The outer membrane lipoprotein GumB from X. campestris was purified and crystallized by the hanging-drop vapour-diffusion method. A native data set was collected to a resolution of 2.54 Å.
Keywords: xanthan gum, exopolysaccharides, outer membrane proteins, lipoproteins
Abstract
GumB is a predicted outer membrane lipoprotein that is involved in the synthesis and/or secretion of xanthan gum. This exopolysaccharide, produced by Xanthomonas campestris, is valuable in industry because of its important rheological properties. Solution of the GumB structure will provide insight into the polymerization and/or secretion mechanisms of xanthan gum. GumB was overexpressed and purified and diffraction-quality crystals of native GumB were obtained. A complete data set was collected to 2.54 Å resolution with an R p.i.m. of 0.034. The crystals belonged to space group P212121, with unit-cell parameters a = 84.4, b = 90.5, c = 120.7 Å.
1. Introduction
In Gram-negative bacteria, macromolecules attached to cells or secreted to the extracellular medium cross both the inner and the outer membranes. Numerous protein-secretion systems that form multiprotein transenvelope complexes are known (Gerlach & Hensel, 2007 ▶). However, the polymerization and secretion of bacterial polysaccharides are poorly understood processes. Three different pathways have been described in Gram-negative bacteria: the Wzy-dependent system (Cuthbertson et al., 2009 ▶), the ABC transporter-dependent system (Cuthbertson et al., 2010 ▶) and a recently described pathway that involves a multiprotein complex that forms a molecular scaffold using TPR domains (Keiski et al., 2010 ▶).
Xanthan gum is a high-molecular-weight exopolysaccharide that is produced by the phytopathogenic bacterium Xanthomonas campestris. It is widely used in industry as a thickening and stabilizing agent (Baird et al., 1983 ▶). It consists of a cellulose backbone with mannose–glucuronic acid–mannose side chains. Xanthan gum is synthesized by the Wzy-dependent mechanism. In this system, synthesis consists of the assembly of pentasaccharide repeating units by specific glycosyltransferases, which occurs on the cytoplasmic side of the inner membrane; the repeating units are translocated by a flippase to the periplasmic side of the inner membrane, where polymerization occurs. Finally, the polymer is secreted into the extracellular medium (Barreras et al., 2008 ▶; Ielpi et al., 1993 ▶; Salinas et al., 2011 ▶).
GumB, a predicted outer membrane lipoprotein, may be involved in the polymerization and/or secretion of xanthan gum. It has been shown that a gumB mutant strain accumulates the repeating units without formation of the polymer, indicating a role of GumB in the polymerization and/or secretion of xanthan gum (Katzen et al., 1998 ▶). To obtain structural details of the mechanism of xanthan-gum polymerization and secretion, we overexpressed, purified and crystallized GumB. Because only three lipoprotein structures involved in polysaccharide secretion are known (Dong et al., 2006 ▶; Keiski et al., 2010 ▶; Sathiyamoorthy et al., 2011 ▶), we believe that the structure of GumB will help us to understand how polysaccharide synthesis and secretion occurs.
2. Materials and methods
2.1. Macromolecule production
The details of GumB cloning and expression are given in Table 1 ▶. The gene gumB (fragment 1336–1971 of the gum region; GenBank accession No. U22511) was cloned into a pQE30 vector by CP Kelco, producing the pQE-Xps#6 plasmid (Patel et al., 2008 ▶). The sequence of the construct was confirmed by DNA sequencing. The GumB signal peptide was removed to direct the protein into the cytoplasm and replaced by an N-terminal His6 tag to allow affinity purification. The protein produced from the construct had a molecular weight of 24.71 kDa. Escherichia coli DH5α cells (pRep4) were transformed with pQE-Xps#6. The transformed cells were grown in LB medium supplemented with 100 µg ml−1 ampicillin and 25 µg ml−1 kanamycin at 310 K until an OD600 of 0.7 was reached. At this point, protein expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactopyranoside (Gold Biotechnology). The cells were grown for an additional 16 h at 293 K, harvested by centrifugation and washed with a solution consisting of 50 mM Tris–HCl pH 8.0, 50 mM MgCl2. The washed cells were suspended in 50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM n-dodecyl β-d-maltopyranoside (DDM; Anatrace), 1 mM phenylmethylsulfonyl fluoride and disrupted by three passages through a French pressure cell at 110 MPa and 277 K. The disrupted cells were incubated with gentle agitation at 277 K for 1 h and centrifuged at 100 000g for 1 h at 277 K. A Jasco FPLC system connected to a UV-1575 Intelligent UV–Vis detector was used to load the supernatant onto a 5 ml Ni–NTA column (GE Healthcare) equilibrated with 50 mM Tris–HCl pH 8.0, 0.15 M NaCl, 0.54 mM DDM (buffer A). The column was washed with buffer A until no absorbance at 280 nm was detected. The bound proteins were eluted with a 60 ml linear gradient of 0–400 mM imidazole at 1 ml min−1. The fractions containing GumB, as identified by SDS–PAGE, were concentrated to 5 mg ml−1 using a Centripep device with a 10 kDa pore size (Amicon Ultra, Millipore). For further purification, the concentrated protein was loaded onto a 10 000–600 000 range Superdex 200 size-exclusion column (GE Healthcare) and eluted with 50 mM Tris–HCl pH 8.0, 0.15 M NaCl, 0.18 mM DDM (buffer B).
Table 1. Macromolecule-production information.
| Source organism | X. campestris FC2, Rifr derivative from wild-type NRRL B-1459 strain |
| Cloning and expression vector | pQE-Xps#6 |
| Expression host | E. coli DH5α (pRep4†) |
| Complete amino-acid sequence of the construct produced | MRGSHHHHHHGSACSLGACSTGPEMASSLPHPDPLAMSTVQPEYRLAPGDLLLVKVFQIDDLERQVRIDQNGHISLPLIGDVKAAGLGVGELEKLVADRYRAGYLQQPQISVFVQESNGRRVTVTGAVDEPGIYPVIGANLTLQQAIAQAKGVSTVASRGNVIVFRMVNGQKMIARFDLTEIEKGANPDPEIYGGDIVVVYRSDARVWLRTMLELTPLVMVWRAYR |
Repressor plasmid; it codes for LacI repressor protein to ensure tight control at the transcriptional level.
2.2. Crystallization
Purified GumB in buffer B was used in the initial crystallization trials. These trials were carried out by the sitting-drop vapour-diffusion method using the commercial kits JBScreen Classic, JBScreen Membrane (Jena Bioscience) and Crystal Screen (Hampton Research). A Honeybee 963 crystallization robot and 96-well CrystalQuick plates (Greiner Bio-One) were used to set up the preliminary screening. The robot dispensed equal volumes of the protein solution and the crystallization solution (0.2 µl each) into the drop well and 50 µl crystallization solution into the reservoir well. The initial crystallization conditions were set up with different protein concentrations (5, 10, 15 and 20 mg ml−1) and the hits were optimized by grid and additive screening. The plates were incubated at 291 K. Optimization was carried out by the hanging-drop vapour-diffusion method, as described in Table 2 ▶. Crystal growth was observed after 30 d of incubation.
Table 2. Crystallization.
| Method | Hanging drop |
| Plate type | VDX plates (Hampton Research) |
| Temperature (K) | 291 |
| Protein concentration (mg ml−1) | 10 |
| Buffer composition of protein solution | 50 mM Tris–HCl pH 8.0, 0.15 M NaCl, 0.18 mM DDM |
| Composition of reservoir solution | 100 mM Tris–HCl pH 8.5, 800 mM LiCl, 8% PEG 4000 and 4% of the additive 1-propanol |
| Volume of drop | 2.2 µl (1 µl protein solution, 1 µl reservoir solution and 0.2 µl additive solution) |
| Volume of reservoir (µl) | 500 |
2.3. Data collection and processing
Prior to data collection, the crystals were transferred into reservoir solution containing 20%(v/v) glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected from a single crystal on the PROXIMA 1 beamline at the SOLEIL synchrotron, Saclay, France. The images were integrated using MOSFLM (Leslie & Powell, 2007 ▶) and scaled using SCALA from the CCP4 program suite (Potterton et al., 2003 ▶; Winn et al., 2011 ▶).
3. Results and discussion
Similarly to other bacterial lipoproteins, GumB is synthesized as a precursor in the cytoplasm. Its processing involves removal of the signal peptide and the binding of a lipid moiety to the N-terminus which anchors the protein to the membrane (Okuda & Tokuda, 2011 ▶). We tried to overexpress GumB with its signal peptide; however, after induction we observed cell lysis in several expression conditions, most likely owing to loss of membrane integrity. However, we succeeded in overexpressing GumB without its signal peptide as a cytoplasmic protein. Purification of GumB was carried out in the presence of DDM to prevent protein aggregation. Purified GumB was obtained by a two-step purification protocol consisting of affinity and size-exclusion chromatography (Fig. 1 ▶). The yield of purified GumB was 20 mg per litre of culture.
Figure 1.
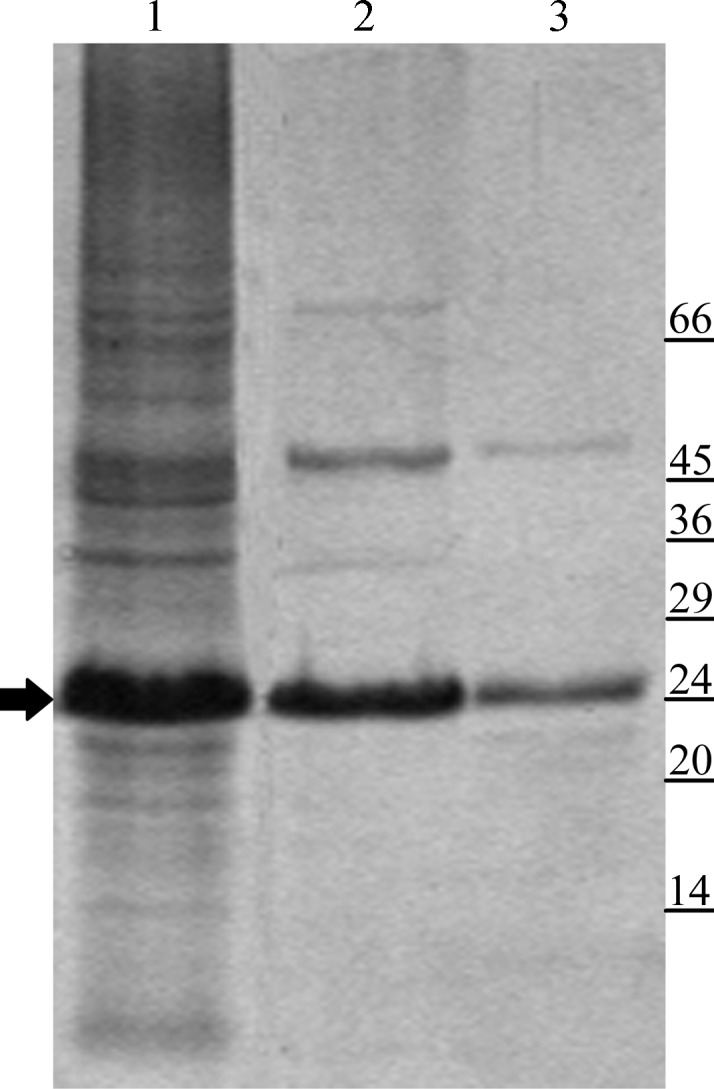
GumB purification. Lane 1, crude extract from DH5α/pQE-Xps#6 after IPTG induction; lane 2, GumB after affinity purification; lane 3, purified GumB after gel-filtration chromatography. The arrow indicates GumB. Molecular-weight markers (labelled in kDa) are indicated on the right.
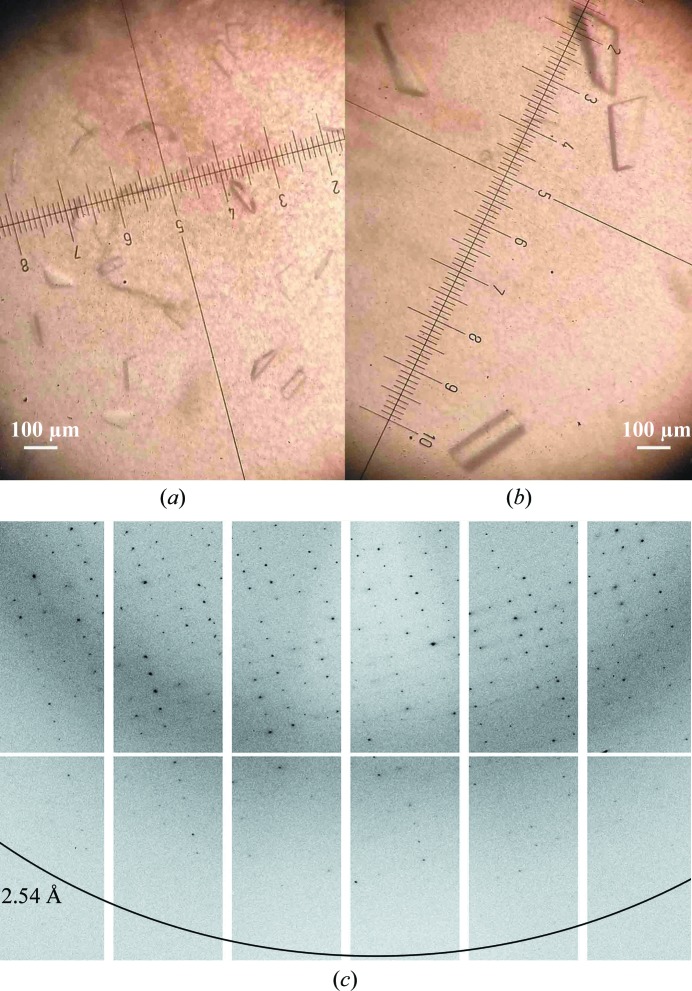
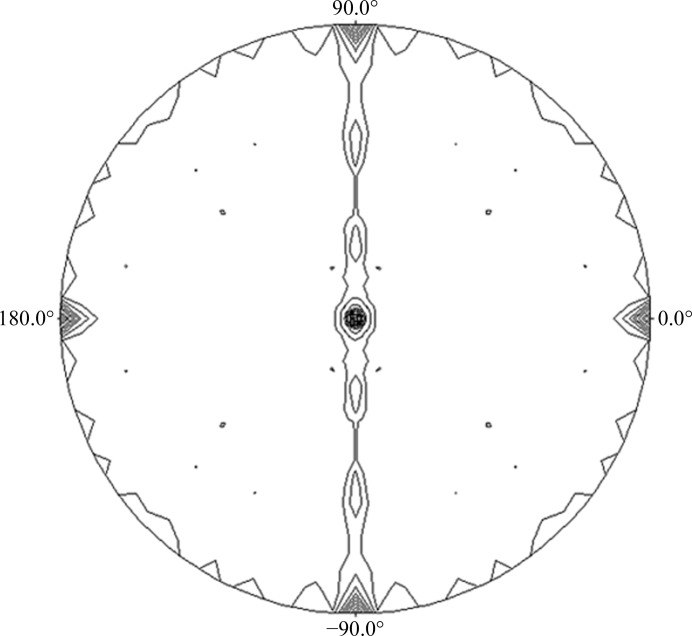
In the initial crystallization trials, 362 commercial screen conditions were tested using the sitting-drop vapour-diffusion method. Precipitate formation and crystal growth were observed in 22 of these conditions, which were then repeated using the hanging-drop vapour-diffusion method. Only four conditions showed crystal growth and were optimized by additive and grid screening. Only one condition (100 mM Tris–HCl pH 8.5, 800 mM LiCl, 8% PEG 4000, 4% of the additive 1-propanol) produced crystals of significant size and quality. The use of 1-propanol as an additive led to larger and more homogenously shaped crystals (Fig. 2 ▶). Analysis of the diffraction data indicated that the crystals belonged to space group P212121, with unit-cell parameters a = 84.4, b = 90.5, c = 120.7 Å, α = β = γ = 90°. The data-processing statistics are given in Table 3 ▶. The diffraction data suggested the presence of four molecules in the asymmetric unit (Matthews coefficient V M of 2.33 Å3 Da−1 and solvent content of 47.23%). The self-rotation function (Fig. 3 ▶) showed four noncrystallographic peaks related by twofold symmetry, confirming the presence of four molecules in the asymmetric unit. Moreover, the presence of three peaks at the ends of mutually perpendicular vectors in the self-rotation function confirmed the 222 symmetry of the crystal.
Figure 2.
GumB crystals grown (a) in 100 mM Tris–HCl pH 8.5, 800 mM LiCl, 8% PEG 4000 and (b) under optimized conditions as in (a) with 1-propanol as an additive. (c) Representative X-ray diffraction pattern of a native GumB crystal.
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | PROXIMA 1 |
| Wavelength (Å) | 0.98011 |
| Temperature (K) | 100 |
| Detector | PILATUS 6M |
| Crystal-to-detector distance (mm) | 486.1 |
| Rotation range per image (°) | 0.2 |
| Total rotation range (°) | 260 |
| Exposure time per image (s) | 0.2 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 84.4, b = 90.5, c = 120.7 |
| Mosaicity (°) | 0.6 |
| Resolution range (Å) | 90.49–2.54 (2.68–2.54) |
| Total No. of reflections | 249621 (35738) |
| No. of unique reflections | 31227 (4482) |
| Completeness (%) | 100 (100) |
| Multiplicity | 8.0 (8.0) |
| 〈I/σ(I)〉 | 11.8 (3.8) |
| R p.i.m. † | 0.034 (0.177) |
| Overall B factor from Wilson plot (Å2) | 62.9 |
Estimated R p.i.m = R merge[1/(N − 1)]1/2, where N is the data multiplicity.
Figure 3.
The self-rotation function was calculated at κ = 180° to identify the twofold rotation angles. The calculations were performed using POLARRFN from CCP4 with data from 9 to 4 Å resolution and an integration radius of 41 Å.
We attempted to resolve the structure of GumB by molecular replacement using Wza (PDB entry 2j58; Dong et al., 2006 ▶), a lipoprotein essential for capsular polysaccharide export in E. coli K30, as the search model. We were not able to find a solution, most likely because the sequence identity between these two proteins was only 29%. At present, the selenomethionine derivative of GumB is being prepared and soaks with several heavy atoms are being performed.
Acknowledgments
We thank Marta Bravo and Soledad Malori (Fundación Instituto Leloir) for their help in DNA sequencing and protein purification, respectively. We acknowledge SOLEIL for provision of synchrotron-radiation facilities and we thank Beatriz Guimaraes and Andrew Thompson for their assistance in using beamline PROXIMA 1. This work was supported by grants PICT 1/2452 from Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT, Argentina) and PIP 399 from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina). MJ and SRS are doctoral fellows of CONICET, MIB is a postdoctoral fellow of ANPCyT and LI is a CONICET Career Investigator.
References
- Baird, J. K., Sandford, P. A. & Cottrell, I. W. (1983). Nature Biotechnol. 1, 778–783.
- Barreras, M., Salinas, S. R., Abdian, P. L., Kampel, M. A. & Ielpi, L. (2008). J. Biol. Chem. 283, 25027–25035. [DOI] [PMC free article] [PubMed]
- Cuthbertson, L., Kos, V. & Whitfield, C. (2010). Microbiol. Mol. Biol. Rev. 74, 341–362. [DOI] [PMC free article] [PubMed]
- Cuthbertson, L., Mainprize, I. L., Naismith, J. H. & Whitfield, C. (2009). Microbiol. Mol. Biol. Rev. 73, 155–177. [DOI] [PMC free article] [PubMed]
- Dong, C., Beis, K., Nesper, J., Brunkan-Lamontagne, A. L., Clarke, B. R., Whitfield, C. & Naismith, J. H. (2006). Nature (London), 444, 226–229. [DOI] [PMC free article] [PubMed]
- Gerlach, R. G. & Hensel, M. (2007). Int. J. Med. Microbiol. 297, 401–415. [DOI] [PubMed]
- Ielpi, L., Couso, R. O. & Dankert, M. A. (1993). J. Bacteriol. 175, 2490–2500. [DOI] [PMC free article] [PubMed]
- Katzen, F., Ferreiro, D. U., Oddo, C. G., Ielmini, M. V., Becker, A., Pühler, A. & Ielpi, L. (1998). J. Bacteriol. 180, 1607–1617. [DOI] [PMC free article] [PubMed]
- Keiski, C.-L., Harwich, M., Jain, S., Neculai, A. M., Yip, P., Robinson, H., Whitney, J. C., Riley, L., Burrows, L. L., Ohman, D. E. & Howell, P. L. (2010). Structure, 18, 265–273. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. J. Read & J. L. Sussman, pp. 41–51. Dordrecht: Springer.
- Okuda, S. & Tokuda, H. (2011). Annu. Rev. Microbiol. 65, 239–259. [DOI] [PubMed]
- Patel, Y., Schneider, J. C., Ielpi, L. & Ielmini, M. V. (2008). US Patent 7439044 B2.
- Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003). Acta Cryst. D59, 1131–1137. [DOI] [PubMed]
- Salinas, S. R., Bianco, M. I., Barreras, M. & Ielpi, L. (2011). Glycobiology, 21, 903–913. [DOI] [PubMed]
- Sathiyamoorthy, K., Mills, E., Franzmann, T. M., Rosenshine, I. & Saper, M. A. (2011). Biochemistry, 50, 5465–5476. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.