (Thompson et al., 1994 ▶). Secondary-structure elements are indicated above the alignment and the consensus is shown below. UBR5 has four extra residues in helix α2, while Smurf2, WWP1, Nedd4 and Huwe1 have an insertion in loop L2. (c) Cartoon representation of the HECT domain of E6AP (orange; PDB entry 1c4z); the superposed C-lobe of UBR5 is shown in light blue. Residues participating in the interaction between loop L2 (green) and the N-lobe of E6AP are shown as sticks in two different orientations with respect to the y axis.
A structure-similarity search using DALI showed that the C-lobe has high Z-scores of 15.0–14.2 (r.m.s.d. of 1.1–1.5 Å) when compared with the HECT domains of E6AP (Huang et al., 1999 ▶), Smurf2 (Ogunjimi et al., 2005 ▶), Nedd4L (Kamadurai et al., 2009 ▶), Huwe1 (Pandya et al., 2010 ▶) and WWP1 (Verdecia et al., 2003 ▶) despite having only 27–34% sequence identity. The most remarkable difference between UBR5 and the other structures is the presence of four additional residues (2708LQFK2711) in helix α2 (Figs. 1 ▶ a and 1 ▶ b). This lengthens helix α2, resulting in a different orientation of loop L1 (Fig. 1 ▶ a). Structural comparison with the UbcH5B–ubiquitin–HECTNedd4L ternary complex (Fig. 2 ▶ a) suggests that the protruding loop L1 in UBR5 may provide additional contacts that specify the E2 enzyme involved in UBR5 catalytic activity. The adjacent loop L3 also has a different conformation in a region of potential contact with the E2 enzyme and ubiquitin (Fig. 2 ▶ a) which may further affect the specificity. In addition, loop L2 of Smurf2, Huwe1, WWP1 and Nedd4L is longer by eight residues (Figs. 1 ▶ b and 1 ▶ c). UBR5 and E6AP lack these additional residues. This extended loop is in the vicinity of the catalytic cysteine; however, its role in affecting the specificity of the HECT domains remains to be elucidated.
Figure 2.
UBR5 HECT C-lobe does not interact with ubiquitin or UBCH4. (a) The overlay shows how the C-lobe is surrounded by the N-lobe, ubiquitin (Ub) and UbcH5B (E2). The unique conformation of loop L1 in UBR5 (dark blue) relative to Nedd4L (magenta; PDB entry 2oni; Structural Genomics Consortium, unpublished work) suggests that UBR5 makes additional contacts with E2 enzymes. Loop L3 contains a conserved Leu (Leu2762 in UBR5) residue that helps to stabilize the interaction with ubiquitin in the UbcH5B–ubiquitin–HECTNedd4L ternary complex; two further residues in helix α4 (Ala2790 and Leu2789 in UBR5) are also conserved. All of these residues are highly conserved in the family of HECT E3 ligases. (b) Overlay and expansion of 15N–1H correlation spectra of 0.15 mM ubiquitin (salmon) titrated with unlabelled UBR5 HECT C-lobe (maximum concentration of 0.5 mM; purple). No chemical shift perturbations were observed at the maximum concentration of titrant used, which indicates no binding. (c) Overlay and expansion of 15N–1H correlation spectra of 0.15 mM UBCH4 (green) titrated with unlabelled UBR5 HECT C-lobe (maximum concentration of 0.5 mM; purple). No chemical shift perturbations were observed, which indicates an absence of binding.
A common feature of HECT-domain crystal structures is the disorder of the last five to seven C-terminal residues. Deletion of the last six residues of E6AP eliminates isopeptide-bond formation between ubiquitin and protein substrates without substantially affecting the formation of the E3 ubiquitin-thioester intermediate (Huibregtse et al., 1995 ▶). Our structure confirms that the C-terminus of the UBR5 HECT domain is disordered in the absence of substrate proteins.
3.3. The UBR5 HECT C-lobe is not involved in noncovalent interactions with ubiquitin and UBCH4
A recent report of the ternary structure of the HECT domain of Nedd4L and UbcH5B covalently linked to ubiquitin showed that the C-lobe of the HECT domain interacts with ubiquitin (Kamadurai et al., 2009 ▶). An overlay of our structure with the UbcH5B–ubiquitin–HECTNedd4L structure (PBD entry 3jw0; Kamadurai et al., 2009 ▶) suggests that most of the residues involved are conserved in the UBR5 C-lobe (Fig. 2 ▶ a). We used two-dimensional HSQC (heteronuclear single-quantum correlation) NMR titrations to test whether the isolated C-lobe of UBR5 can interact with ubiquitin in solution. The addition of unlabelled C-lobe to 15N-labelled ubiquitin did not cause any chemical shift perturbations at C-lobe concentrations of up to 0.5 mM (Fig. 2 ▶ b). Since NMR is sensitive to even small substoichiometric interactions, the results suggest that the isolated C-lobe does not contain a ubiquitin-binding site with an affinity higher than 0.5 mM.
The UbcH5B–ubiquitin–HECTNedd4L structure also shows extensive contacts between the Nedd4L C-lobe and UbcH5B (Kamadurai et al., 2009 ▶). We wanted to evaluate whether this interaction could be observed with the isolated C-lobe. We used NMR titrations with 15N-labelled UBCH4 to test binding of the UBR5 C-lobe. The excellent quality 1H–15N correlation spectrum of UBCH4 is similar to that reported for other E2 enzymes (Hamilton et al., 2000a ▶,b ▶; Fig. 2 ▶ c). As observed in the ubiquitin titration experiment, addition of the unlabelled C-lobe did not result in any changes in chemical shifts, indicating a lack of interaction in solution (Fig. 2 ▶ c). Taken together, these results and observations from the UbcH5B–ubiquitin–HECTNedd4L structure suggest that the interactions in the ternary complex are triggered by the close spatial proximity of the C-lobe to the corresponding E2 and ubiquitin surfaces. The activity relies on the cooperativity of multiple binding sites which are too weak to be detected when isolated. The C-lobe of the HECT domain not only contains the active cysteine for ubiquitin ligation, but possesses recognition elements that contribute to stabilize the ternary complex.
Recently, the structure of the Nedd4 HECT domain in complex with ubiquitin showed that the N-lobe of the HECT domain contains a noncovalent ubiquitin-binding surface (Kim et al., 2011 ▶; Maspero et al., 2011 ▶). This interaction helps to retain the ubiquitin moieties in close proximity to build a chain on the substrate (Kim et al., 2011 ▶; Maspero et al., 2011 ▶). This surface has also been identified for the HECT domain of Smurf2 (Ogunjimi et al., 2010 ▶). It is currently unclear whether this is a general mechanism of ubiquitin recognition by HECT-family E3 ligases; however, these studies suggest that the N- and C-lobes cooperate with each other and possibly with additional domains within the E3 ligase to assure specificity and efficiency of the ubiquitination process.
Interestingly, UBR5 contains a UBA domain that binds ubiquitin. This UBA domain shows a slight preference for Lys63-linked over Lys48-linked polyubiquitin chains, possibly owing to the more open conformation of these types of chains (Kozlov et al., 2007 ▶). It is plausible that the UBA domain facilitates protein ubiquitination by the UBR5 HECT domain. While accumulating evidence suggests that the Lys63 polyubiquitin linkage serves a nonproteolytic role in different cellular processes including DNA-damage responses, in vitro studies show that proteins conjugated by Lys63 ubiquitin chains can still be efficiently degraded by the proteasome (Hofmann & Pickart, 2001 ▶). The role of UBR5 in DNA-damage signalling via degradation of TopBP1 (Honda et al., 2002 ▶) and in translational control via degradation of Paip2 (Yoshida et al., 2006 ▶) would hint at the involvement of UBR5 in forming Lys63 ubiquitin chains, although this has yet to be verified experimentally.
3.4. The UBR5 HECT C-lobe is catalytically active
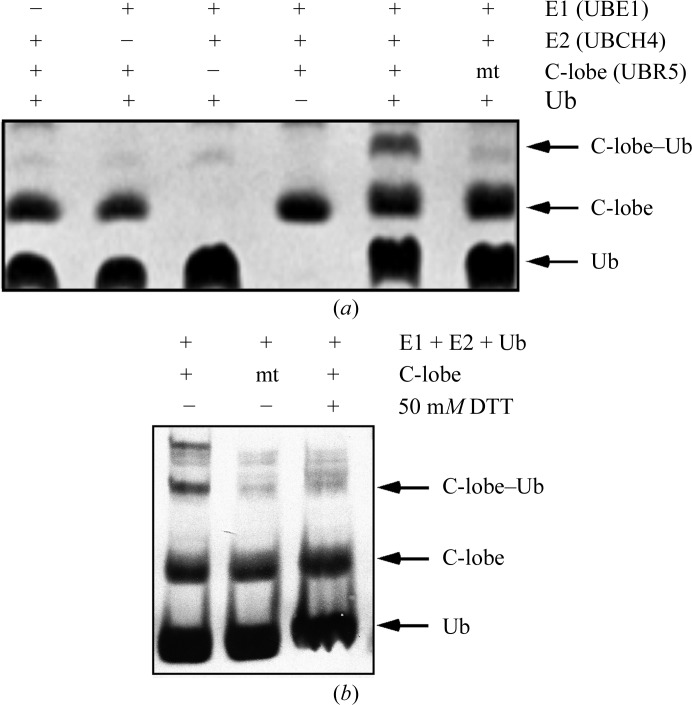
We investigated whether the isolated C-lobe is capable of forming the ubiquitin-thioester intermediate using an in vitro ubiquitination assay of C-lobe self-ubiquitination. The C-lobe UBR5 was able to incorporate ubiquitin covalently, as corroborated by mass-spectrometric analysis, indicating that the C-lobe is able to retain its catalytic activity independent of the N-lobe (Fig. 3 ▶). Mutation of Cys2768 to alanine blocked thioester formation. Similar results were obtained when the C-lobe–ubiquitin complex was challenged with 50 mM DTT (Fig. 3 ▶), confirming that the linkage is a thioester.
Figure 3.
Formation of the ubiquitin thioester. Top: in vitro formation of the ubiquitin-thioester intermediate with the UBR5 HECT C-lobe in the presence of UBE1, UBCH4 and ubiquitin (Ub) shows that the UBR5 C-lobe is catalytically active. Mutation of the UBR5 catalytic cysteine to alanine (mt) abolishes the formation of the C-lobe–ubiquitin intermediate. The products were resolved by SDS–PAGE and visualized using Coomassie Blue. Bottom: disruption of the thioester C-lobe–ubiquitin bond in the presence of 50 mM DTT.
In summary, we report the high-resolution crystal structure of the HECT C-lobe of UBR5 ubiquitin ligase. To our knowledge, this is the first structure of a catalytic HECT C-lobe in isolation from other domains. Information regarding contacts with the N-lobe and potentially other domains of the ubiquitin ligase may be lacking; however, the structure provides insight into the intrinsic structure of the C-lobe when it is not in contact with the N-lobe, ubiquitin or a protein substrate. Furthermore, the structure reveals unique features of the UBR5 HECT domain that are likely to contribute to the function of this enigmatic protein in multiple pathways.
Supplementary Material
PDB reference: C-terminal lobe of human UBR5 HECT domain, 3pt3
Acknowledgments
We thank Jordan Vergara for technical assistance and Yogita Patel for critical reading of the manuscript.
References
- Ardley, H. C., Hung, C.-C. & Robinson, P. A. (2005). FEBS Lett. 579, 571–576. [DOI] [PubMed]
- Arnold, J., Dawson, S., Fergusson, J., Lowe, J., Landon, M. & Mayer, R. J. (1998). Prog. Brain Res. 117, 23–34. [DOI] [PubMed]
- Bartels, C., Xia, T. H., Billeter, M., Güntert, P. & Wüthrich, K. (1995). J. Biomol. NMR, 6, 1–10. [DOI] [PubMed]
- Choi, W. S., Jeong, B.-C., Joo, Y. J., Lee, M.-R., Kim, J., Eck, M. J. & Song, H. K. (2010). Nature Struct. Mol. Biol. 17, 1175–1181. [DOI] [PubMed]
- Clancy, J. L. et al. (2003). Oncogene, 22, 5070–5081. [DOI] [PubMed]
- Cojocaru, M., Bouchard, A., Cloutier, P., Cooper, J. J., Varzavand, K., Price, D. H. & Coulombe, B. (2010). J. Biol. Chem. 286, 5012–5022. [DOI] [PMC free article] [PubMed]
- Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995). J. Biomol. NMR, 6, 277–293. [DOI] [PubMed]
- Deo, R. C., Sonenberg, N. & Burley, S. K. (2001). Proc. Natl Acad. Sci. USA, 98, 4414–4419. [DOI] [PMC free article] [PubMed]
- Eblen, S. T., Kumar, N. V., Shah, K., Henderson, M. J., Watts, C. K., Shokat, K. M. & Weber, M. J. (2003). J. Biol. Chem. 278, 14926–14935. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fuja, T. J., Lin, F., Osann, K. E. & Bryant, P. J. (2004). Cancer Res. 64, 942–951. [DOI] [PubMed]
- Hamilton, K. S., Ellison, M. J. & Shaw, G. S. (2000a). J. Biomol. NMR, 16, 351–352. [DOI] [PubMed]
- Hamilton, K. S., Ellison, M. J. & Shaw, G. S. (2000b). J. Biomol. NMR, 18, 319–327. [DOI] [PubMed]
- Hay-Koren, A., Caspi, M., Zilberberg, A. & Rosin-Arbesfeld, R. (2010). Mol. Biol. Cell, 22, 399–411. [DOI] [PMC free article] [PubMed]
- Henderson, M. J., Munoz, M. A., Saunders, D. N., Clancy, J. L., Russell, A. J., Williams, B., Pappin, D., Khanna, K. K., Jackson, S. P., Sutherland, R. L. & Watts, C. K. (2006). J. Biol. Chem. 281, 39990–40000. [DOI] [PubMed]
- Henderson, M. J., Russell, A. J., Hird, S., Muñoz, M., Clancy, J. L., Lehrbach, G. M., Calanni, S. T., Jans, D. A., Sutherland, R. L. & Watts, C. K. (2002). J. Biol. Chem. 277, 26468–26478. [DOI] [PubMed]
- Hershko, A. & Ciechanover, A. (1986). Prog. Nucleic Acid Res. Mol. Biol. 33, 19–56. [DOI] [PubMed]
- Hershko, A. & Ciechanover, A. (1998). Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed]
- Hofmann, R. M. & Pickart, C. M. (2001). J. Biol. Chem. 276, 27936–27943. [DOI] [PubMed]
- Honda, Y., Tojo, M., Matsuzaki, K., Anan, T., Matsumoto, M., Ando, M., Saya, H. & Nakao, M. (2002). J. Biol. Chem. 277, 3599–3605. [DOI] [PubMed]
- Hu, G., Wang, X., Saunders, D. N., Henderson, M., Russell, A. J., Herring, B. P. & Zhou, J. (2010). J. Biol. Chem. 285, 11800–11809. [DOI] [PMC free article] [PubMed]
- Huang, L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P. M., Huibregtse, J. M. & Pavletich, N. P. (1999). Science, 286, 1321–1326. [DOI] [PubMed]
- Huibregtse, J. M., Scheffner, M., Beaudenon, S. & Howley, P. M. (1995). Proc. Natl Acad. Sci. USA, 92, 2563–2567. [DOI] [PMC free article] [PubMed]
- Kamadurai, H. B., Souphron, J., Scott, D. C., Duda, D. M., Miller, D. J., Stringer, D., Piper, R. C. & Schulman, B. A. (2009). Mol. Cell, 36, 1095–1102. [DOI] [PMC free article] [PubMed]
- Kim, H. C. & Huibregtse, J. M. (2009). Mol. Cell. Biol. 29, 3307–3318. [DOI] [PMC free article] [PubMed]
- Kim, H. C., Steffen, A. M., Oldham, M. L., Chen, J. & Huibregtse, J. M. (2011). EMBO Rep. 12, 334–341. [DOI] [PMC free article] [PubMed]
- Kozlov, G., Nguyen, L., Lin, T., De Crescenzo, G., Park, M. & Gehring, K. (2007). J. Biol. Chem. 282, 35787–35795. [DOI] [PubMed]
- Kozlov, G., Trempe, J.-F., Khaleghpour, K., Kahvejian, A., Ekiel, I. & Gehring, K. (2001). Proc. Natl Acad. Sci. USA, 98, 4409–4413. [DOI] [PMC free article] [PubMed]
- Mäkiniemi, M., Hillukkala, T., Tuusa, J., Reini, K., Vaara, M., Huang, D., Pospiech, H., Majuri, I., Westerling, T., Mäkelä, T. P. & Syväoja, J. E. (2001). J. Biol. Chem. 276, 30399–30406. [DOI] [PubMed]
- Maspero, E., Mari, S., Valentini, E., Musacchio, A., Fish, A., Pasqualato, S. & Polo, S. (2011). EMBO Rep. 12, 342–349. [DOI] [PMC free article] [PubMed]
- Matta-Camacho, E., Kozlov, G., Li, F. F. & Gehring, K. (2010). Nature Struct. Mol. Biol. 17, 1182–1187. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ogunjimi, A. A., Briant, D. J., Pece-Barbara, N., Le Roy, C., Di Guglielmo, G. M., Kavsak, P., Rasmussen, R. K., Seet, B. T., Sicheri, F. & Wrana, J. L. (2005). Mol. Cell, 19, 297–308. [DOI] [PubMed]
- Ogunjimi, A. A., Wiesner, S., Briant, D. J., Varelas, X., Sicheri, F., Forman-Kay, J. & Wrana, J. L. (2010). J. Biol. Chem. 285, 6308–6315. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pandya, R. K., Partridge, J. R., Love, K. R., Schwartz, T. U. & Ploegh, H. L. (2010). J. Biol. Chem. 285, 5664–5673. [DOI] [PMC free article] [PubMed]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed]
- Tasaki, T., Mulder, L. C. F., Iwamatsu, A., Lee, M. J., Davydov, I. V., Varshavsky, A., Muesing, M. & Kwon, Y. T. (2005). Mol. Cell. Biol. 25, 7120–7136. [DOI] [PMC free article] [PubMed]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed]
- Verdecia, M. A., Joazeiro, C. A., Wells, N. J., Ferrer, J.-L., Bowman, M. E., Hunter, T. & Noel, J. P. (2003). Mol. Cell, 11, 249–259. [DOI] [PubMed]
- Wang, M. & Pickart, C. M. (2005). EMBO J. 24, 4324–4333. [DOI] [PMC free article] [PubMed]
- Yoshida, M., Yoshida, K., Kozlov, G., Lim, N. S., De Crescenzo, G., Pang, Z., Berlanga, J. J., Kahvejian, A., Gehring, K., Wing, S. S. & Sonenberg, N. (2006). EMBO J. 25, 1934–1944. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C-terminal lobe of human UBR5 HECT domain, 3pt3