Abstract
Sinorhizobium fredii strain USDA191 forms N-fixing nodules on the soybean (Glycine max L. Merr.) cultivars (cvs) McCall and Peking, but S. fredii strain USDA257 nodulates only cv Peking. We wondered whether specificity in this system is conditioned by the release of unique flavonoid signals from one of the cultivars or by differential perception of signals by the strains. We isolated flavonoids and used nodC and nolX, which are nod-box-dependent and -independent nod genes, respectively, to determine how signals activate genes in the microsymbionts. Seeds of cv McCall and cv Peking contain the isoflavones daidzein, genistein, and glycitein, as well as their glucosyl and malonylglucosyl glycosides. Roots exude picomolar concentrations of daidzein, genistein, glycitein, and coumestrol. Amounts are generally higher in cv Peking than in cv McCall, and the presence of rhizobia markedly influences the level of specific signals. Nanomolar concentrations of daidzein, genistein, and coumestrol induce expression of nodC and nolX in strain USDA257, but the relative nolX-inducing activities of these signals differ in strain USDA191. Glycitein and the conjugates are inactive. Strain USDA257 deglycosylates daidzin and genistin into daidzein and genistein, respectively, thereby converting inactive precursors into active inducers. Although neither soybean cultivar contains unique nod-gene-inducing flavonoids, strain- and cultivar-specific interactions are characterized by distinct patterns of signal release and response.
The symbiotic interaction between legume plants and rhizobia from the soil is of immense significance, both in agricultural and in native ecosystems. Symbiotic bacteria become internalized within root nodules, where they fix atmospheric N and make it available for plant growth. The process of nodulation depends on mutual recognition of the symbiotic partners and is characterized by varying degrees of selectivity. Although some strains of rhizobia and species of legumes have nonstringent requirements for symbiotic partners, nodulating associations are for the most part fairly specific (Pueppke, 1996). Such specificity is determined early in the interaction, prior to infection, and is based on the exchange of molecular cues between the growing plant root and rhizobia in the rhizosphere (Long, 1996).
Most of the known plant substances used as signals to rhizobia are flavonoids (Phillips, 1992). Leguminous species synthesize vast and diverse arrays of these compounds (Bisby et al., 1994), and many of them are released into the root zone, where they enjoy unrestricted access to microorganisms. Flavonoids serve as chemoattractants, influence bacterial growth, and selectively activate expression of the nodulation (nod) genes of symbiotic rhizobia (Schlaman et al., 1992; Pueppke, 1996). This latter process requires the regulatory protein NodD, which interacts with the nod box, a conserved, cis-acting promoter element that governs expression of the inducible nod genes. When the appropriate flavonoid signal(s) from the plant is perceived by NodD, the DNA helix in the vicinity of the nod box is deformed so that the adjacent nod genes can be transcribed (Fisher and Long, 1993). Most of these genes in turn function to direct the synthesis and export of Nod factors, return signals that complete the circuit, and stimulate plant responses that ultimately lead to nodule morphogenesis.
Sinorhizobium fredii is an Asian species with unusual specificity for legumes (Keyser et al., 1982). In addition to its original host, soybean (Glycine max), this symbiont nodulates more than 50 diverse legumes (S.G. Pueppke and W.J. Broughton, unpublished data). Therefore, in one sense it is a broad-host-range symbiont, but on the other hand, some S. fredii strains are highly specific for certain soybean cultivars (Keyser et al., 1982; Heron and Pueppke, 1984). Strain USDA257 typifies this group. Although it forms N-fixing nodules on a few soybean cultivars, including Peking, most soybeans are not nodulated or respond with abnormal root proliferations (Balatti and Pueppke, 1992). We have examined one such cultivar, McCall, and know that infection by strain USDA257 ceases prior to the appearance of the infection thread in root hairs (Chatterjee et al., 1990). Other S. fredii strains, including USDA191, are not cultivar specific, forming typical N-fixing nodules on cv McCall and all other soybean cultivars that have been tested (Keyser et al., 1982; Heron and Pueppke, 1984).
The inability of strain USDA257 to nodulate cv McCall is controlled by a complex genetic locus, nolXWBTUV, which contains at least six genes (Heron et al., 1989; Meinhardt et al., 1993; Kovács et al., 1995). Insertional inactivation of any of these genes allows mutant bacteria to form N-fixing nodules on cv McCall, and levels of fixed N are comparable to those recorded for wild-type strain USDA191 (Chatterjee et al., 1990). Expression of nolX and the nolBTUV transcriptional unit is controlled by flavonoid signals released by the host and is under the jurisdiction of NodD, even though nod-box promoters are absent (Meinhardt et al., 1993; Kovács et al., 1995; Bellato et al., 1996, 1997a, 1997b).
Here we address two major hypotheses to explain the differential nodulation phenotype of the soybean cvs McCall and Peking in response to S. fredii strains USDA191 and USDA257. The first is that specificity depends on differential production of unique nod-gene-activating flavonoid signals by one or the other cultivar. The second is that specificity depends on the differential abilities of the two bacteria to perceive flavonoids. Together, these hypotheses predict that incompatibility between cv McCall and strain USDA257 is due to an inadequate signaling environment. We have identified flavonoids from seeds of cvs McCall and Peking, quantified signal molecules in the root exudates of both cultivars and several lines of Glycine soja, and measured the impact of strains USDA191 and USDA257 on the flavonoid composition of such exudates. In parallel, we have quantified the capacities of flavonoids from cvs McCall and Peking to induce expression of nod genes in strains USDA191 and USDA257 and assessed the ability of strain USDA257 to convert flavonoid conjugates into functional nod-gene inducers.
MATERIALS AND METHODS
Biological Materials
Seeds of the soybean (Glycine max [L.] Merr.) cvs Peking and McCall were obtained from Dr. S.C. Anand (Delta Center, University of Missouri, Portageville) and Eric Pueppke (Erie, ND), respectively. Dr. Randall Nelson (U.S. Department of Agriculture/Agricultural Research Service, University of Illinois, Urbana) provided lines of G. max and Glycine soja Sieb. & Zucc. Seeds were germinated aseptically and nodulation was assessed in hydroponic culture vessels (Pueppke, 1983; Krishnan and Pueppke, 1991).
Sinorhizobium fredii strain USDA191 was obtained from the collection of the U.S. Department of Agriculture (Beltsville, MD; Keyser et al., 1982). Strain 257S1, a kanamycin-resistant derivative of strain USDA257, retains the symbiotic phenotype of the parental strain and was used in nodulation experiments (Heron and Pueppke, 1984; Heron et al., 1989). The three gene fusions used as reporter constructions are designated 257nodC, 257nolX, and 191nolX. Each contains a copy of minimu that had been transferred by homologous recombination into the coding region of a single gene; therefore, expression can be measured as β-galactosidase activity. 257nodC corresponds to 257B17 and contains the transposon in nodC (Krishnan and Pueppke, 1991). 191nolX corresponds to RfCB26, a strain with the transposon at nucleic acid position +646 with respect to the translational start site of nolX (Bellato et al., 1997b). 257nolX corresponds to mutant 411mu78 of Meinhardt et al. (1993) and contains the minimu element at nucleic acid position +439 with respect to the translational start site of nolX. Bacteria were stored and cultured in yeast extract-mannitol medium (Krishnan and Pueppke, 1991).
Isolation of Flavonoids
Seeds of cvs McCall and Peking (50 g of each) were extracted overnight in 50 mL of 50% aqueous ethanol with shaking. Each seed extract was then dried under vacuum and dissolved in 15 mL of 10% aqueous ethanol. Aliquots of 5 mL were loaded onto C18 Sep-Pak cartridges (Waters). After the samples were washed with 3 mL of 10% aqueous ethanol, flavonoids were eluted with 5 mL of 50% aqueous ethanol, dried, redissolved in 2 mL of 50% aqueous ethanol, passed through 0.2-μm filters, and stored at −70°C.
Root exudates were collected from pregerminated seedlings growing in 2.3-mL test tubes containing 1 mm CaCl2 in 5 mm Mes buffer at a final pH of 6.8 (Bolaños-Vásquez and Werner, 1997). Each tube contained a 1.0- × 6.5-cm strip of cellulose acetate filter (type OE66, Schleicher & Schuell) and received either a single cv McCall or cv Peking seedling or four G. soja seedlings. Cells of S. fredii strain USDA191 or 257S1 were included in some tubes with cvs McCall and Peking, at a concentration of 107 cells mL−1. Filter strips were removed after incubation for 48 h and rinsed, and the flavonoids were eluted quantitatively and then stored at −20°C (Bolaños-Vásquez and Werner, 1997). Seedlings from treatments containing bacteria were aseptically transferred to plastic growth pouches (Pueppke, 1983) and grown for 2 weeks to confirm symbiotic phenotypes and to ensure that the uninoculated controls had remained free of S. fredii.
Purification and Analytical Characterization of Flavonoids
Seed extract (20–100 μL) was loaded onto an HLPC system (Gilson, Middleton, WI) fitted with an analytical C18 reverse-phase 10-μm Nucleosil column (7.5 × 250 mm; Aldrich). The column was eluted isocratically for 5 min with 25% aqueous methanol, for 20 min with a linear gradient from 25 to 55% methanol, and then for 5 min with a linear gradient from 55 to 75% methanol. The gradient was subsequently increased to 100% methanol, and the column was washed for 5 min in this solvent. The flow rate was 2.0 mL min−1, and the eluant was monitored at 262 nm with an absorbance detector (Shimadzu, Columbia, MD). UV-absorbing compounds were collected and samples pooled and rechromatographed as necessary prior to further analysis.
Electrospray MS was performed with a quadrupole instrument (VG-Trio 2000, Fisons VG, Manchester, UK). The electrostatic spray ion source was operated at atmospheric pressure and 4 kV, with an extraction cone value of 45 V. Dried HPLC fractions were dissolved in 100 μL of 50% aqueous acetonitrile or 50% aqueous ethanol prior to analysis. Compounds were eluted at a rate of 10 μL min−1 with an isocratic solvent system of 50% aqueous acetonitrile containing 2% formic acid.
Carbohydrates were determined after hydrolysis of fractions for 4 h in 4 n trifluoroacetic acid at 110°C. After evaporation of the acid, flavonoids were removed by organic extraction with dichloromethane. The residual aqueous fraction was dried, and glycosyl components were trimethylsilylated with Tri-Sil (Pierce) and analyzed by GC (series 30, Girdel, France) on an instrument fitted with an OV1 column (0.32 mm × 12 m, 0.1 μm, Spiral Biotech, Bethesda, MD) and a flame-ionization detector. The temperature gradient was 2°C min−1 from 100 to 250°C.
Flavonoids in root exudates were fractionated with an HPLC system (model 2152, LKB, Cambridge, UK) fitted with an ODS column (5 μm, 250 × 4 mm; Hypersil, Hewlett-Packard). Solvent A was water that was adjusted to pH 3.3 with acetic acid, and solvent B was a mixture of acetonitrile and methanol (20:25, v/v). The gradient began at 30% solvent B, was increased over 25 min to 60% solvent B, and ended with a 5-min isocratic elution, all at a flow rate of 1.0 mL min−1. The eluant was monitored from 195 to 365 nm with a diode-array detector (Spectra Focus, Spectra-Physics, San Jose, CA), and spectra were analyzed using the associated software (SpectraSYSTEM). The integrator function of the software package was used to determine flavonoid concentrations on the basis of standard curves that had been prepared with authentic compounds (Bolaños-Vásquez and Werner, 1997).
Flavonoid standards were from the following sources: coumestrol was from Eastman Kodak; daidzein, genistein, and quercetin were from Roth Chemicals (Karlsruhe, Germany); daidzin, formononetin, and kaempferol were from Apin Chemicals (Abingdon, UK); and genistin was from Sigma. Isoliquiritigenin (Kape et al., 1992) and glycitein (Naim et al., 1973) were purified from soybean roots and seeds, respectively, and their identities were confirmed by MS.
nod-Gene-Inducing Activities of Flavonoids
The capacities of flavonoids to induce the expression of nodC-lacZ and nolX-lacZ gene fusions were assessed by the protocol of Krishnan and Pueppke (1991). β-Galactosidase activities were measured by the method of Miller (1972). HPLC fractions from seed extracts were diluted in ethanol to give an Amax = 0.5, and 20-μL aliquots were added to 4-mL cultures. HPLC fractions from the root exudates of individual soybean seedlings were dissolved in 50 μL of methanol and dispensed into 3.0-mL bacterial cultures. Methanolic solutions of authentic flavonoid standards were delivered to achieve final concentrations ranging from 25 to 2000 nm, depending on the compound analyzed.
The time course for induction of nolX in strain USDA257 by daidzein and daidzin was measured with a series of 5.0-mL cultures supplemented with 100 nm flavonoid. Controls received either methanol alone or flavonoid, but no bacteria. Cultures were harvested in duplicate after incubation for 12, 24, 48, 72, or 96 h. Turbidity was measured immediately as A625, and bacteria were pelleted from 0.5-mL aliquots and stored at −20°C for later assay of β-galactosidase activity. Three milliliters of supernatant solution from each culture was frozen and later extracted twice with equal volumes of ethyl acetate. The organic fractions were then dried, and concentrations of daidzein were quantified by HPLC as described above.
RESULTS
Flavonoids in cv McCall and cv Peking Seeds
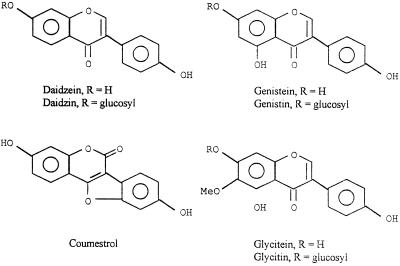
Using HPLC, we detected 10 UV-absorbing fractions in aqueous ethanolic extracts of cv Peking seeds (Table I). Approximately equivalent amounts of nine of these fractions were also present in cv McCall seed extracts, but a 10th peak, fraction 4, was found only in cv Peking. The first two rapidly eluting fractions were unresolved mixtures without nod-gene-inducing activity, and we did not attempt to purify them further. cv Peking-specific fraction 4 also lacked activity and was not additionally investigated. All of the other peaks represented flavonoids or flavonoid conjugates (Fig. 1), as determined by MS and GC analysis. Fraction 3 was a mixture of three substituted isoflavone glucosides: malonyldaidzin, malonylgenistin, and malonylglycitin. The (M + H)+ molecular ions of these compounds appeared at m/z 503, 519, and 533, respectively. Fractions 5, 6, and 7 contained the three nonmalonated glucosides: daidzin (m/z 417), glycitin (m/z 447), and genistin (m/z 433), respectively. The corresponding free aglycones then eluted from the column in the same relative order as the glucosides. Fraction 8 was daidzein (m/z 255), fraction 9 was glycitein (m/z 285), and fraction 10 was genistein (m/z 271). Only two of the fractions, fraction 8 (daidzein) and fraction 10 (genistein), showed measurable nod-gene-inducing activity, and both were active with nodC and nolX (Table I).
Table I.
HPLC analysis of nod-gene-inducing flavonoids from cv Peking seeds
| Fraction | Retention Time | Identitya | Inductionb
|
|
|---|---|---|---|---|
| nolX | nodC | |||
| min | Miller units | |||
| 1 | 3.3 | Unknown mixture | 1.0 ± 0.1 | 1.0 ± 0.1 |
| 2 | 4.1–7.8 | Unknown mixture | 1.0 ± 0.1 | 1.1 ± 0.2 |
| 3 | 9.5–12.3 | Malonylisoflavone glucosides | 1.0 ± 0.1 | 1.3 ± 0.2 |
| 4 | 18.8 | Unknown | 1.0 ± 0.1 | 0.9 ± 0.1 |
| 5 | 23.0 | Daidzin | 1.1 ± 0.1 | 1.6 ± 0.4 |
| 6 | 24.1 | Glycitin | 1.0 ± 0.2 | 1.1 ± 0.1 |
| 7 | 27.1 | Genistin | 1.1 ± 0.1 | 1.2 ± 0.1 |
| 8 | 33.6 | Daidzein | 4.7 ± 0.6 | 8.4 ± 0.1 |
| 9 | 34.2 | Glycitein | 1.2 ± 0.1 | 1.4 ± 0.1 |
| 10 | 36.0 | Genistein | 6.3 ± 0.7 | 8.9 ± 0.8 |
As determined by MS and GC analysis. Copies of chromatograms and mass spectra are available from the corresponding author upon request.
Data are means from two independent experiments ± sd, each with two replicates. HPLC fractions were diluted in ethanol to Amax = 0.5, and 20-μL aliquots were added to 4-mL cultures of 257nolX or 257nodC.
Figure 1.
Structures of flavonoids from seeds and roots of cvs McCall and Peking.
S. fredii Converts Isoflavone Glucosides into nod-Gene Inducers
The inability of genistin and daidzin from seeds to elevate expression of nod genes in S. fredii was intriguing, because these glucosides are nod-gene inducers in a distantly related symbiotic bacterium, Bradyrhizobium japonicum (Smit et al., 1992). Repeated β-galactosidase assays with authentic genistin and daidzin nevertheless confirmed the absence of both nolX- and nodC-inducing activity after the standard 24-h incubation period. We used 257nolX and daidzin to determine whether S. fredii can deglycosylate such conjugates after prolonged incubation, thereby releasing nod-gene-inducing aglycones. Table II shows that the aglycone daidzein induces expression of nolX after just a 12-h incubation period. Activity increases very rapidly thereafter and reaches a plateau, about 700 Miller units, after 72 h. Each bacterial culture originally contained 500 pmol of daidzein in these experiments, and between 75 and 90% of the inducer could be recovered from the medium, depending on culture age.
Table II.
Relationship between daidzin deglycosylation and induction of nolX expression in S. fredii strain USDA257
| Incubation Time | Daidzein Treatment
|
Daidzin Treatment
|
||
|---|---|---|---|---|
| Daidzein recoverya | nolX activityb | Daidzein recoverya | nolX activityb | |
| h | pmol | Miller units | pmol | Miller units |
| 12 | 413 ± 49 | 57 ± 5 | 2 ± 3 | 0 |
| 24 | 432 ± 15 | 124 ± 16 | 7 ± 1 | 0 |
| 48 | 456 ± 8 | 475 ± 15 | 13 ± 3 | 15 ± 4 |
| 72 | 374 ± 8 | 700 ± 31 | 18 ± 4 | 122 ± 1 |
| 96 | 373 ± 31 | 727 ± 64 | 20 ± 4 | 279 ± 1 |
Values are means ± sd.
Daidzein and daidzin were supplied at an initial concentration of 100 nm. The mean recovery of daidzein from bacteria-free, daidzein-containing control tubes was 458 ± 90 pmol. Daidzein was not recovered from bacteria-free, daidzin-containing control tubes, except at 72 h, when 3.7 pmol was detected.
Values from methanol-treated controls have been subtracted from those given in the table. These controls yielded an average of 8, 15, 28, 28, and 25 Miller units of β-galactosidase activity, respectively, after 12, 24, 48, 72, and 96 h of incubation.
As expected, nolX was not expressed in 12- and 24-h-old cultures that had been induced with 100 nm daidzin (Table II). nolX activity was first detected after 48 h, and it increased substantially thereafter; by 96 h, β-galactoside levels in daidzin-treated cultures were nearly 40% of those in daidzein-treated cultures. Traces of the aglycone could be detected in daidzin-treated cultures at all times, but its concentration did not exceed 10 nm until 48 h, when low levels of nolX expression first became apparent. With the exception of the 72-h control, which contained 3.7 pmol of free daidzein, none of the bacteria-free, daidzin-treated tubes contained aglycone as a breakdown product. Genistin-treated cultures also lacked nolX-inducing activity after 24 h (1.0- to 2.1-fold induction of gene expression after exposure to 250 nm genistin; n = 3). As was the case with daidzin, HPLC-detectable genistein appeared and gene expression was elevated as the cultures aged (data not shown).
Flavonoid Levels in Root Exudates Depend on Cultivar and Strain
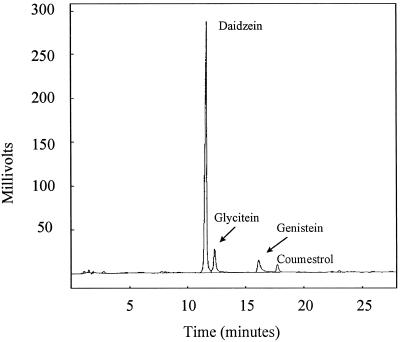
The arrays of flavonoids released by individual cv McCall and cv Peking roots were qualitatively indistinguishable from one another and considerably less complex than those of seeds. Although very minor peaks were inevitably present, only four compounds were detected in measurable quantity (Fig. 2). Three of them, daidzein, glycitein, and genistein, correspond to isoflavones that had been detected in seeds. The fourth, coumestrol, is a coumestan with the basic hydroxylation pattern of daidzein (Fig. 1).
Figure 2.
HPLC profile of flavonoids exuded from the roots of a single cv Peking seedling. A250 was measured.
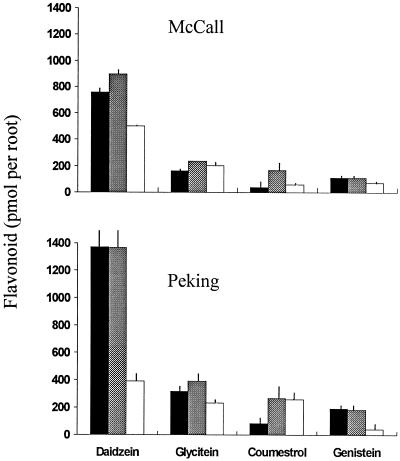
Levels of these four compounds in root exudates of the two types of soybean are shown in Figure 3. Although roots of both cvs McCall and Peking released all four flavonoids, there was a striking difference between the cultivars. Flavonoid concentrations in bacteria-free cv Peking exudates were nearly twice that of those in cv McCall exudates. This differential was maintained in seedlings that had been inoculated with rhizobia, but differences were statistically significant only with daidzein. Daidzein was by far the dominant isoflavonoid in root exudates of both cultivars, whether or not rhizobia were present. Indeed, with the exception of the interaction between cv Peking and strain 257S1, the concentration of daidzein was always greater than the sum of the remaining three compounds combined. Rhizobia influenced isoflavonoid concentrations, often significantly. Strain 257S1 reduced daidzein levels by 72% in cv McCall and by 34% in cv Peking, but strain USDA191 had little or no effect. The influence of rhizobia on the other flavonoids was not so clear-cut. However, both strains significantly elevated coumestrol levels in cv Peking, and strain 257S1 decreased genistein in this cultivar.
Figure 3.
Flavonoid content of cv McCall and cv Peking root exudates in the absence of bacteria (black bars) or in the presence of strain USDA191 (gray bars) or strain 257S1 (white bars). The bars represent means of measurements with individual seedlings (n = 10 to 16, depending on treatment), and the vertical lines represent se values.
We assayed the nod-gene-inducing activities of fractions collected from the HPLC column with both 257nolX and 257nodC. Included were the tiny peaks that are barely visible in chromatograms (Fig. 2) but were nonetheless potential inducers. Only daidzein and coumestrol yielded measurable activity. The daidzein fractions led to 5.7- to 11.6-fold induction of nolX expression (n = 7) and 5.4- to 14.1-fold induction of nodC expression (n = 5). Induction by coumestrol averaged 57 ± 9 and 55 ± 8% of these levels, respectively.
Soybean Flavonoids Differentially Activate nodX and nodC
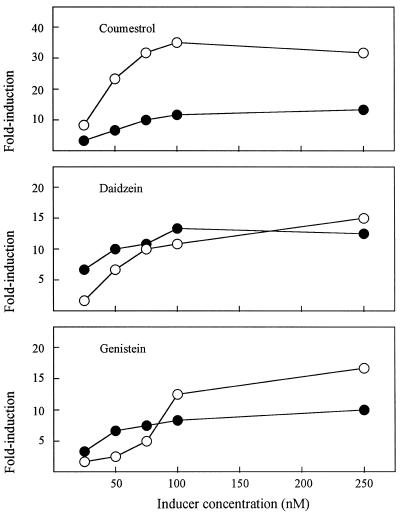
The availability of 257nolX and 257nodC allowed us to assess directly and quantitatively the activities of signals released by roots of the two soybean cultivars against a nod-box-independent and a nod-box-dependent gene (Fig. 4). The responsiveness of the two genes to coumestrol differed greatly, especially at concentrations greater than 50 nm, where nolX is activated to a much higher level than nodC. The greater than 30-fold induction of this gene at coumestrol concentrations of 75 nm and higher was more than twice the response to either of the isoflavones from root exudates. In contrast, the dose-response curves for induction of the two genes by daidzein paralleled one another fairly closely (Fig. 4). The genistein curves represent yet a third pattern: nodC was more highly inducible at lower concentrations but nolX was more responsive at higher concentrations. All three inducers from root exudates were active at the lowest concentration tested, 25 nm, when induction was as great as 8.1-fold (coumestrol against nolX). All responses approached saturation at or near 100 nm, i.e. elevation of the inducer concentration to 250 nm inevitably led to little or no additional activity. Active concentrations all were well within the range of flavonoid concentrations expected in the rhizosphere (Fig. 3).
Figure 4.
Induction of the expression of nolX (○) and nodC (•) by flavonoids detected in cv Peking and cv McCall root exudates. Data are means from two to four independent experiments. βGalactosidase activity in methanol controls was 5.2 to 9.4 Miller units for nodC and 25.4 to 41.4 Miller units for nolX. sd values were calculated but are less than the width of the symbols.
We compared the nod-gene-inducing activities of flavonoids from root exudates to those of four other potential signals from soybean. Three of these compounds have been detected only in leaves: the isoflavone formononetin (Osman and Fett, 1983), the flavonol kaempferol (Porter et al., 1985), and the hydroxyflavonol quercetin (Porter et al., 1985). We also examined isoliquiritigenin, a trihydroxychalcone from root exudates of soybean cv Maple Arrow and a potent nod-gene inducer in B. japonicum (Kape et al., 1992). Although these four flavonoids all induce expression of both nod genes, they are significantly less active than the signals from root exudates. In replicate experiments kaempferol and quercetin were both active in the same concentration range as the root signals, but maximum levels of induction fell in the range of 5- to 8-fold. Formononetin and isoliquiritigenin at 25 to 100 nm triggered no significant expression of either gene, but at micromolar concentrations, both compounds became inducers of one or both genes. Isoliquiritigenin at 2000 nm, for example, elevated expression of nolX to levels comparable to that achieved with 75 nm coumestrol.
We used all seven compounds to determine whether 257nolX and 191nolX differ in their inherent responsiveness to flavonoids produced by soybean. Each compound was assayed in both strains at a concentration of 100 nm in replicate experiments. Previously constructed dose-response curves with 257nolX (Fig. 4; data not shown) predicted that the relative inducer activities of coumestrol, daidzein, genistein, quercetin, kaempferol, formononetin, and isoliquiritigenin would be approximately 100:45:40:30:20:10:5, respectively, under these conditions. The actual relative potencies (n = 6) were 100:55:34:23:8:7:7, in good agreement with the expected values. The corresponding values for the strain USDA191 genetic background (n = 6) were 100:190:67:8:27:6:4, indicating that on a relative scale daidzein and, to a lesser extent, genistein are significantly more active inducers of nolX in strain USDA191 than in strain USDA257.
Flavonoids in Root Exudates of Cultivar-Specific and -Nonspecific G. soja
The symbiotic responses of only a few wild soybean lines to S. fredii have been documented (Keyser et al., 1982; Heron and Pueppke, 1984; Keyser and Cregan, 1984). We wished to examine strain specificity of G. soja more broadly and then relate it to release of signals by roots as independent verification of our observations with cvs Peking and McCall. Thirty-six randomly chosen G. soja lines were assayed with strain 257S1. Twenty formed normal nodules: PI378685, 378686B, 378893A, 407075, 407158, 407285, 407320, 424033, 424124, 464939B, 479751, 507597, 507606, 507613, 507624, 507734, 507786, 507803, 507830B, and 518281. The remaining 16 lines were either nodule free or formed abnormal, nonfixing structures as described by Balatti and Pueppke (1992): PI245331, 339371A, 398220, 407055, 407272, 407283, 407297, 423991, 424022A, 424119, 479753B, 479767, 507625, 512279A, 518280, and 522196A. We used the two sets of germ plasm to test the hypothesis that the flavonoids released by G. soja lines capable of nodulation with strain USDA257 differ quantitatively or qualitatively from those of nonnodulating lines.
Root exudates from a total of 105 samples, each collected from seedlings of a single G. soja line, were fractionated by HPLC (most lines were represented by three samples, but the range was two to five). We then analyzed the relative concentrations of flavonoids released by nodulating versus nonnodulating genotypes. Daidzein, coumestrol, and genistein made up 69.6 ± 1.2, 4.3 ± 1.0, and 6.8 ± 0.5%, respectively, of the total in the nonnodulating lines (means ± se, n = 49). The corresponding values for nodulating lines (n = 56) were 69.9 ± 1.2, 3.6 ± 0.4, and 7.6 ± 0.4%, respectively. In each case, the residual percentage represents the noninducer glycitein. The remarkable agreement between the two groups indicates that the relative proportions of released nod-gene inducers are the same.
Most of the HPLC spectra from G. soja resembled those of soybean (Fig. 2), but about one-third of the samples contained additional, comparatively small peaks eluting between 24 and 26 min after injection. The peaks were visible in each of the root exudate samples of 11 lines. Seven of them nodulated and four did not, and thus the presence of the compounds was not related to strain specificity. The long retention times of these peaks suggested that they are hydrophobic and might represent the phytoalexin glyceollin (Parniske et al., 1991). Although an authentic mixture of glyceollin isomers (Kape et al., 1992) eluted as two peaks with retention times between 25.6 and 27.4 min, the UV-absorption spectra of the unknowns (data not shown) did not match those of known glyceollin isomers (Lyne et al., 1976; Parniske et al., 1991).
DISCUSSION
S. fredii nodulates cvs McCall and Peking, but the interaction is specific. Strain USDA191 fixes N with both cultivars, but strain USDA257 enters into symbiosis only with cv Peking (Heron and Pueppke, 1984). Selective activation of nod genes by flavonoid signals is known to regulate symbiotic interactions at the level of bacterial and legume species (Schlaman et al., 1992; Pueppke, 1996); therefore we wondered whether similar sorts of cues from the plant govern the finer degree of specificity that characterizes the soybean-S. fredii system, and, if so, how might such control be imposed? One hypothesis is that the types of flavonoids produced by cv Peking, a dark-seeded cultivar, differ from those of cv McCall, a light-seeded cultivar, in a manner analogous to the known variability in flavonoid synthesis by black-and-white-seeded beans (Hungria et al., 1991; Hungria and Phillips, 1993). A second hypothesis is that the bacterial strains differ fundamentally in their capacities to perceive and respond to flavonoid signals. This possibility seemed reasonable, because strains USDA191 and USDA257 originate from different geographical regions of China (Keyser et al., 1982) and both contain nod-box-dependent genes such as nodABC (Krishnan and Pueppke, 1991) and nod-box-independent genes such as nolX (Heron et al., 1989; Bellato et al., 1997a).
Elaboration of Flavonoid Signals by Soybean
Legume seeds serve as abundant reservoirs of flavonoids (Bisby et al., 1994), some of which can be released into the soil during germination (Graham, 1991; Smit et al., 1992), so we began by assessing the potential contributions of these signals to cultivar specificity in the soybean-S. fredii system. The pigments of dark-seeded soybeans are alcohol-soluble anthocyanidin glycosides (Buzzell et al., 1987; Todd and Vodkin, 1993). Related flavonoids are the dominant nod-gene inducers in black-seeded beans, which are much more effective than white-seeded cultivars in signaling to Rhizobium leguminosarum bv phaseoli (Hungria et al., 1991; Hungria and Phillips, 1993). In preliminary experiments we nevertheless detected equivalent total levels of nolX-inducing activities in seed extracts of cv Peking and cv McCall soybeans. These data, in conjunction with the observed insensitivity of strain USDA257 to flavonoid glycosides, make it unlikely that such compounds serve as cv Peking-specific signals.
Indeed, with the exception of a single peak that is present in cv Peking but not in cv McCall extracts, the HPLC profiles of flavonoids from the two types of seeds are virtually identical. Both cultivars accumulate daidzein, genistein, and glycitein, their glucosides, and the corresponding malonylglucosides. Various combinations of these derivatives have been isolated previously from seeds of other soybean varieties (Ohta et al., 1979; Graham, 1991; Kudou et al., 1991; Smit et al., 1992; Bisby et al., 1994), and neither the minor or unresolved peaks nor the cv Peking-specific peak had significant nod-gene-inducing activity. It therefore seems unlikely that the seed flavonoids of cvs Peking and McCall are unique or different from one another to the extent that they control cultivar specificity.
We also quantified the exuded signals of cvs McCall and Peking and, in parallel, the flavonoids of cv McCall-like and cv Peking-like lines of the wild progenitor G. soja, a species with root exudates of unknown composition. In contrast to earlier analyses, which often were made on batches of seedlings after incubation times of 2 weeks or longer (d'Arcy-Lameta, 1986; Kape et al., 1992; Smit et al., 1992), we examined large numbers of individual seedlings and used short incubation times that correspond to the period required for infection of root hairs by the bacterium (Turgeon and Bauer, 1982). Our data therefore are quantitative and suitable for statistical analysis. In spite of differences in the cultivar, incubation time, nutrient conditions, and growth media, our observations are in remarkable agreement with the more limited data of others (d'Arcy-Lameta, 1986; Schmidt et al., 1994; Bolaños-Vásquez and Werner, 1997).
During a 2-d incubation period, individual cv McCall and cv Peking roots released coumestrol and the three isoflavones present in seeds, including glycitein (Naim et al., 1973), an isoflavone not previously detected in soybean root extracts. Isoflavonoid glycosides were never present, although they have been detected in other cultivars after more prolonged incubation times (Smit et al., 1992; Rao and Cooper, 1995). The relative proportions of the three nod-gene inducers are fairly uniform in cv McCall, cv Peking, and G. soja (our data) and in several other soybean cultivars (Kape et al., 1992; Schmidt et al., 1994), but the flavonoid levels in cv Peking root exudates are remarkably high; daidzein concentrations, for example, averaged 1371 pmol seedling−1 in root exudates of this cultivar.
Inoculation of cvs Peking or McCall with S. fredii leads to the appearance of neither degradation products nor novel flavonoids. This contrasts with vetch (van Brussel et al., 1990; Recourt et al., 1991), alfalfa (Dakora et al., 1993), and subterranean clover (Lawson et al., 1996), in which inoculation triggers the appearance of new nod-gene inducers. S. fredii can nevertheless reduce levels of flavonoids in the root zone, as is the case with strain USDA257 and daidzein, apparently by attenuating their release. B. japonicum has just the opposite effect—it elevates daidzein, coumestrol, and genistein concentrations, as well as that of the pterocarpan phytoalexin glyceollin (Schmidt et al., 1992, 1994).
Some flavonoids are detectable only under special conditions or are present at low concentrations (Graham and Graham, 1996). The phytoalexin glyceollin, for example, appears in root exudates of cv Preston soybean following inoculation with a very high concentration of B. japonicum (Schmidt et al., 1992). Isoliquiritigenin has also been identified as a minor component in cv Preston root exudates that had been collected batchwise and analyzed by procedures similar to ours. Its concentration was estimated to be about one-third that of coumestrol (Kape et al., 1992; Rao and Cooper, 1995). We were nevertheless unable to detect this chalcone in exudates from individual cv McCall and cv Peking seedlings under conditions in which coumestrol was readily identified. Although we cannot exclude the possibility that it is present at a very low level, the high isoliquiritigenin concentration required for significant nod-gene-inducing activity in S. fredii argues against a role in nodulation of cvs Peking or McCall.
Our analysis of flavonoids in plants allows us to rule out two explanations for the differential specificity of S. fredii with cvs Peking and McCall: the constitutive presence of unique nod-gene inducers in root exudates of one of the two cultivars and the induction of new flavonoid signals specifically in response to one of the strains.
Responsiveness of nolX and nodC of S. fredii to Flavonoid Signals from Soybean
Apparently unique among rhizobia, S. fredii places some of its nod genes under the control of conventional nod boxes but regulates others independently of this element (Sadowsky et al., 1988; Krishnan and Pueppke, 1991; Meinhardt et al., 1993; Boundy-Mills et al., 1994; Bellato et al., 1996). Genistein and daidzein are inducers of both nodC and nolX in strain USDA257, and these two isoflavones also function as signals from soybean to the distantly related N-fixing symbiont B. japonicum (Kosslak et al., 1987; Banfalvi et al., 1988; Göttfert et al., 1988; Sutherland et al., 1990; Krishnan and Pueppke, 1991; Smit et al., 1992; Meinhardt et al., 1993; Bellato et al., 1996). We sought to systematically provide answers to four key questions about the response of strains USDA191 and USDA257 to these and other flavonoids: (a) which other flavonoids in cvs Peking and McCall are inducers and how do their potencies compare with daidzein and genistein?; (b) do the exuded signals preferentially activate nodC or nolX?; (c) does the response of the cultivar-nonspecific strain USDA191 differ from that of strain USDA257?; and (d) can S. fredii convert isoflavone glucosides from seeds into nod-gene inducers?
We found that three of the flavonoids in root exudates—daidzein, genistein, and coumestrol—are potent activators of both nod genes in strain USDA257. The responsiveness of nolX and nodC to genistein and daidzein is approximately similar, but there is a strong bias in favor of nolX with coumestrol. This coumestan was not previously recognized to activate expression of this gene, and although its levels are lower than those of daidzein, its contribution to nolX induction by root exudates must nevertheless be disproportionately great. This sort of differential is the first clear indication that soybean roots can selectively influence expression of the nod-box-independent nod genes in S. fredii and forms the basis for more detailed experiments that appear elsewhere (Bellato et al., 1997b). Genistein and daidzein are relatively more potent inducers of nolX in strain USDA191 than in strain USDA257; therefore, the genetic background of the microbe clearly fine-tunes responsiveness to this gene. The promoters of nolX from strains USDA191 and USDA257 are in fact structurally identical (Meinhardt et al., 1993; Bellato et al., 1997a). Selective activation of strain USDA191 therefore must reflect symbiotically significant physiological differences such as signal uptake (Hubac et al., 1993).
Although minor flavonoids represent possible sources of inducers in root exudates, their low abundance makes it unlikely that they are symbiotically significant. However, isoflavone glycosides are abundant in seeds and thus represent potentially significant reservoirs of signaling molecules. Although there is evidence that S. fredii strain HH103 can degrade genistin and daidzin into unidentified metabolites (Rao and Cooper, 1995), neither glycoside itself induces expression of nod genes in S. fredii unless incubation is prolonged. We used quantitative HPLC to prove that nolX expression is elevated soon after daidzein levels begin to increase in daidzin-treated cultures and that no further degradation products appear during a 4-d incubation period. Therefore, S. fredii can liberate signals from inactive isoflavonoid glucosides of seeds, and these conjugates must be viewed as sources of nod-gene inducers in the S. fredii-soybean system. This interplay is precisely the opposite in B. japonicum, which cannot hydrolyze flavonoid glycosides but responds to them directly as inducers of nodABC (Smit et al., 1992).
In summary, our data lead us to reject the first hypothesis to explain cultivar-specific nodulation in the S. fredii-soybean system: that cv Peking or cv McCall releases unique nod-gene inducers. These two cultivars nevertheless do present the bacteria with unique signaling environments, but differences are quantitative and regulated in part by the bacterium itself. Our data provide support for the second hypothesis, that strains USDA191 and USDA257 respond differentially to cues from the plant. It is therefore almost certain that nodX and nodC are expressed differently in strain USDA191 than in strain USDA257, depending on whether the microbe is in the cv Peking rhizosphere or in the cv McCall rhizosphere.
Footnotes
This research was supported by the Food for the 21st Century Program, University of Missouri (Columbia), and by the National Science Foundation (grant no. 96112441 awarded to S.G.P.). This is journal series no. 12,745 of the Missouri Agricultural Experiment Station.
LITERATURE CITED
- Balatti PA, Kovács LG, Krishnan HB, Pueppke SG. Mol Plant-Microbe Interact. 1996;8:693–699. doi: 10.1094/mpmi-9-0457. [DOI] [PubMed] [Google Scholar]
- Balatti PA, Pueppke SG. Identification of North American soybean lines that form nitrogen-fixing nodules with Rhizobium fredii USDA257. Can J Plant Sci. 1992;72:49–55. [Google Scholar]
- Banfalvi Z, Nieuwkoop A, Schell M, Besl L, Stacey G. Mol Gen Genet. 1988;214:420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- Bellato C, Balatti PA, Pueppke SG, Krishnan HB. Proteins from cells of Rhizobium fredii bind to DNA sequences preceding nolX, a flavonoid-inducible nod gene that is not associated with a nod box. Mol Plant-Microbe Interact. 1996;9:457–463. doi: 10.1094/mpmi-9-0457. [DOI] [PubMed] [Google Scholar]
- Bellato C, Krishnan HB, Cubo T, Temprano F, Pueppke SG. Microbiology. 1997a;143:1381–1388. doi: 10.1099/00221287-143-4-1381. [DOI] [PubMed] [Google Scholar]
- Bellato C, Pueppke SG, Krishnan HB. Regulation of the expression of the nod box-independent nodulation gene, nolX, in Sinorhizobium fredii, a nitrogen-fixing symbiont of legume plants. FEMS Microbiol Lett. 1997b;157:13–18. [Google Scholar]
- Bisby FA, Buckingham J, Harbourne JB, Zarucchi JL, Polhill RM, Adams BR, Lock JM, White RJ, Bowes I, Hollis S and others. Phytochemical Dictionary of the Leguminosae, Vol 1: Plants and Their Constituents. London: Chapman and Hall; 1994. [Google Scholar]
- Bolaños-Vásquez MC, Werner D. Mol Plant-Microbe Interact. 1997;10:339–346. [Google Scholar]
- Boundy-Mills KL, Kosslak RM, Tully RE, Pueppke SG, Lohrke S, Sadowsky MJ. Induction of the Rhizobium fredii nod box-independent nodulation gene nolJ requires a functional nodD1 gene. Mol Plant-Microbe Interact. 1994;7:305–308. [Google Scholar]
- Buzzell RI, Buttery BR, MacTavish DC. Biochemical genetics of black pigmentation of soybean seed. J Hered. 1987;78:53–54. [Google Scholar]
- Chatterjee A, Balatti PA, Gibbons W, Pueppke SG. Interaction of Rhizobium fredii USDA257 and nodulation mutants derived from it with the agronomically improved soybean cultivar McCall. Planta. 1990;180:303–311. doi: 10.1007/BF00198781. [DOI] [PubMed] [Google Scholar]
- Dakora FD, Joseph CM, Phillips DA. Plant Physiol. 1993;101:819–824. doi: 10.1104/pp.101.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Arcy-Lameta A. Study of soybean and lentil root exudates. II. Identification of some polyphenolic compounds, relation with plantlet physiology. Plant Soil. 1986;92:113–123. [Google Scholar]
- Fisher RF, Long SR. Interactions of NodD at the nod box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J Mol Biol. 1993;233:336–348. doi: 10.1006/jmbi.1993.1515. [DOI] [PubMed] [Google Scholar]
- Göttfert M, Weber J, Hennecke H. Induction of a nodA-lacZ fusion in Bradyrhizobium japonicum by an isoflavone. J Plant Physiol. 1988;132:394–397. [Google Scholar]
- Graham TL. Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol. 1991;95:594–603. doi: 10.1104/pp.95.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TL, Graham MY. Signaling in soybean phenylpropanoid responses. Dissection of primary, secondary, and conditioning effects of light, wounding, and elicitor treatments. Plant Physiol. 1996;110:1123–1133. doi: 10.1104/pp.110.4.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heron DS, Érsek T, Krishnan HB, Pueppke SG. Nodulation mutants of Rhizobium fredii USDA257. Mol Plant-Microbe Interact. 1989;2:4–10. [Google Scholar]
- Heron DS, Pueppke SG. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J Bacteriol. 1984;160:1061–1066. doi: 10.1128/jb.160.3.1061-1066.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubac C, Ferran J, Guerrier D, Trémolières A, Kondorosi A. Luteolin absorption in Rhizobium meliloti wild-type and mutant strains. J Gen Microbiol. 1993;139:1571–1578. [Google Scholar]
- Hungria M, Joseph CM, Phillips DA. Anthocyanidins and flavonols, major nod gene inducers from seeds of a black-seeded common bean (Phaseolus vulgaris L.) Plant Physiol. 1991;97:751–758. doi: 10.1104/pp.97.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungria M, Phillips DA. Effects of a seed color mutation on rhizobial nod-gene-inducing flavonoids and nodulation in common bean. Mol Plant-Microbe Interact. 1993;6:418–422. [Google Scholar]
- Kape R, Parniske M, Brandt S, Werner D. Isoliquiritigenin, a strong nod gene- and glyceollin resistance-inducing flavonoid from soybean root exudate. Appl Environ Microbiol. 1992;58:1705–1710. doi: 10.1128/aem.58.5.1705-1710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser HH, Bohlool BB, Hu TS, Weber DF. Fast-growing rhizobia isolated from root nodules of soybean. Science. 1982;215:1630–1632. doi: 10.1126/science.215.4540.1631. [DOI] [PubMed] [Google Scholar]
- Keyser HH, Cregan PB. Interactions of selected Glycine soja Sieb. & Zucc. genotypes with fast- and slow-growing soybean rhizobia. Crop Sci. 1984;24:1059–1062. [Google Scholar]
- Kosslak RM, Bookland R, Barkei J, Paaren HE, Appelbaum ER. Proc Natl Acad Sci USA. 1987;84:7428–7432. doi: 10.1073/pnas.84.21.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács LG, Balatti PA, Krishnan HB, Pueppke SG. Transcriptional organization and expression of nolXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1995;17:923–933. doi: 10.1111/j.1365-2958.1995.mmi_17050923.x. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Pueppke SG. Sequence and analysis of the nodABC region of Rhizobium fredii USDA257, a nitrogen-fixing symbiont of soybean and other legumes. Mol Plant-Microbe Interact. 1991;4:512–520. doi: 10.1094/mpmi-4-512. [DOI] [PubMed] [Google Scholar]
- Kudou S, Fleury Y, Welti D, Magnolato D, Uchida T, Kitamura K, Okubo K. Malonyl isoflavone glycosides in soybean seeds (Glycine max Merrill) Agric Biol Chem. 1991;55:2227–2233. [Google Scholar]
- Lawson CGR, Rolfe BG, Djordjevic MA. Aust J Plant Physiol. 1996;23:93–101. [Google Scholar]
- Long SR. Rhizobium symbiosis: Nod factors in perspective. Plant Cell. 1996;8:1885–1898. doi: 10.1105/tpc.8.10.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne RL, Mulheirn LJ, Leworthy DP (1976) New pterocarpinoid phytoalexins of soybean. J Chem Soc Chem Commun 497–498
- Meinhardt LW, Krishnan HB, Balatti PA, Pueppke SG. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Naim M, Gestetner B, Kirson I, Birk Y, Bondi A. A new isoflavone from soya beans. Phytochemistry. 1973;12:169–170. [Google Scholar]
- Ohta N, Kuwata G, Akahori H, Watanabe T. Isoflavonoid constituents of soybeans and isolation of a new acetyl daidzin. Agric Biol Chem. 1979;43:1415–1419. [Google Scholar]
- Osman SF, Fett WF. Isoflavone glucoside stress metabolites of soybean leaves. Phytochemistry. 1983;22:1921–1923. [Google Scholar]
- Parniske M, Ahlborn B, Werner D. Isoflavonoid-inducible resistance to the phytoalexin glyceollin in soybean rhizobia. J Bacteriol. 1991;173:3432–3439. doi: 10.1128/jb.173.11.3432-3439.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DA (1992) Flavonoids: plant signals to soil microbes. In HA Stafford, RK Ibrahim, eds, Phenolic Metabolism in Plants. Plenum Press, New York, pp 201–231
- Porter PM, Banwart WL, Hassett JJ. HPLC isolation and GC-MS identification of genistein, daidzein, and coumestrol from unhydrolyzed soybean root extracts. Environ Exp Bot. 1985;25:229–232. [Google Scholar]
- Pueppke SG. Rhizobium infection threads in root hairs of Glycine max (L.) Merr., Glycine soja Sieb. & Zucc., and Vigna unguiculata (L.) Walp. Can J Microbiol. 1983;29:69–76. [Google Scholar]
- Pueppke SG. The genetic and biochemical basis for nodulation of legumes by rhizobia. Crit Rev Biotechnol. 1996;16:1–51. doi: 10.3109/07388559609146599. [DOI] [PubMed] [Google Scholar]
- Rao JR, Cooper JE. Soybean nodulating rhizobia modify nod gene inducers daidzein and genistein to yield aromatic products that can influence gene-inducing activity. Mol Plant-Microbe Interact. 1995;8:855–862. [Google Scholar]
- Recourt K, Schripsema J, Kijne JW, van Brussel AAN, Lugtenberg BJJ. Inoculation of Vicia sativa subsp. nigra roots with Rhizobium leguminosarum biovar viciae results in release of nod gene activating flavanones and chalcones. Plant Mol Biol. 1991;16:841–852. doi: 10.1007/BF00015076. [DOI] [PubMed] [Google Scholar]
- Sadowsky MJ, Olson ER, Foster VE, Kosslak RM, Verma DPS. Two host-inducible genes of Rhizobium fredii and characterization of the inducing compound. J Bacteriol. 1988;170:171–178. doi: 10.1128/jb.170.1.171-178.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaman HRM, Okker RJH, Lugtenberg BJJ. Regulation of nodulation gene expression by NodD in rhizobia. J Bacteriol. 1992;174:5177–5184. doi: 10.1128/jb.174.16.5177-5182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PE, Broughton WJ, Werner D. Nod factors of Bradyrhizobium japonicum and Rhizobium sp. NGR234 induce flavonoid accumulation in soybean root exudate. Mol Plant-Microbe Interact. 1994;7:384–390. [Google Scholar]
- Schmidt PE, Parniske M, Werner D. Production of the phytoalexin glyceollin I by soybean roots in response to symbiotic and pathogenic infection. Bot Acta. 1992;105:18–25. [Google Scholar]
- Smit G, Puvanesarajah V, Carlson RW, Barbour WM, Stacey G. Bradyrhizobium japonicum nodD1 can be specifically induced by soybean flavonoids that do not induce the nodYABCSUIJ operon. J Biol Chem. 1992;267:310–318. [PubMed] [Google Scholar]
- Sutherland TD, Bassam BJ, Schuller LJ, Gresshoff PM. Early nodulation signals of the wild type and symbiotic mutants of soybean (Glycine max) Mol Plant-Microbe Interact. 1990;3:122–128. [Google Scholar]
- Todd JJ, Vodkin LO. Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiol. 1993;102:663–670. doi: 10.1104/pp.102.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon BG, Bauer WD. Early events in the infection of soybean by Rhizobium japonicum. Time course and cytology of the initial infection process. Can J Bot. 1982;60:152–161. [Google Scholar]
- van Brussel AAN, Recourt K, Pees E, Spaink HP, Tak T, Wijffelman CA, Kijne JW, Lugtenberg BJJ. J Bacteriol. 1990;172:5394–5401. doi: 10.1128/jb.172.9.5394-5401.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]