The crystal structure of AkMan, a gastropoda β-1,4-mannanase prepared from the common sea hare Aplysia kurodai, was determined at 1.05 Å resolution. This is the first report on the three-dimensional structure of gastropoda β-1,4-mannanase.
Keywords: mannanases, hemicellulose, secretory proteins
Abstract
β-1,4-Mannanase (EC 3.2.1.78) catalyzes the hydrolysis of β-1,4-glycosidic bonds within mannan, a major constituent group of the hemicelluloses. Bivalves and gastropods possess β-1,4-mannanase and may degrade mannan in seaweed and/or phytoplankton to obtain carbon and energy using the secreted enzymes in their digestive systems. In the present study, the crystal structure of AkMan, a gastropod β-1,4-mannanase prepared from the common sea hare Aplysia kurodai, was determined at 1.05 Å resolution. This is the first report of the three-dimensional structure of a gastropod β-1,4-mannanase. The structure was compared with bivalve β-1,4-mannanase and the roles of residues in the catalytic cleft were investigated. No obvious binding residue was found in subsite +1 and the substrate-binding site was exposed to the molecular surface, which may account for the enzymatic properties of mannanases that can digest complex substrates such as glucomannan and branched mannan.
1. Introduction
Hemicelluloses are structural polysaccharides of the plant cell wall and are the second most abundant type of heteropolymer present in nature (Moreira & Filho, 2008 ▶). Mannan is a major constituent group of the hemicelluloses and comprises linear mannan, galactomannan, glucomannan and galactoglucomannan (Moreira & Filho, 2008 ▶; Zahura et al., 2010 ▶). β-1,4-Mannanase (CAZy GH5 and GH26; EC 3.2.1.78) catalyzes the hydrolysis of β-1,4-glycosidic bonds within mannan (Cantarel et al., 2009 ▶; Dhawan & Kaur, 2007 ▶). It is widely distributed in microorganisms, plants and invertebrate animals such as arthropods and molluscs (Dhawan & Kaur, 2007 ▶; Zahura et al., 2010 ▶; Apweiler et al., 2004 ▶). Among the molluscs, bivalves and gastropods possess a β-1,4-mannanase classified into subfamily 10 of CAZy GH5 (Cantarel et al., 2009 ▶; Zahura et al., 2011 ▶). To date, the protein sequences of one bivalve β-1,4-mannanase (from the blue mussel Mytilus edulis; Xu, Sellos et al., 2002 ▶) and of four gastropod β-1,4-mannanases (from the common sea hare Aplysia kurodai, the freshwater snail Biomphalaria glabrata, the disc abalone Haliotis discus discus and the Japanese abalone H. discus hannai; Zahura et al., 2011 ▶; Vergote et al., 2005 ▶; Ootsuka et al., 2006 ▶) have been deposited in the UniProtKB (UniProtKB/Swiss-Prot + UniProtKB/TrEMBL) database (Apweiler et al., 2004 ▶). Since all of the proteins have typical N-terminal secretion signals, bivalves and gastropods may degrade mannan in seaweed and/or phytoplankton to obtain carbon and energy using the secreted enzymes in their digestive systems (Zahura et al., 2010 ▶). Zahura and coworkers reported that β-1,4-mannanase from the digestive fluid of A. kurodai preferably degrades linear mannan from the green alga Codium fragile, thus producing mannotriose and mannobiose as the major end products. Bivalves and gastropods also possess cellulases and other enzymes in their digestive fluid that can degrade polysaccharides in seaweed and/or phytoplankton (Zahura et al., 2010 ▶; Kumagai & Ojima, 2010 ▶; Rahman et al., 2010 ▶; Sakamoto et al., 2009 ▶; Sakamoto & Toyohara, 2009 ▶).
Recently, β-1,4-mannanase has attracted great attention in food and pharmaceutical science. For example, prebiotic mannooligosaccharides, which are hydrolysis products of mannan degradation, are believed to promote the selective growth and proliferation of beneficial human intestinal microflora (Dhawan & Kaur, 2007 ▶). In addition, mannan-degrading enzymes have found application in the food, feed, paper and energy industries, such as the clarification of fruit juice and wine, viscosity reduction of coffee extract for instant coffee production, extraction of vegetable oil from leguminous seeds, improvement of the nutritional value of poultry feed, pulp bleaching and saccharification of plant biomass for bioethanol production (Moreira & Filho, 2008 ▶; Dhawan & Kaur, 2007 ▶).
Of the invertebrate β-1,4-mannanases, the crystal structure of bivalve β-1,4-mannanase prepared from M. edulis (MeMan) has been reported (Larsson et al., 2006 ▶). The structure has the (α/β)8-barrel fold, as do known bacterial and plant β-1,4-mannanases (Larsson et al., 2006 ▶; Dhawan & Kaur, 2007 ▶; Tailford et al., 2009 ▶; Bourgault et al., 2005 ▶). In the present study, the crystal structure of gastropod β-1,4-mannanase prepared from the common sea hare A. kurodai (AkMan) was determined at 1.05 Å resolution. On the basis of this structure, we compare the structures of two invertebrate β-1,4-mannanases, AkMan and MeMan, and the catalytic mechanisms of these enzymes.
2. Materials and methods
2.1. Purification of AkMan
Sea hare digestive fluid was fractionated by ammonium sulfate precipitation (60% saturation). The precipitate was dissolved in 20 mM Tris–HCl buffer pH 7.0 and dialyzed against the same buffer. Following centrifugation at 15 000g for 10 min at 277 K, the supernatant was applied onto a CM-Sepharose column equilibrated with 20 mM Tris–HCl buffer pH 7.0. β-1,4-Mannanase was eluted using a linear gradient of NaCl (0–0.4 M) in the same buffer. The fraction possessing β-1,4-mannanase activity was applied onto a HiLoad 16/10 phenyl Sepharose column (GE Healthcare, Wauwatosa, Wisconsin, USA) equilibrated with 20 mM Tris–HCl buffer pH 7.0 containing 1 M ammonium sulfate. The enzyme was eluted using a linear gradient of ammonium sulfate (1–0 M) in the same buffer. The fraction exhibiting enzyme activity was concentrated by ultrafiltration, applied onto a Sephacryl S-200 column (GE Healthcare) and eluted with 20 mM Tris–HCl buffer pH 7.0 containing 0.1 M NaCl. The purified sample was concentrated to 23.7 mg ml−1 using VivaSpin 20 10 000 molecular-weight cutoff PES centrifugal concentrators (Sartorius, Aubagne, France). The buffer was changed to 20 mM Tris–HCl on the filter unit. The purified AkMan was found to be almost pure by PAGE analysis. The enzyme activity was assayed at 301 K using locust bean gum as a substrate. The reducing sugar liberated by the hydrolysis of locust bean gum was determined by the method of Nelson and Somogyi (Zahura et al., 2010 ▶; Nelson, 1944 ▶). The sequence database reference code for AkMan is UniProtKB/Swiss-Prot E5RSM0 (Zahura et al., 2011 ▶). All chemicals were guaranteed grade from Wako Pure Chemical Industries (Osaka, Japan), Nacalai Tesque (Kyoto, Japan) and Sigma–Aldrich Chemicals (St Louis, Missouri, USA).
2.2. Crystallization
The concentrated purified AkMan sample (23.7 mg ml−1 in 20 mM Tris–HCl pH 8.0) was crystallized using the hanging-drop vapour-diffusion method. The solution in the crystallization drop was prepared on a siliconized cover glass by mixing identical volumes (1 µl + 1 µl) of protein solution and precipitant solution (20% PEG 3350, 0.1 M sodium sulfate, 100 mM Tris–HCl pH 6.5). The droplets were equilibrated against 1.0 ml precipitant solution at 293 K. Colourless plate-shaped or cube-shaped crystals were obtained.
2.3. Data collection and processing
The crystal was mounted in a nylon CryoLoop (Hampton Research, Aliso Viejo, California, USA) and placed in liquid nitrogen. Diffraction data were collected at 100 K (in a cold nitrogen gas stream) on an R-AXIS V image-plate detector (Rigaku/MSC, Woodlands, Texas, USA) using synchrotron radiation of wavelength 0.80 Å at the BL26B1 station, SPring-8, Hyogo, Japan. The resulting data set was processed, merged and scaled using HKL-2000 (HKL Research, Charlottesville, Virginia, USA) to a resolution of 1.05 Å (Table 1 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution bin.
| Diffraction data | |
| X-ray source | BL26B1, SPring-8 |
| Wavelength (Å) | 0.80 |
| Detector | Rigaku/MSC R-AXIS V image plate |
| Crystal system | Orthorhombic |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 110.09, b = 114.10, c = 51.14 |
| Resolution limits (Å) | 50–1.05 (1.07–1.05) |
| Measured reflections | 1266976 (59042) |
| Unique reflections | 292160 (13911) |
| Completeness (%) | 98.5 (93.7) |
| Multiplicity | 4.3 (4.2) |
| R merge (%) | 7.0 (32.9) |
| Mean I/σ(I) | 25.8 (3.6) |
| Refinement | |
| Program used | phenix.refine |
| Resolution range (Å) | 24.95–1.05 (1.062–1.050) |
| No. of reflections used | 291356 (9257) |
| Completeness (%) | 97.4 (94.0) |
| Protein molecules per asymmetric unit | 2 |
| Contents of asymmetric unit | |
| Residues | 702 |
| Waters | 1179 |
| Tris | 2 |
| Sulfates | 14 |
| Average B factor (Å2) | |
| Protein | 8.47 |
| Waters | 21.25 |
| Tris | 8.45 |
| Sulfates | 19.01 |
| Bond-length r.m.s. (Å) | 0.006 |
| Bond-angle r.m.s. (°) | 1.252 |
| R work (%) | 13.67 (17.11) |
| R free (%) | 15.65 (20.24) |
2.4. Structure refinement
The initial model was obtained by molecular replacement using MOLREP from the CCP4 package (Winn et al., 2011 ▶). The protein portion of the structure of MeMan (PDB entry 2c0h; Larsson et al., 2006 ▶) was used as a model. Two copies of the protein molecule were found in the asymmetric unit. Rigid-body refinement was carried out using REFMAC5 from CCP4. Several rounds of restrained maximum-likelihood refinement at 1.05 Å resolution followed by manual model building using Coot (WinCoot; Emsley & Cowtan, 2004 ▶) were carried out using REFMAC5 and phenix.refine (Adams et al., 2002 ▶). Two Tris molecules and 14 sulfate ions, which were identified from a clear difference electron-density (F o − F c) map, were included in the model in an early round of refinement (Table 1 ▶). The final refinement with individual anisotropic B factors was performed using phenix.refine and the refinement statistics are given in Table 1 ▶. The atomic coordinates for AkMan have been deposited in the Protein Data Bank (accession code 3vup).
2.5. Energy-minimization calculation of mannanase–mannopentaose complex
The initial model of the mannanase–mannopentaose complex was made using the structures of fungal mannanase (Trichoderma reesei MAN5A; PDB entry 1qnr) with mannobiose (subsites +1 and +2; Sabini et al., 2000 ▶), bacterial mannanase (Thermobifida fusca Man5; PDB entry 3man) with mannotriose (subsites −4, −3 and −2; Hilge et al., 1998 ▶) and AkMan with a Tris molecule (subsite −1). The structures were superimposed using the SSM algorithm in Coot and five mannose residues were placed at the corresponding positions (subsites −3, −2, −1, +1 and +2). Energy-minimization calculation for the mannopentaose was performed using phenix.refine in the absence of X-ray diffraction data (wxc_scale = 0).
3. Results and discussion
3.1. Quality of the final model
The initial crystallization conditions found using a commercial crystallization-screening kit (Hampton Research PEG/Ion screen) were optimized by adding Tris–HCl buffer pH 6.5. Good-quality crystals of AkMan were obtained and X-ray diffraction data were collected to 1.05 Å resolution. The space group of the crystal was P21212 and it was estimated that two protein molecules were contained in the asymmetric unit of the crystal. Other data-collection statistics are summarized in Table 1 ▶. Molecular-replacement calculations were performed using the crystal structure of MeMan and modelling and refinement calculations were performed. The cDNA of AkMan reported by Zahura and coworkers consists of 1392 base pairs and encodes 369 amino-acid residues (Zahura et al., 2011 ▶). The SignalP 3.0 server (Bendtsen et al., 2004 ▶) estimated that the 18 N-terminal residues act as a secretion signal and that cleavage would take place between Ser18 and Arg19. In the present X-ray crystallographic study, electron density was not observed for the first 18 residues and was observed for the mature protein starting from Arg19, which coincided with the estimation. The asymmetric unit contained two molecules and all 351 residues of each molecule were included in the final model. The root-mean-square deviation (r.m.s.d.) between molecules A and B was 0.19 Å for all Cα atoms. Noncrystallographic symmetry restraints for the two molecules were not applied during refinement. One Tris molecule and seven sulfate ions were added in each molecule. Relevant refinement statistics are given in Table 1 ▶. In the Ramachandran plot (Ramakrishnan & Ramachandran, 1965 ▶) of the main-chain torsion angles, 84.7% of the residues are in the most favoured regions and 97.7% of the residues are within the generously allowed regions as defined in PROCHECK (Winn et al., 2011 ▶). In the plot, one nonglycine residue (Ile65) lies outside the allowed regions of conformational space as in MeMan (Ile82); it is in a γ-turn.
3.2. Overall organization of the structure
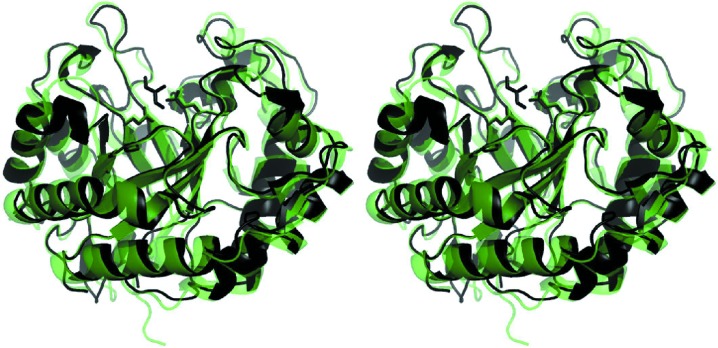
Fig. 1 ▶ displays the overall structure of AkMan (chain A in the asymmetric unit) as a cartoon representation. The structure has the (α/β)8-barrel fold, as do other known mannanases, and the overall structure is similar to that of MeMan (Fig. 1 ▶). The fifth and sixth barrel α-helices are very short (3–4 residues) and an α-helix perpendicular to the fourth barrel α-helix is present between the fifth α-helix and the sixth β-strand. Three additional β-strands at the N-terminal end of the sequence form a motif capping the barrel at the opposite end to the active site, as in MeMan (Larsson et al., 2006 ▶). A disulfide bond (Cys177–Cys244) with a bond distance of 2.04 Å was found in the structure of AkMan; this disulfide bond (Cys192–Cys259) was not observed in MeMan. Larsson and coworkers reported that the absence of the disulfide bond in MeMan might be a consequence of X-ray radiation (Larsson et al., 2006 ▶). An electron-density map revealed that a free cysteine (Cys109) in AkMan was not modified by any processes such as oxidation (data not shown). The structure of MeMan had the highest Z-score (56.2) using the DaliLite (Holm et al., 2008 ▶; Holm & Park, 2000 ▶) structure-similarity search server; other mannanases, together with β-galactosidases, β-mannosidases and β-glucosidases, exhibited much lower Z-scores (below 25.1). The r.m.s.d. calculated using the SSM superposition in Coot was 1.07 Å between AkMan and MeMan for the Cα atoms of 347 corresponding residues.
Figure 1.
Overall structure of A. kurodai β-1,4-mannanase. Crystal structures of AkMan and MeMan (PDB entry 2c0h; Larsson et al., 2006 ▶) are shown in cartoon form in black and transparent green, respectively. A Tris molecule in the active site of AkMan is also shown in black as a stick model. The Cα atoms of the structures were superimposed using the SSM algorithm implemented in Coot. The figure was produced using PyMOL (Schrödinger).
3.3. Mannanase subsite structures
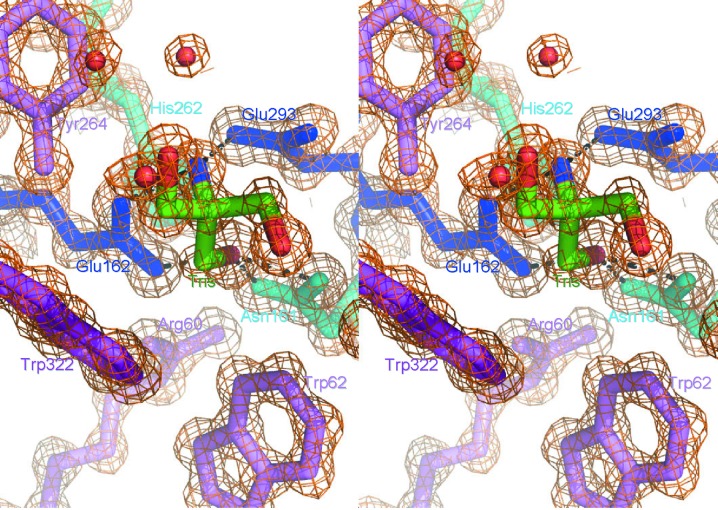
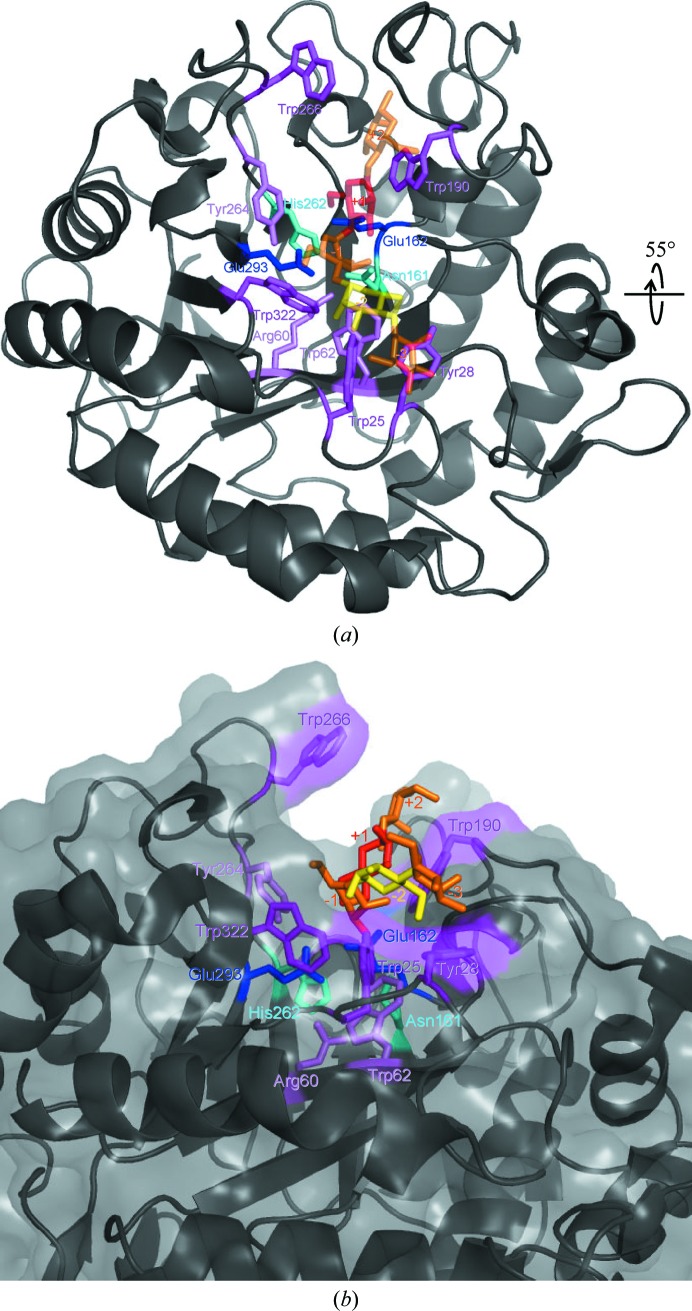
One bound Tris molecule was found in the active-site cleft formed in the barrel (Fig. 2 ▶). An N atom of the Tris molecule was hydrogen bonded to one of the catalytic residues, Glu162, and one of the O atoms was hydrogen bonded to the other catalytic residue, Glu293, as well as to Asn161. The Tris molecule is a well known saccharide analogue in glycoside hydrolases (Sabini et al., 2000 ▶). A series of oligomannose-soaking experiments did not succeed (data not shown); this failure might be attributable to the bound Tris molecule and the orientation of two molecules in the asymmetric unit (the active sites of the two molecules face each other). Therefore, energy minimization was calculated for the mannanase–mannopentaose complex (the initial model of the complex was obtained as described in §2). The mannopentaose structure after the calculation and the 12 residues in the active-site cleft (Trp25, Tyr28, Arg60, Trp62, Asn161, Glu162, Trp190, His262, Tyr264, Trp266, Glu293 and Trp322) that are conserved between AkMan and MeMan are shown in Fig. 3 ▶. Among these residues, Glu162 and Glu293 may act as catalytic residues and Asn161 and His262 may support their roles (Larsson et al., 2006 ▶). Residues Tyr28, Trp25, Trp322 and Trp190 may interact with mannose residues of substrates in subsites −3, −2, −1 and +2, respectively. Residues Trp62, Arg60 and Tyr264 may help to maintain the positions of these residues in the subsites. No obvious interaction was found between the substrate and protein in subsite +1 and the substrate-binding site is exposed to the molecular surface (Fig. 3 ▶ b); this may coincide with the enzymatic properties of AkMan, which can digest complex substrates such as glucomannan and branched mannan. Although the substrate specificity of MeMan has not been clearly elucidated, MeMan can also digest branched mannans such as locust bean gum (Xu, Hägglund et al., 2002 ▶). The similarity of the substrate-binding sites of AkMan and MeMan may account for the enzymatic properties of these mannanases. Larsson and coworkers suggested that Trp284 in MeMan (Trp266 in AkMan) might be flexible and might interact with the substrate at subsite +2 (Larsson et al., 2006 ▶). However, this residue is not conserved in the mannanases from disc abalone (H. discus discus; UniProt B6VEZ3) or freshwater snail (B. glabrata; UniProt Q5DL76) (Vergote et al., 2005 ▶). Therefore, the corresponding residue may not play an important role in substrate binding. Larsson and coworkers also reported that MeMan characteristically has a large number (19) of His residues (Larsson et al., 2006 ▶). Although AkMan has a comparable number (17) of His residues, only three of these residues are conserved between MeMan and AkMan. These His residues, except for the His residue (His262) in the active site, might not have important roles in the mannanase activity. The present paper is the first to elucidate the crystal structure of a gastropod β-1,4-mannanase. In spite of the low sequence identity between gastropod and bivalve β-1,4-mannanases (47.1% between AkMan and MeMan), it has been revealed that they have similar overall structures and substrate-binding sites. The accumulated knowledge obtained from the high-resolution structures (AkMan and MeMan at 1.05 and 1.60 Å resolution, respectively) may be employed to improve the enzymatic properties of β-1,4-mannanases, such as alteration of substrate specificity, in the field of enzyme engineering.
Figure 2.
Electron-density map around the Tris molecule bound in the active site of AkMan. A Tris molecule bound in putative subsite −1 of the active site (standard colours), the catalytic residues Glu293 and Glu162 (blue), the substrate-binding residue Trp322 (magenta) and the auxiliary residues Asn161, His262 (cyan), Trp62, Arg60 and Tyr264 (magenta) are shown as stick models. The electron-density map (2F o − F c contoured at 1.5σ) is shown as an orange mesh.
Figure 3.
Model of AkMan complexed with a mannopentaose molecule bound in the catalytic cleft. (a) The mannopentaose model after energy minimization is shown in orange (subsites −3, −1 and +2), red (+1) and yellow (−2) as a stick model. The overall protein structure is shown in black in cartoon form. The catalytic residues Glu293 and Glu162 (blue), the substrate-binding residues Tyr28, Trp25, Trp322, Trp190 and Trp266 (magenta) and the auxiliary residues Asn161, His262 (cyan), Trp62, Arg60 and Tyr264 (violet) are shown as stick models. (b) The molecule viewed from a different direction (55° rotation along the horizontal axis as illustrated) is shown in the same manner as in (a) and the molecular surface is added in transparent grey.
Supplementary Material
PDB reference: AkMan, 3vup
Acknowledgments
The synchrotron-radiation experiments were performed on BL26B1 at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) and RIKEN (2010B1469).
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., Martin, M. J., Natale, D. A., O’Donovan, C., Redaschi, N. & Yeh, L.-S. L. (2004). Nucleic Acids Res. 32, D115–D119. [DOI] [PMC free article] [PubMed]
- Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. (2004). J. Mol. Biol. 340, 783–795. [DOI] [PubMed]
- Bourgault, R., Oakley, A. J., Bewley, J. D. & Wilce, M. C. (2005). Protein Sci. 14, 1233–1241. [DOI] [PMC free article] [PubMed]
- Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V. & Henrissat, B. (2009). Nucleic Acids Res. 37, D233–D238. [DOI] [PMC free article] [PubMed]
- Dhawan, S. & Kaur, J. (2007). Crit. Rev. Biotechnol. 27, 197–216. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hilge, M., Gloor, S. M., Rypniewski, W., Sauer, O., Heightman, T. D., Zimmermann, W., Winterhalter, K. & Piontek, K. (1998). Structure, 6, 1433–1444. [DOI] [PubMed]
- Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Holm, L. & Park, J. (2000). Bioinformatics, 16, 566–567. [DOI] [PubMed]
- Kumagai, Y. & Ojima, T. (2010). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155, 138–144. [DOI] [PubMed]
- Larsson, A. M., Anderson, L., Xu, B., Muñoz, I. G., Usón, I., Janson, J. C., Stålbrand, H. & Ståhlberg, J. (2006). J. Mol. Biol. 357, 1500–1510. [DOI] [PubMed]
- Moreira, L. R. S. & Filho, E. X. F. (2008). Appl. Microbiol. Biotechnol. 79, 165–178. [DOI] [PubMed]
- Nelson, N. (1944). J. Biol. Chem. 153, 375–380.
- Ootsuka, S., Saga, N., Suzuki, K., Inoue, A. & Ojima, T. (2006). J. Biotechnol. 125, 269–280. [DOI] [PubMed]
- Rahman, M. M., Inoue, A., Tanaka, H. & Ojima, T. (2010). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 157, 317–325. [DOI] [PubMed]
- Ramakrishnan, C. & Ramachandran, G. N. (1965). Biophys. J. 5, 909–933. [DOI] [PMC free article] [PubMed]
- Sabini, E., Schubert, H., Murshudov, G., Wilson, K. S., Siika-Aho, M. & Penttilä, M. (2000). Acta Cryst. D56, 3–13. [DOI] [PubMed]
- Sakamoto, K. & Toyohara, H. (2009). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 152, 390–396. [DOI] [PubMed]
- Sakamoto, K., Uji, S., Kurokawa, T. & Toyohara, H. (2009). Gene, 435, 72–79. [DOI] [PubMed]
- Tailford, L. E., Ducros, V. M. A., Flint, J. E., Roberts, S. M., Morland, C., Zechel, D. L., Smith, N., Bjornvad, M. E., Borchert, T. V., Wilson, K. S., Davies, G. J. & Gilbert, H. J. (2009). Biochemistry, 48, 7009–7018. [DOI] [PubMed]
- Vergote, D., Bouchut, A., Sautière, P. E., Roger, E., Galinier, R., Rognon, A., Coustau, C., Salzet, M. & Mitta, G. (2005). Int. J. Parasitol. 35, 215–224. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Xu, B., Hägglund, P., Stålbrand, H. & Janson, J. C. (2002). J. Biotechnol. 92, 267–277. [DOI] [PubMed]
- Xu, B., Sellos, D. & Janson, J. C. (2002). Eur. J. Biochem. 269, 1753–1760. [DOI] [PubMed]
- Zahura, U. A., Rahman, M. M., Inoue, A., Tanaka, H. & Ojima, T. (2010). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 157, 137–143. [DOI] [PubMed]
- Zahura, U. A., Rahman, M. M., Inoue, A., Tanaka, H. & Ojima, T. (2011). Comp. Biochem. Physiol. B Biochem. Mol. Biol., 159, 227–235. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: AkMan, 3vup