The first crystallization of a fibrillar procollagen C-propeptide domain, that from human procollagen III, is described. Present throughout the Metaozoa in the form of ∼90 kDa trimers, this domain plays crucial roles in tissue homeostasis and disease.
Keywords: procollagen III, C-propeptide domain
Abstract
The C-propeptide domains of the fibrillar procollagens, which are present throughout the Metazoa in the form of ∼90 kDa trimers, play crucial roles in both intracellular molecular assembly and extracellular formation of collagen fibrils. The first crystallization of a C-propeptide domain, that from human procollagen III, is described. Following transient expression in mammalian 293T cells of both the native protein and a selenomethionine derivative, two crystal forms of the homotrimer were obtained: an orthorhombic form (P212121) that diffracted to 1.7 Å resolution and a trigonal form (P321) that diffracted to 3.5 Å resolution. Characterization by MALDI-TOF mass spectrometry allowed the efficiency of selenomethionine incorporation to be determined.
1. Introduction
Fibrillar collagens (types I, II, III, V and XI) account for approximately 25% of protein mass in the body, where they occur in the form of banded fibrils with a characteristic 64–67 nm periodicity (Kadler et al., 2007 ▶; Ricard-Blum, 2011 ▶). These collagens are synthesized in a precursor form, procollagen (∼450 kDa), as rod-like molecules (∼300 nm in length) with globular N- and C-terminal propeptide extensions (∼50 and 90 kDa, respectively). Each procollagen molecule consists of three polypeptide chains.
Inside the cell, procollagen molecular assembly is initiated by the trimerization of C-propeptide domains (Boudko et al., 2011 ▶), followed by zipper-like folding towards the N-terminal end. Trimerization is a highly specific process leading to the correct association of chains into heterotrimers (procollagens I, V and XI) or homotrimers (procollagens I and II) and also preventing incorrect association of different genetic types. There is a region in the amino-acid sequence of the C-propeptide that controls specificity (Lees et al., 1997 ▶), although the structural basis of chain recognition has yet to be elucidated. The importance of the C-propeptides in procollagen trimerization is underlined by the large number of heritable disorders of connective tissue that are characterized by mutations in this region of the molecule (Bateman et al., 2009 ▶). These affect several tissues, including bone (Chessler et al., 1993 ▶; Pace et al., 2002 ▶), cartilage (Nishimura et al., 2005 ▶), skin (De Paepe et al., 1997 ▶) and blood vessels (Pickup & Pollanen, 2011 ▶).
Outside the cell (or during secretion), collagen fibril formation is triggered by proteolytic removal of the N- and C-terminal propeptides (Colige et al., 2005 ▶; Kronenberg et al., 2010 ▶; Muir & Greenspan, 2011 ▶; Vadon-Le Goff et al., 2011 ▶) and is controlled by numerous interactions with other extracellular-matrix and cell-surface components (Kadler et al., 2008 ▶). Solubility is assured by the presence of the propeptides, particularly the C-propeptides, which increase solubility by 1000-fold (Kadler et al., 1987 ▶). The C-propeptides also interact with integrins (Davies et al., 1997 ▶), with such interactions being involved in feedback regulation of biosynthesis (Mizuno et al., 2000 ▶; Wu et al., 1991 ▶) as well as angiogenesis and tumour progression (Palmieri et al., 2008 ▶; Vincourt et al., 2010 ▶). In addition, the C-propeptides are involved in mineralization (Kirsch & Pfäffle, 1992 ▶; Lee et al., 1996 ▶; Lindahl et al., 2011 ▶).
Despite their obvious biomedical importance, high-resolution three-dimensional structures of the fibrillar procollagen C-propeptides have thus far remained elusive. Low-resolution structural analysis of the C-propeptide trimer from procollagen III by small-angle X-ray scattering revealed a tri-lobed structure (Bernocco et al., 2001 ▶), suggesting that the three polypeptide chains associate at their N-termini and then separate into distinct globular domains. This was reinforced by amino-acid sequence analysis, which identified a coiled-coil-like motif in this N-terminal region (McAlinden et al., 2003 ▶). Relatively extended structures have also been proposed based on model building (Malone et al., 2005 ▶). To gain further information, high-resolution structural data are clearly urgently required. Here, we describe the large-scale production, purification and crystallization of the C-propeptide trimer from human procollagen III in both native and selenomethionine-labelled forms, resulting in X-ray diffraction data to 1.7 Å resolution and thus heralding the first molecular structure determination for this important protein domain.
2. Methods
2.1. Expression and purification
DNA encoding the C-propeptide trimer from human procollagen III (CPIIIHis) was amplified from a previous construct in pBAC3 (Vadon-Le Goff et al., 2011 ▶) using the forward primer 5′-ACCGGTCATCATCACCACCATCACTCC-3′ and the reverse primer 5′-TCTAGACTCGAGTTATAAAAAGCAAACAGG-3′ and then inserted into the AgeI and XhoI sites of the pHLsec vector (Aricescu et al., 2006 ▶). The corresponding protein is mutated at the single N-glycosylation site (N185Q) and contains an N-terminal His6 tag followed by a Ser-Ala sequence just before the start of the native protein (Vadon-Le Goff et al., 2011 ▶; see Fig. 1 ▶). Plasmid DNA was amplified in Escherichia coli XL1 Blue cells and then purified using the Endofree Plasmid Giga kit (Qiagen). For protein expression, mammalian HEK 293T cells were cultured in six 555 ml Hyperflasks (Corning) in a CompacT SelecT automated cell-culture system (TAP Biosystems) in DMEM (high glucose) supplemented with 1× non-essential amino acids (PAA) and 10% foetal bovine serum (FBS, Invitrogen). The cells were transfected at 90% confluence with DNA–PEI transfection mixture. This latter was prepared, for each Hyperflask, by pre-incubating 1 mg plasmid DNA with 2 mg branched polyethyleneimine (PEI MW 25 000; Aldrich) for 10 min at room temperature in 100 ml serum-free DMEM. After the DNA–PEI mixture was delivered into each Hyperflask, DMEM containing 2% FBS (455 ml) was used to top up the flask (Zhao et al., 2011 ▶).
Figure 1.
Amino-acid sequence of the expressed protein. The arrow indicates the start of the secreted protein after cleavage of the signal peptide. The His6 sequence is shown in red and the arrowhead shows the start of the corresponding native protein after cleavage from procollagen by BMP-1 and tolloid-like proteinases. The Asn185Q mutation in the N-glycosylation site is boxed.
After 4 d, conditioned medium (3.3 l) was collected and was then clarified by centrifugation and filtration through a Steritop vacuum-driven filtration system (Millipore). The supernatant was then dialyzed over 2 d (with one change) against phosphate-buffered saline (PBS) at 277 K. Dialyzed medium was then applied onto a 5 ml column of Talon Co2+-affinity resin (Clontech) followed by washing with four column volumes of phosphate-buffered saline (PBS) and two column volumes of PBS containing 20 mM imidazole. Bound protein was eluted with 250 mM imidazole in 20 mM Tris–HCl pH 8.0, 150 mM NaCl. Fractions were then pooled, microconcentrated (molecular-weight cutoff 30 kDa) and further purified on a Superdex 200 16/60 column eluted with 20 mM HEPES pH 7.4, 150 mM NaCl. Finally, fractions were analyzed by SDS–PAGE and further concentrated to ∼30 mg ml−1.
For production of the selenomethionine derivative, cell culture and transfection were performed as above using 12 roller bottles (Greiner) instead of Hyperflasks. After 24–48 h transfection in normal DMEM containing 2% FCS (250 ml per roller bottle), the culture medium was removed, the cell layer was washed twice with PBS, and fresh methionine-free DMEM (MP Biomedicals) containing 3% dialyzed FBS medium was then added together with l-Gln, non-essential amino acids and 30 mg ml−1 l-selenomethionine (SeMet; Eburon Organics). After 4 d of culture, the medium was collected and the SeMet derivative was purified as for CPIIIHis (above).
2.2. Mass spectrometry
Mass spectrometry was performed using a Voyager-DE Pro MALDI-TOF mass spectrometer (Applied Biosystems) equipped with a nitrogen UV laser (λ = 337 nm, 3 ns pulse). The instrument was operated in the positive linear mode (mass accuracy 0.05%) with an accelerating potential of 20 kV. Typically, mass spectra were obtained by the accumulation of 600 laser shots for each analysis and were processed using Data Explorer 4.0 software (AB Sciex). For analysis of reduced proteins, 10 µl CPIIIHis and its SeMet derivative (at ∼3 mg ml−1) were first reduced with 30 µl dithiothreitol (DTT; 10 mM in ammonium bicarbonate buffer) for 45 min at 328 K with constant stirring. After desalting using C4 ZipTips (Millipore), samples were mixed with sinapinic acid (saturated solution in 30% acetonitrile and 0.3% trifluoroacetic acid), deposited on the MALDI target and air-dried before analysis.
2.3. Crystallization and data collection
Initial screening and optimization of crystallization conditions were carried out using a Cartesian Technologies pipetting robot in 96-well sitting-drop vapour-diffusion plates (Greiner). Crystallization kits from Hampton Research (Index and SaltRx), Molecular Dimensions (PACTpremier) and the Oxford Protein Production Facility (Blocks 1–3; http://www.oppf.ox.ac.uk) were used. Crystals of forms I (SeMet) and II (native) were obtained directly using the Block 3 and PACTpremier crystallization kits, respectively, as detailed in Table 1 ▶. Crystals of form III (native) were initially obtained using the Block 3 kit in 10% PEG 6000, 0.1 M MES pH 6.0 and were optimized to the conditions shown in Table 1 ▶. Images of crystallization drops were taken automatically at regular intervals using a TAP Biosystems storage vault (Walter et al., 2005 ▶) maintained at 294 K. Further information is provided in Table 1 ▶.
Table 1. Crystallization conditions.
| Form I (SeMet) | Form II (native) | Form III (native) | |
|---|---|---|---|
| Method | Sitting drop | Sitting drop | Sitting drop |
| Plate type | 96-well Greiner | 96-well Greiner | 96-well Greiner |
| Temperature (K) | 294 | 294 | 294 |
| Protein concentration (mg ml−1) | 30 | 30 | 30 |
| Buffer composition of protein solution | 20 mM HEPES pH 7.4, 0.15 M NaCl | 20 mM HEPES pH 7.4, 0.15 M NaCl | 20 mM HEPES pH 7.4, 0.15 M NaCl |
| Composition of reservoir solution | 20% PEG 3350, 0.2 M NaCl | 20% PEG 3350, 0.1 M bis-Tris propane pH 6.5, 0.2 M KNO3 | 8% PEG 6000, 60 mM HEPES pH 7.0, 40 mM MES pH 6.0 |
| Volume (nl) and ratio of drop | 100, 1:1 | 100, 1:1 | 100, 1:1 |
| Volume of reservoir (µl) | 100 | 100 | 100 |
For data collection, crystals were flash-cooled in liquid nitrogen. Form I crystals were cryoprotected by gradually increasing the glycerol concentration in the mother liquor to 30%(v/v), form II crystals were immersed in oil (perfluoropolyether PFO-X125/03; Alfa Aesar) and form III crystals were cryoprotected with 25% ethylene glycol. Diffraction data were collected at the Diamond Light Source synchrotron (Didcot, England). Data were processed using xia2 (form I) and MOSFLM (forms II and III) and were scaled with SCALA from the CCP4 program suite (Winn et al., 2011 ▶). Each data set was collected from a single crystal. Data-collection and processing statistics are given in Table 2 ▶.
Table 2. Data collection and processing.
Values in parentheses are for the outer shell.
| Form I (SeMet) | Form II (native) | Form III (native) | |
|---|---|---|---|
| Diamond beamline | I03 | I04 | I04 |
| Wavelength (Å) | 0.9795 | 0.9763 | 0.9763 |
| Temperature (K) | 100 | 100 | 100 |
| Detector | ADSC Q315r | PILATUS 6M-F | PILATUS 6M-F |
| Crystal-to-detector distance (mm) | 288 | 320 | 553 |
| Rotation range per image (°) | 1.0 | 0.1 | 0.1 |
| Total rotation range (°) | 360 | 360 | 360 |
| Exposure time per image (°) | 0.5 | 0.1 | 0.1 |
| Space group | P212121 | P212121 | P321 |
| Unit-cell parameters (Å, °) | a = 83.9, b = 89.3, c = 101.5, α = β = γ = 90 | a = 76.5, b = 90.4, c = 102.4, α = β = γ = 90 | a = b = 86.1, c = 73.0, α = β = 90, γ = 120 |
| Molecules in the asymmetric unit | 3 | 3 | 1 |
| Mosaicity (°) | 0.40 | 0.75 | 1.61 |
| Resolution range (Å) | 101.5–2.2 (2.27–2.21) | 61.3–1.7 (1.73–1.68) | 43.0–3.5 (3.69–3.50) |
| Total No. of reflections | 557946 | 271723 | 32977 |
| No. of unique reflections | 38676 | 78019 | 4149 |
| Completeness (%) | 100 (100) | 96.2 (95.6) | 99.7 (99.8) |
| Redundancy | 14.4 (14.7) | 3.5 (3.6) | 7.9 (8.2) |
| Anomalous completeness (%) | 100 (100) | n.a. | n.a. |
| Anomalous redundancy | 7.6 | n.a. | n.a. |
| 〈I/σ(I)〉 | 19.4 (4.0) | 4.7 (2.2) | 10.7 (3.2) |
| R r.i.m. (%) | 11.7 | 10.2 | 10.5 |
| Overall B value from Wilson plot (Å2) | 34.9 | 21.5 | 93.6 |
3. Results and discussion
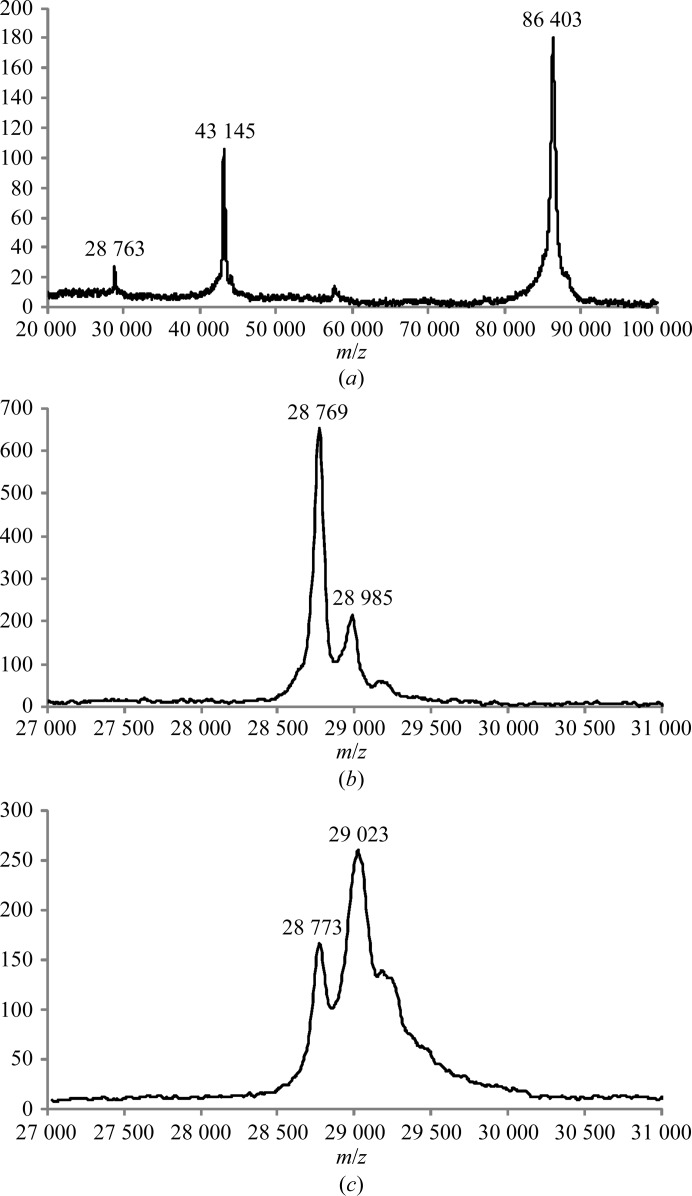
Recombinant C-propeptide trimer from human procollagen III (CPIIIHis; where each chain is mutated at the single N-glycosylation site and carries an N-terminal His6 tag; Vadon-Le Goff et al., 2011 ▶) was expressed by large-scale transient transfection in HEK 293T cells (Aricescu et al., 2006 ▶). Approximately 18 mg CPIIIHis was obtained from 3 l conditioned medium. Purity was at least 99% as assessed by SDS–PAGE (Fig. 2 ▶), which showed a single band (under nonreducing conditions) migrating at approximately the mass expected for a trimer. We also checked for purity and trimerization ability by mass spectrometry under both reducing and nonreducing conditions. By MALDI-TOF, the [M + H]+ ion for trimeric CPIIIHis was observed at an m/z value of 86 403 ± 43 (Fig. 3 ▶ a). After reduction with DTT, the [M + H]+ ion for monomeric CPIIIHis was observed at m/z = 28 769 ± 14 (Fig. 3 ▶ b). These figures compared favourably with the expected molecular masses of 86 320 and 28 776 Da, respectively. Thus, both the SDS–PAGE and the mass-spectrometric data showed that the presence of the N-terminal His6 tag did not interfere with the trimerization of CPIIIHis.
Figure 2.

SDS–PAGE analysis. 5 µg purified CPIIIHis was analyzed on a 10% acrylamide gel under nonreducing conditions followed by Coomassie Blue staining. Migration positions of marker proteins are indicated in kDa.
Figure 3.
Mass-spectrometric analysis. (a) MALDI-TOF spectrum of nonreduced CPIIIHis showing the major singly charged [M + H]+ ion at m/z = 86 403 as expected for the trimer, as well as doubly [M + 2H]2+ and triply [M + 3H]3+ charged ions at m/z = 43 145 and 28 763, respectively. (b) MALDI-TOF spectrum of reduced CPIIIHis showing a major peak at m/z = 28 769 as expected for the monomeric form. The minor [M + H]+ ion at m/z = 28 985 probably corresponds to a noncovalent adduct with one molecule of sinapinic acid matrix. (c) MALDI-TOF spectrum of the CPIIIHis SeMet derivative showing a major [M + H]+ ion at m/z = 29 023 corresponding to the incorporation of approximately five SeMet residues per monomer. The minor [M + H]+ ion at m/z = 28 773 corresponds to unlabelled CPIIIHis.
For structure determination, as there are no homologues of the fibrillar procollagen C-propeptides in the Protein Data Bank, heavy-atom labelling or selenium substitution is required in order to solve the phase problem. Since the transient expression system used here has previously been shown to efficiently incorporate free selenomethionine from the culture medium (Aricescu et al., 2006 ▶), we also produced the SeMet derivative of CPIIIHis (see §2), resulting in good, although somewhat reduced, yields (∼6 mg).
Analysis of the SeMet derivative of CPIIIHis by MALDI-TOF mass spectrometry showed a minor [M + H]+ ion signal at m/z = 28 773 ± 14 and a broader major [M + H]+ ion signal at m/z = 29 023 ± 15 (Fig. 3 ▶ c). The first peak corresponds to unlabelled monomer (representing ∼25% of the total), while the broader peak (∼75% of the total) indicates a mixed population of selenomethionine-labelled protein species with an average mass corresponding to approximately five selenomethionine residues incorporated into each CPIIIHis monomer (the mass increment per SeMet residue is 47). This compares with six methionine residues per chain observed in the amino-acid sequence of CPIIIHis. The incorporation of the SeMet label into CPIIIHis was therefore highly efficient.
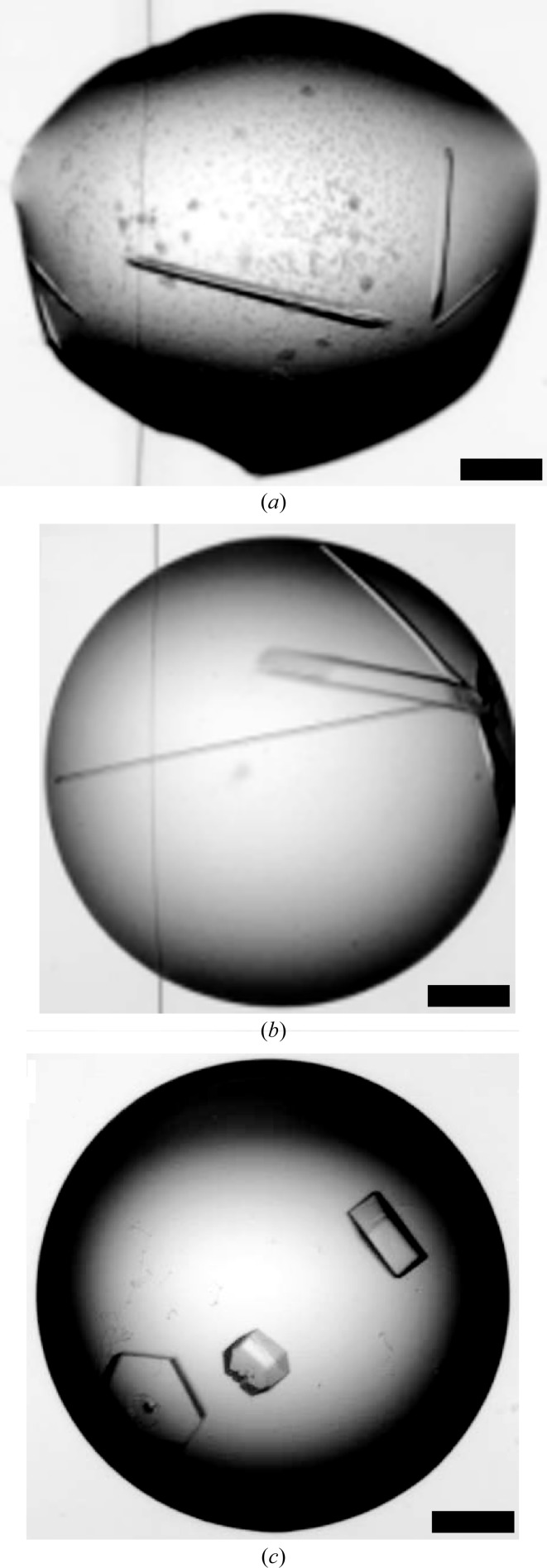
After concentration to approximately 30 mg ml−1, crystallization trials were set up for the unlabelled and SeMet-labelled proteins. After a few days, crystals began to appear in several conditions (Table 1 ▶). In general, the crystals were of two types: needle-like or hexagonal plates (Fig. 4 ▶). X-ray diffraction screening of crystals grown under different conditions resulted in the identification of two crystal forms that diffracted X-rays to 1.7 and 3.5 Å resolution in space groups P212121 and P321, respectively (Table 2 ▶). For the SeMet derivative, screening using the most promising conditions identified for the unlabelled protein also rapidly resulted in high-quality crystals (space group P212121). These diffracted to 2.2 Å resolution and the anomalous signal in the data collected allowed phasing to be carried out. Structure determination using single-wavelength anomalous dispersion is described elsewhere (Bourhis et al., 2012 ▶).
Figure 4.
Crystallization. Crystals of the SeMet derivative of CPIIIHis (form I) are shown (a) as well as those of the two native forms II (b) and III (c). Scale bars = 200 µm.
Acknowledgments
This work was funded by the Fondation de France, the Agence National de la Recherche (SCARFREE and TOLLREG projects), the European Commission (P-CUBE project), the Centre National de la Recherche Scientifique, the Université Lyon 1 and Lyinbiopôle. We thank Annie Chaboud and Isabelle Grosjean of the Protein Production and Analysis Facility (SFR Biosciences Gerland-Lyon Sud UMS3444/US8) as well as the staff of Diamond Light Source (Didcot, England) for technical support.
References
- Aricescu, A. R., Lu, W. & Jones, E. Y. (2006). Acta Cryst. D62, 1243–1250. [DOI] [PubMed]
- Bateman, J. F., Boot-Handford, R. P. & Lamandé, S. R. (2009). Nature Rev. Genet. 10, 173–183. [DOI] [PubMed]
- Bernocco, S., Finet, S., Ebel, C., Eichenberger, D., Mazzorana, M., Farjanel, J. & Hulmes, D. J. S. (2001). J. Biol. Chem. 276, 48930–48936. [DOI] [PubMed]
- Boudko, S. P., Engel, J. & Bachinger, H. P. (2011). Int. J. Biochem. Cell Biol. 44, 21–32. [DOI] [PubMed]
- Bourhis, J.-M., Marinaro, N., Zhao, Y., Harlos, K., Exposito, J.-Y., Jones, E. Y., Moali, C., Aghajari, N. & Hulmes, D. J. S. (2012). Nature Struct. Mol. Biol In the press. [DOI] [PMC free article] [PubMed]
- Chessler, S. D., Wallis, G. A. & Byers, P. H. (1993). J. Biol. Chem. 268, 18218–18225. [PubMed]
- Colige, A., Ruggiero, F., Vandenberghe, I., Dubail, J., Kesteloot, F., Van Beeumen, J., Beschin, A., Brys, L., Lapière, C. M. & Nusgens, B. (2005). J. Biol. Chem. 280, 34397–34408. [DOI] [PubMed]
- Davies, D., Tuckwell, D. S., Calderwood, D. A., Weston, S. A., Takigawa, M. & Humphries, M. J. (1997). Eur. J. Biochem. 246, 274–282. [DOI] [PubMed]
- De Paepe, A., Nuytinck, L., Hausser, I., Anton-Lamprecht, I. & Naeyaert, J. M. (1997). Am. J. Hum. Genet. 60, 547–554. [PMC free article] [PubMed]
- Kadler, K. E., Baldock, C., Bella, J. & Boot-Handford, R. P. (2007). J. Cell Sci. 120, 1955–1958. [DOI] [PubMed]
- Kadler, K. E., Hill, A. & Canty-Laird, E. G. (2008). Curr. Opin. Cell Biol. 20, 495–501. [DOI] [PMC free article] [PubMed]
- Kadler, K. E., Hojima, Y. & Prockop, D. J. (1987). J. Biol. Chem. 262, 15696–15701. [PubMed]
- Kirsch, T. & Pfäffle, M. (1992). FEBS Lett. 310, 143–147. [DOI] [PubMed]
- Kronenberg, D., Bruns, B. C., Moali, C., Vadon-Le Goff, S., Sterchi, E. E., Traupe, H., Böhm, M., Hulmes, D. J. S., Stöcker, W. & Becker-Pauly, C. (2010). J. Invest. Dermatol. 130, 2727–2735. [DOI] [PubMed]
- Lee, E. R., Smith, C. E. & Poole, R. (1996). J. Histochem. Cytochem. 44, 433–443. [DOI] [PubMed]
- Lees, J. F., Tasab, M. & Bulleid, N. J. (1997). EMBO J. 16, 908–916. [DOI] [PMC free article] [PubMed]
- Lindahl, K. et al. (2011). Hum. Mutat. 32, 598–609. [DOI] [PMC free article] [PubMed]
- Malone, J. P., Alvares, K. & Veis, A. (2005). Biochemistry, 44, 15269–15279. [DOI] [PubMed]
- McAlinden, A., Smith, T. A., Sandell, L. J., Ficheux, D., Parry, D. A. & Hulmes, D. J. S. (2003). J. Biol. Chem. 278, 42200–42207. [DOI] [PubMed]
- Mizuno, M., Fujisawa, R. & Kuboki, Y. (2000). Calcif. Tissue Int. 67, 391–399. [DOI] [PubMed]
- Muir, A. & Greenspan, D. S. (2011). J. Biol. Chem. 286, 41905–41911. [DOI] [PMC free article] [PubMed]
- Nishimura, G., Haga, N., Kitoh, H., Tanaka, Y., Sonoda, T., Kitamura, M., Shirahama, S., Itoh, T., Nakashima, E., Ohashi, H. & Ikegawa, S. (2005). Hum. Mutat. 26, 36–43. [DOI] [PubMed]
- Pace, J. M., Chitayat, D., Atkinson, M., Wilcox, W. R., Schwarze, U. & Byers, P. H. (2002). J. Med. Genet. 39, 23–29. [DOI] [PMC free article] [PubMed]
- Palmieri, D., Astigiano, S., Barbieri, O., Ferrari, N., Marchisio, S., Ulivi, V., Volta, C. & Manduca, P. (2008). Exp. Cell Res. 314, 2289–2298. [DOI] [PubMed]
- Pickup, M. J. & Pollanen, M. S. (2011). Forensic Sci. Med. Pathol. 7, 192–197. [DOI] [PubMed]
- Ricard-Blum, S. (2011). Cold Spring Harb. Perspect. Biol. 3, a004978. [DOI] [PMC free article] [PubMed]
- Vadon-Le Goff, S., Kronenberg, D., Bourhis, J.-M., Bijakowski, C., Raynal, N., Ruggiero, F., Farndale, R. W., Stöcker, W., Hulmes, D. J. S. & Moali, C. (2011). J. Biol. Chem. 286, 38932–38938. [DOI] [PMC free article] [PubMed]
- Vincourt, J.-B., Etienne, S., Cottet, J., Delaunay, C., Malanda, C. B., Malanda, B., Lionneton, F., Sirveaux, F., Netter, P., Plénat, F., Mainard, D., Vignaud, J.-M. & Magdalou, J. (2010). Cancer Res. 70, 4739–4748. [DOI] [PubMed]
- Walter, T. S. et al. (2005). Acta Cryst. D61, 651–657. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, C. H., Walton, C. M. & Wu, G. Y. (1991). J. Biol. Chem. 266, 2983–2987. [PubMed]
- Zhao, Y., Bishop, B., Clay, J. E., Lu, W., Jones, M., Daenke, S., Siebold, C., Stuart, D. I., Jones, E. Y. & Aricescu, A. R. (2011). J. Struct. Biol. 175, 209–215. [DOI] [PMC free article] [PubMed]