Abstract
The anaphase-promoting complex/cyclosome bound to CDC20 (APC/CCDC20) initiates anaphase by ubiquitylating B-type cyclins and securin. During chromosome bi-orientation, CDC20 assembles with MAD2, BUBR1 and BUB3 into a mitotic checkpoint complex (MCC) which inhibits substrate recruitment to the APC/C. APC/C activation depends on MCC disassembly, which has been proposed to require CDC20 auto-ubiquitylation. Here we characterized APC15, a human APC/C subunit related to yeast Mnd2. APC15 is located near APC/C’s MCC binding site, is required for APC/CMCC-dependent CDC20 auto-ubiquitylation and degradation, and for timely anaphase initiation, but is dispensable for substrate ubiquitylation by APC/CCDC20 and APC/CCDH1. Our results support the view that MCC is continuously assembled and disassembled to enable rapid activation of APC/CCDC20 and that CDC20 auto-ubiquitylation promotes MCC disassembly. We propose that APC15 and Mnd2 negatively regulate APC/C coactivators, and report the first generation of recombinant human APC/C.
The spindle assembly checkpoint (SAC) delays chromosome segregation until all chromosomes are bi-oriented on the mitotic or meiotic spindle (reviewed in ref. 1). Defects in this surveillance mechanism can lead to chromosome mis-segregation and aneuploidy, a condition associated with congenital trisomies, tumorigenesis and ageing. The checkpoint is activated in prometaphase by chromosomes whose kinetochores are not or incorrectly attached to microtubules2,3. The checkpoint inhibits CDC20, a protein which associates with and activates the anaphase promoting complex/cyclosome (APC/C) in mitosis4-8. APC/C initiates chromosome segregation by ubiquitylating securin, an inhibitor of the protease separase, and B-type cyclins, activators of cyclin-dependent kinase 1 (CDK1). The subsequent degradation of these proteins by the 26S proteasome leads to activation of separase, which destroys sister chromatid cohesion and thereby initiates anaphase (reviewed in ref. 9).
The APC/C is a 1.5 MDa protein complex composed of three structural domains, called the ‘arc lamp’, the ‘platform’ and the ‘catalytic core’10,11. The latter domain contains the cullin APC2 and the RING finger subunit APC11 (refs. 10,12). APC11 interacts with ubiquitin conjugating (E2) enzymes bound to activated ubiquitin residues which are transferred to APC/C substrates13-15. Substrates contain APC/C recognition sequences, called the destruction box (D box)16 and KEN box17. These ‘degrons’ are thought to be simultaneously recognized by the APC/C subunit APC10 and either CDC20 or CDH1, a CDC20 related coactivator protein that associates with the APC/C in late mitosis and G1 phase12,18-20.
The SAC inhibits CDC20 by promoting its association with three other proteins, MAD2, BUB3 and BUBR1 (also known as BUB1B, and in fission yeast as Mad3), leading to the formation of a mitotic checkpoint complex, MCC21. A rate limiting step in MCC assembly is the binding of CDC20 to MAD2. This process requires conversion of MAD2 from an open (MAD2°) to a closed conformation (MAD2C) in which MAD2C stably embraces CDC20 via a ‘safety belt’ mechanism22,23. The generation of CDC20-MAD2C complexes occurs at checkpoint-active kinetochores to which MAD2C is recruited by binding to MAD1. This MAD1-MAD2C complex promotes conversion of MAD2° to MAD2C and binding of the latter to CDC20. This process is thought to require the transient formation of a conformational MAD2°-MAD2C heterodimer, in which MAD2° is recruited to the MAD2C subunit of the kinetochore associated MAD1-MAD2C complex24,25.
MCC can associate with the APC/C, but unlike APC/C bound to CDC20 (APC/CCDC20), APC/C associated with MCC (APC/CMCC) is unable to bind and ubiquitylate securin and B-type cyclins11. MCC might inhibit CDC20 through multiple mechanisms: a KEN box in Mad3 occupies the KEN box receptor site on Cdc20 (refs. 20,26), another Mad3 domain might partially block a putative D box receptor site on Cdc20 (ref. 20), and CDC20 occupies different positions on the APC/C in the presence or absence of the other MCC subunits11. To allow APC/C activation, MCC therefore has to be replaced by CDC20. Several studies have observed that this depends on MCC disassembly (see below). MCC disassembly is an energy dependent process27,28, possibly because spontaneous release of CDC20 from MAD2 would be too slow to allow rapid APC/C activation in metaphase29.
Several mechanisms have been discussed for how MCC may be disassembled. Reddy et al. proposed that CDC20 auto-ubiquitylation by APC/CMCC leads to MCC disassembly27. However, this view has been challenged by Nilsson et al. who observed that CDC20 is continuously synthesized and degraded during prometaphase and proposed that ubiquitin-mediated CDC20 degradation is required to keep CDC20 levels below a threshold that would override the checkpoint30. Several studies have shown that MCC disassembly depends on CUEDC2 (ref. 31) and p31(comet)27,29,32-34 and that CDK1 mediated phosphorylation of these proteins and of CDC20 promotes MCC disassembly31,35. CUEDC2’s mechanism of action is unknown, but p31(comet) structurally mimics MAD2° and can therefore bind to MAD2C (refs. 36,37). This interaction might facilitate dissociation of MAD2C from CDC20.
Here we have characterized the function of C11ORF51, a protein required for mitotic progression38 and associated with the APC/C39. We show that C11ORF51 is a subunit of APC/C’s platform domain and is located near the MCC binding site on the APC/C11. As recently reported by Mansfeld et al.40 we find that depletion of C11ORF51, which has been renamed APC15, reduces the rate of MCC disassembly. In addition, we show for the first time that APC15 is required for efficient ubiquitylation of CDC20 as part of APC/CMCC and for CDC20 degradation in prometaphase, even though APC15 is not required for substrate ubiquitylation by APC/CCDC20 or APC/CCDH1. Our results support the view27,29 that CDC20 auto-ubiquitylation contributes to MCC disassembly, and that continuous CDC20 synthesis and degradation facilitates rapid APC/C activation in metaphase. In this study, we also report the first generation and characterization of recombinant human APC/C, assembled from 14 core subunits and either CDC20 or CDH1.
Results
C11ORF51/APC15 is a subunit of APC/C’s platform domain
As part of the MitoCheck project we identified two previously unknown APC/C interactors, chromosome 10 open reading frame 104 (C10ORF104; ref. 41) and chromosome 11 open reading frame 51 (C11ORF51; ref. 39). C10ORF104 is an 11.7 kDa subunit of APC/C’s arc lamp domain and is now called APC16 (refs. 41,42). C11ORF51 is a 14.3 kDa protein which was only reported by Hubner et al.39, whereas this protein had been excluded from the analysis in Hutchins et al.41 because only a single tryptic C11ORF51 peptide had been identified in the APC/C samples analyzed in this study. C11ORF51 had therefore not met the criteria for a specific ‘prey’ protein that had been used in this study.
To address if C11ORF51 is a subunit of the APC/C we raised antibodies to this protein. In addition we used recombineering of a bacterial artificial chromosome (BAC) to tag the human C11ORF51 gene with a sequence that encodes a C-terminal localization affinity purification (LAP) tag43,44 and generated a HeLa cell pool that stably expresses C11ORF51-LAP. When APC/C was immunoprecipitated from these cells with CDC27 antibodies both endogenous and LAP-tagged C11ORF51 could be detected by immunoblotting (Fig. 1a). During sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) these proteins migrate corresponding to molecular masses of 20 kDa and 55 kDa, respectively. Depletion of endogenous C11ORF51 with four different siRNAs confirmed that the 20 kDa band recognized by our antibodies represents C11ORF51 (see, for example, Fig. 3a below).
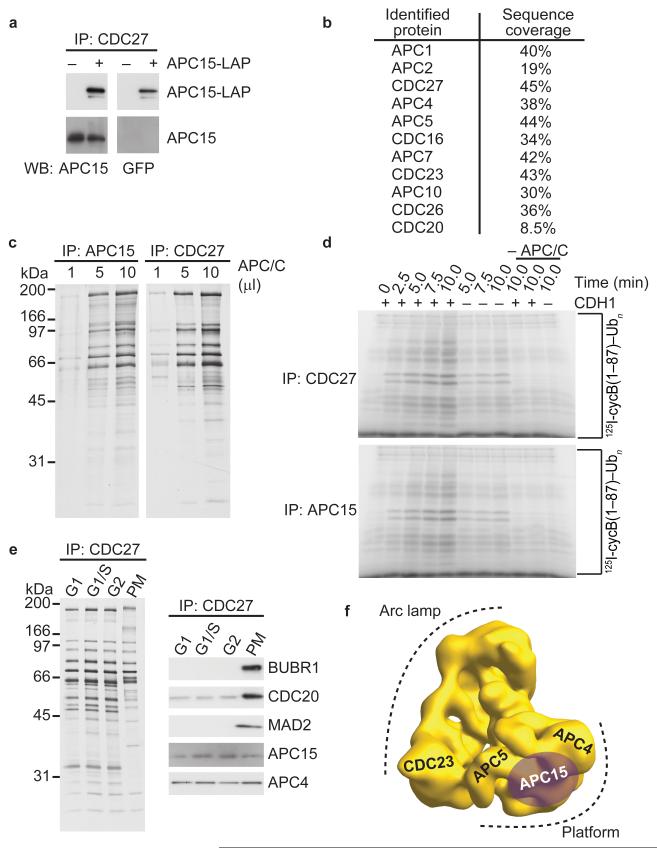
Figure 1.
C11ORF51/APC15 is a subunit of APC/C’s platform domain. (a) Western blot showing APC/C immunoprecipitations from asynchronous cells or cells expressing APC15-LAP. IP, immunoprecipitation. WB, western blot. (b) Table showing APC/C subunits identified by mass spectrometry after tandem affinity purification of APC15-LAP. Peptide sequence coverage of each subunit in percent is listed. (c) Silver stain showing APC/C immunoprecipitations using APC15 or CDC27 antibodies. (d) Autoradiography after in vitro ubiquitylation of [I125]-labeled human cyclin-B1 fragment (1-87) by APC/C. APC/C concentration used in this assay was normalized based on APC/C subunit silver staining intensity shown in c. (e) Silver stain and western blot analysis after APC/C immunoprecipitation from cells arrested in different cell cycle stages. G1, gap 1 phase. S, synthesis phase. G1/S, G1–S transition. G2, gap 2 phase. PM, prometaphase. (f) Three-dimensional model of human APC/C obtained by electron microscopy11 showing the location of APC15 as determined by antibody labeling.
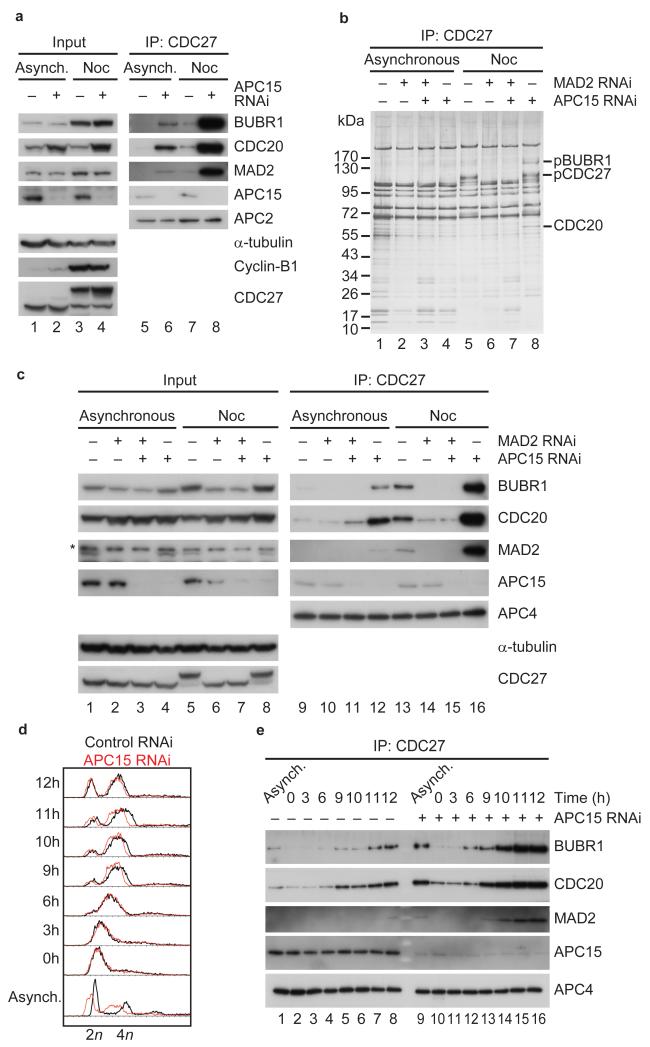
Figure 3.
APC15 is required for the turnover of BUBR1, CDC20 and MAD2 proteins on APC/C during prometaphase in the presence of an active checkpoint. (a) Western blot showing APC/C immunoprecipitations from asynchronous (Asynch.) or nocodazole (Noc) arrested HeLa cells treated with control or APC15 siRNAs. (b) Silver stain showing APC/C immunoprecipitations from asynchronous or nocodazole arrested HeLa cells treated with control RNAi or APC15 or/and MAD2 RNAi. (c) Western blot showing APC/C immunoprecipitations from asynchronous or nocodazole arrested HeLa cells treated with control RNAi or APC15 or/and MAD2 RNAi. Asterisk indicates MAD2 antibody cross-reaction with an unknown protein. (d) FACS profile of control or APC15 depleted HeLa cells arrested in S phase (0h) and released from S phase for up to 12 hours. (e) Western blot showing APC/C immunoprecipitations from extracts of FACS sorted cells shown in d.
When C11ORF51-LAP was isolated by tandem affinity purification from HeLa cells and analyzed by mass spectrometry, peptides from ten APC/C subunits and CDC20 were identified, confirming that C11ORF51 is specifically associated with the APC/C (Fig. 1b). In addition, SDS-PAGE and silver staining showed that immunoprecipitates obtained with C11ORF51 antibodies contained a similar set of proteins as APC/C samples isolated with CDC27 antibodies (Fig. 1c), and these C11ORF51 and CDC27 immunoprecipitates were able to support cyclin-B1 ubiquitylation reactions similarly well (Fig. 1d). Immunoblotting with C11ORF51 antibodies revealed further that C11ORF51 is associated with APC/C in cells synchronized in G1, S, G2 and prometaphase (Fig. 1e). These observations indicate that C11ORF51 is a bona fide subunit of the human APC/C.
We therefore tested if C11ORF51 is related to subunits of APC/C that have so far only been identified in lower eukaryotes, such as Apc9 or Mnd2/Apc15 in budding yeast45,46. The sequence of C11ORF51 is well conserved among metazoans, ranging from cnidarians to mammals. We used a Hidden Markov model (HMM) obtained by multiple sequence alignment of these sequences to search for related proteins in fungal and plant proteomes. We identified sequences significantly related to metazoan C11ORF51 in plants and fungi, with the closest relative of C11ORF51 in S. pombe being Apc15, an ortholog of S. cerevisiae Mnd2 (Supplementary Fig. 1a and b). A reciprocal search in the human, mouse and A. thaliana proteomes with a HMM derived from an alignment of S. pombe and Agaricomycetes Apc15 confirmed that C11ORF51 in metazoans is related to Mnd2 in fungi. It is therefore possible that C11ORF51 and Mnd2 are derived from a common ancestral APC/C subunit. Like Mansfeld et al.40 we will refer to C11ORF51 as APC15.
To analyze where APC15 is located within the APC/C we labeled purified human APC/C with APC15 antibodies under conditions that induce antibody-dependent APC/C dimerization10-12, isolated the APC/C dimers by density gradient centrifugation and analyzed them by negative staining electron microscopy (Supplementary Fig. 1c and d). The angular orientation of APC/C monomers in the antibody induced dimers indicates that APC15 is part of APC/C’s platform domain, where it might be located in vicinity to APC4, APC5 and APC1 (Fig. 1f). This location is consistent with the possibility that human C11ORF51 is related to yeast Mnd2 because Hall et al.46 had observed in an in vitro transcription-translation system that Mnd2 interacts with Apc1 and Apc5, which are components of the platform domain, and with Cdc23, which is located close to the platform11,47.
APC15 is required for anaphase
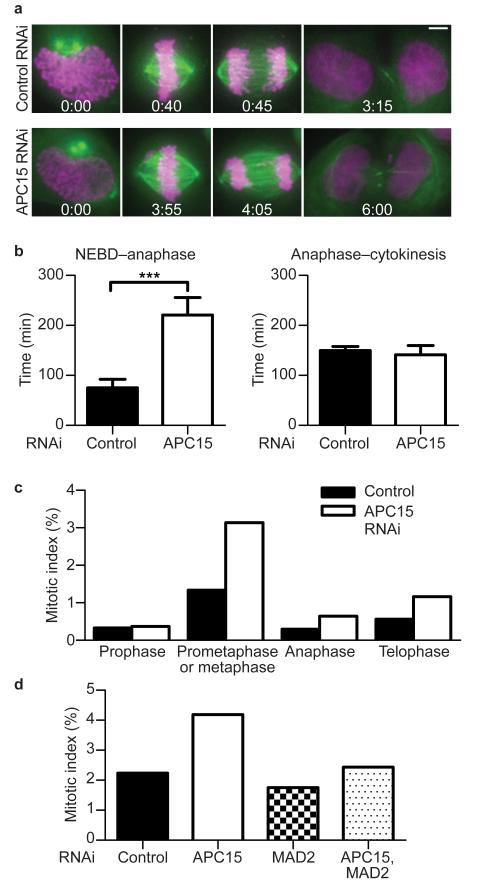
A previous genome wide RNA interference (RNAi) screen had shown that depletion of C11ORF51/APC15 delays mitotic progression38. To analyze the function of APC15 further we depleted this protein by RNAi from HeLa cells stably expressing the histone H2B tagged with mCherry (H2B-mCherry) and β-tubulin tagged with EGFP (TUBB-LAP) and analyzed cells by time-lapse fluorescence microscopy (Fig. 2a). In APC15 depleted cells progression from nuclear envelope breakdown (NEBD) to anaphase onset took on average more than twice as long as in control transfected cells, whereas progression from anaphase to cytokinesis was not significantly delayed (Fig. 2b). Similar results were obtained when the APC/C subunit CDC16 was depleted (data not shown). An increase in the number of prometaphase and metaphase cells following APC15 depletion was also observed when fixed cells were analyzed by immunofluorescence microscopy, consistent with a delay during these stages of mitosis (Fig. 2c and Supplementary Fig. 2). In this experiment also an increase in the frequency of cells in anaphase and telophase could be observed, perhaps because larger numbers of cells were analyzed (n>8600) than by live cell imaging (n=25). The increase in mitotic index that was observed after APC15 depletion was partly restored by co-depletion of MAD2, consistent with the notion that APC15 depletion delays mitotic progression by compromising APC/C function (Fig. 2d).
Figure 2.
APC15 depletion causes delay in mitosis. (a) Still images of time-lapse microscopy experiments showing representative cells at nuclear envelope breakdown (NEBD), metaphase, anaphase and telophase (from left to right). Time of progression for control or APC15 RNAi treated cells after NEBD (h:min) is indicated for the respective cell cycle stages. Size bar equals 5 μm. H2B-mCherry shown in magenta. β-tubulin shown in green. (b) Histogram showing quantification of time-lapse microscopy experiments. Average time of progression of control or APC15 RNAi treated cells from NEBD to anaphase and anaphase to cytokinesis (n=25; error bars denote s.e.m.; asterisks denote P-value of 0.0005 from a two-tailed t test). (c) Histogram showing quantification of mitotic stages after control or APC15 RNAi treatment (n>8600 per condition). CDC20, BUB1, α-tubulin and DNA staining were used to distinguish different mitotic stages. (d) Histogram showing quantification of mitotic index after control RNAi or APC15 or/and MAD2 RNAi (n=5000 per condition). Phospho-H3Ser10, α-tubulin and DNA staining were used to distinguish mitotic and interphase cells.
APC15 is required for APC/CMCC disassembly
Because APC/C is activated by CDC20 and inhibited by MAD2 and BUBR1, we analyzed if APC15 depletion might alter APC/C function by affecting the steady state levels of these proteins on the APC/C. Interestingly, SDS-PAGE followed by immunoblotting (Fig. 3a) or silver staining (Fig. 3b) revealed that the levels of all three proteins were strongly increased on the APC/C following depletion of APC15, indicating that APC15 depletion leads to accumulation of MCC on the APC/C. This effect was particularly evident when APC/C was isolated from cells which had been synchronized in prometaphase by nocodazole treatment (Fig. 3a, lane 8), but could also be observed to a lesser extent in APC/C samples from asynchronous cell populations (Fig. 3a, lane 6). When MAD2 was co-depleted from cells at the same time as APC15, neither CDC20 nor BUBR1 were able to accumulate on APC/C, indicating that the increased binding of these proteins to the APC/C depends on the SAC (Fig. 3b, compare lanes 7 and 8; Fig. 3c, compare lanes 15 and 16). To further test if the accumulation of CDC20, MAD2 and BUBR1 on the APC/C occurs specifically in mitosis we released APC15 depleted cells from a thymidine induced arrest in S phase, monitored cell cycle progression by fluorescence activated cell sorting (FACS) of cells whose DNA had been stained with propidium iodide (Fig. 3d), and analyzed the amounts of these proteins on the APC/C by immunoblotting (Fig. 3e). The strongest increase in the levels of CDC20, MAD2 and BUBR1 on the APC/C was observed when cells entered mitosis (Fig. 3e, lanes 13 to 16) indicating that APC15 depletion leads to accumulation of MCC on the APC/C when the SAC becomes active.
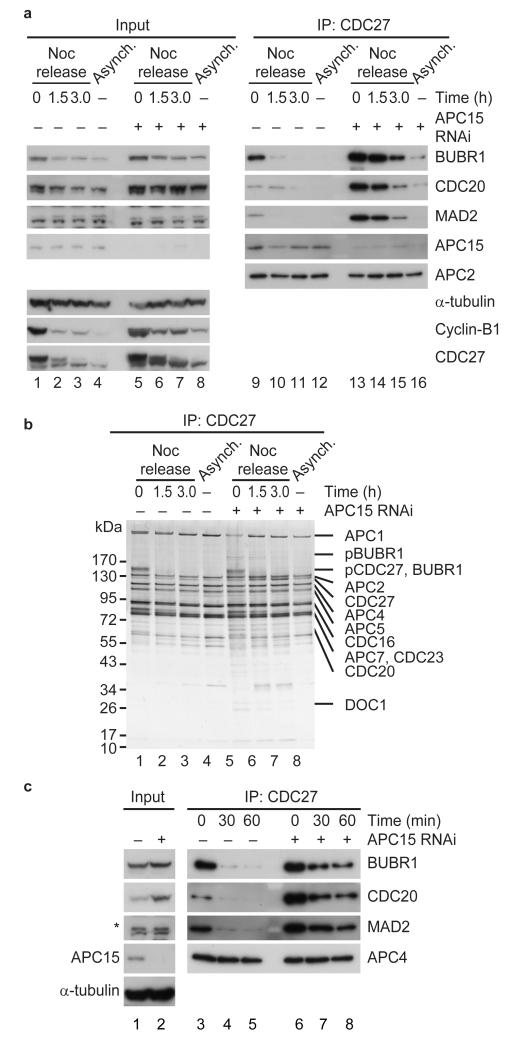
Because Reddy et al.27 had found that MCC disassembly in cell extracts depends on APC/C activity, and because APC15 could be required for APC/C activity, we next analyzed if APC15 depletion might decrease MCC disassembly. To test this possibility we released APC15 depleted HeLa cells from a nocodazole arrest and followed the levels of CDC20, MAD2 and BUBR1 on the APC/C over time. Immunoblotting and silver staining experiments revealed that in control cells the majority of MAD2 and BUBR1 had been removed from the APC/C 90 minutes after the release (Fig. 4a, lane 10; Fig. 4b, lane 2). At this time, cells had begun to exit mitosis, as judged by a decrease in cyclin-B1 levels and an increase in the electrophoretic mobility of CDC27, indicative of CDC27 dephosphorylation (Fig. 4a, lane 2; Fig. 4b, lane 2). In contrast, the levels of CDC20, MAD2 and BUBR1 remained higher on the APC/C 90 minutes after the release when APC15 had been depleted (Fig. 4a, compare lanes 13 and 14; Fig 4b, lanes 5 and 6), and cyclin-B1 degradation and CDC27 dephosphorylation were reduced, although not abolished (Fig. 4a, lane 6; Fig. 4b, lane 6). Following APC15 depletion, all three MCC subunits could still be detected on the APC/C even 180 minutes after the release, even though now at greatly reduced levels (Fig. 4a, lane 15). These results indicate that APC15 is required for rapid MCC disassembly.
Figure 4.
APC15 is required for MCC disassembly in vivo and in vitro. (a) Western blot showing MCC disassembly in vivo. Control and APC15 depleted HeLa cells were arrested in prometaphase and released from nocodazole arrest (Noc arrest) for indicated times. APC/C immunoprecipitates from cells released from nocodazole arrest are shown on the right. (b) Silver stain of APC/C immunoprecipitates from cells released from nocodazole arrest. pBUBR1, phosphorylated BUBR1. pCDC27, phosphorylated CDC27. (c) Western blot showing in vitro MCC disassembly using APC/C immunoprecipitated from cells arrested in prometaphase. APC/C immobilized on CDC27-antibody beads was incubated with a ubiquitylation mixture for indicated times, washed and eluted using antigenic peptides.
To confirm this notion we analyzed MCC disassembly in vitro by incubating immunopurified APC/CMCC with ubiquitin, the ubiquitin activating enzyme E1 and the E2 enzyme UBCH10. Also under these conditions MCC disassembly was strongly decreased when APC/CMCC was isolated from APC15 depleted cells (Fig. 4c, lanes 6 to 8). These observations indicate that APC15 promotes progression through mitosis at least in part by facilitating MCC disassembly. We presently do not know if the reduced rate of MCC disassembly that we have observed in our assays is due to residual amounts of APC15, or if MCC disassembly can also proceed slowly in the absence of APC15.
MCC disassembly depends on p31(comet)29,32-34 and proteasome activity32,48-50. We therefore compared the effects of APC15 depletion to the effects of p31(comet) depletion and proteasome inhibition. We released HeLa cells depleted of APC15 or p31(comet) or both from a nocodazole arrest in either the presence or absence of the proteasome inhibitor MG132, and followed the levels of CDC20, MAD2 and BUBR1 on the APC/C over time (Supplementary Fig. 3). These experiments revealed that APC15 depletion resulted in accumulation of much more MCC on APC/C than p31(comet) depletion, despite the fact that both treatments delayed MCC disassembly to a similar extent. It is possible that APC15 and p31(comet) were depleted to different degrees in our experiments, or that p31(comet) acts catalytically in MCC disassembly and is therefore less sensitive to partial depletion than APC15, which as an APC/C subunit is expected to function stoichiometrically. However, the difference between the effects of APC15 and p31(comet) depletion is also consistent with the possibility that p31(comet) depletion affects MCC steady state levels through two distinct mechanisms, by ‘capping’ the MAD1-MAD2C template at unattached kinetochores and thereby preventing MCC formation, and by binding to MAD2C as part of MCC and thereby promoting MCC disassembly. In contrast, APC15 depletion might reduce MCC disassembly without slowing down MCC formation, thus leading to a more pronounced accumulation of MCC on the APC/C.
These experiments also confirmed that proteasome inhibition delays MCC disassembly. Like p31(comet) depletion, MG132 treatment led to only a small increase in MCC levels on the APC/C. However, in this case the difference to depleting APC15 is presumably due to the fact that the proteasome had only been inhibited during the last three hours of the experiment, whereas cells had been gradually depleted of APC15 for a much longer period of time (48 hours) during which MCC was able to accumulate. These observations indicate that MCC disassembly depends on at least three components, APC15, p31(comet), and proteasome activity.
APC15 is required for CDC20 degradation in prometaphase
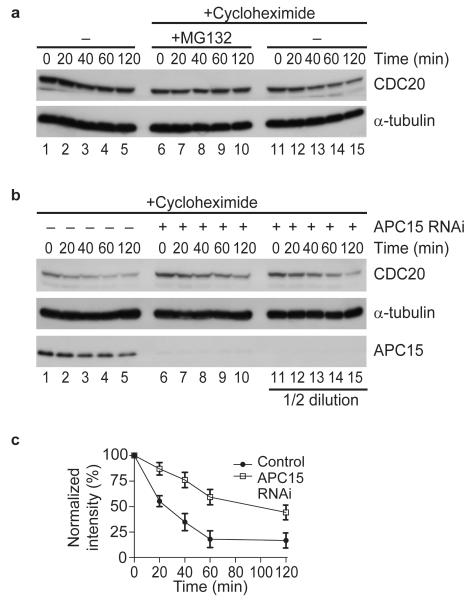
APC15 depletion did not only increase the amounts of CDC20 that were associated with the APC/C, but also increased CDC20 steady state levels in total cell lysates (Fig. 3a, lanes 2 and 4). This was not, or only to a much lesser degree, the case for MAD2 and BUBR1, implying that APC15 depletion might have two different effects: (i) a change in the equilibrium between APC/C associated and non-associated MCC subunits, and (ii) an increase in the cellular concentration of CDC20. Because CDC20 is continuously synthesized and degraded during prometaphase30 we analyzed if APC15 depletion might stabilize CDC20. As reported30, CDC20 levels decreased in prometaphase arrested cells when protein synthesis was inhibited by cycloheximide (Fig. 5a, lanes 11 to 15). This effect was reversed by MG132, indicating that in cycloheximide treated cells CDC20 levels decrease due to proteolysis (Fig. 5a, lanes 6 to 10). Interestingly, the rate of this proteolysis was reduced in cells depleted of APC15, indicating that APC15 is required for rapid degradation of CDC20 in cells with an active SAC (Fig. 5b, lanes 6 to 10; quantified in Fig. 5c).
Figure 5.
CDC20 turnover in prometaphase depends on APC15. (a) Western blot showing CDC20 degradation in control cells arrested in prometaphase using nocodazole. Cells were treated with or without cycloheximide to block protein synthesis in the presence or absence of the proteasome inhibitor MG132 for indicated times. (b) Western blot showing CDC20 degradation in prometaphase arrested control and APC15 RNAi treated cells. Cells were incubated with cycloheximide to block protein synthesis for indicated times. (c) Diagram showing mean CDC20 protein levels in cycloheximide treated control and APC15 depleted HeLa cells (n=4; error bars denote s.e.m.). Signal intensity was normalized to α-tubulin levels.
APC15 is dispensable for APC/CCDC20 and APC/CCDH1 activity
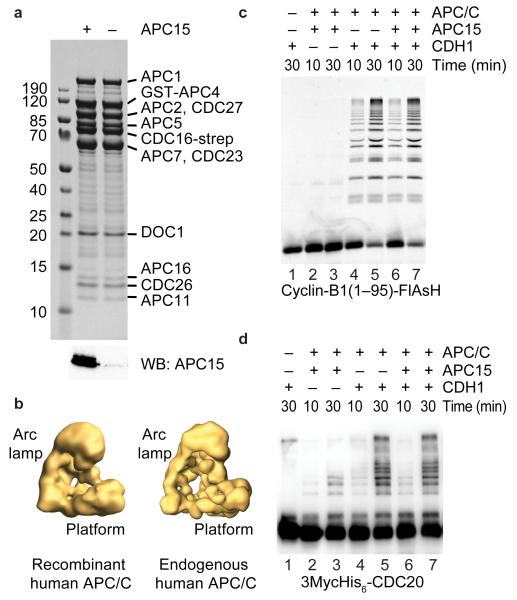
One possible interpretation of the above results is that APC15 depletion would compromise APC/C activity, that this defect would reduce CDC20 ubiquitylation, and that as a result MCC disassembly would be delayed. To test this possibility we generated Baculoviruses expressing all 14 subunits of the human APC/C, co-infected insect cells with these and isolated recombinant APC/C. SDS-PAGE and Coomassie staining showed that the recombinant complexes contained a subunit pattern that is reminiscent of APC/C purified from HeLa cells (Fig. 6a). Single particle electron microscopy and angular reconstitution revealed that at the resolution obtained the overall shape of the recombinant complexes is indistinguishable from the 3D models we have previously obtained for endogenous human APC/C (Fig. 6b; ref. 11,12). Furthermore, the recombinant complexes were able to ubiquitylate an N-terminal fragment of cyclin-B1 in a CDH1-dependent manner (Fig. 6c, lanes 6 and 7). Recombinant APC/C lacking APC11 was unable to mediate ubiquitylation reactions, confirming that this activity was attributable to human APC/C (Supplementary Fig. 4). These results indicate that the recombinant human APC/C which we have generated here for the first time is functional and similar in its properties to endogenous APC/C.
Figure 6.
APC15 is not essential for cyclin-B1 and CDC20 ubiquitylation by APC/CCDH1. (a) Coomassie stain (top) showing recombinant human APC/C as either wild-type complex or lacking the APC15 subunit (bottom, APC15 western blot). Identities of APC/C proteins and sizes of molecular weight markers (kDa) are indicated. (b) 3D models of recombinant and endogenous human apo-APC/C derived by negative-staining single particle electron microscopy. (c) SDS-PAGE showing direct fluorescence of FlAsH-labeled N-terminal fragment of cyclin-B1 and cyclin-B1–ubiquitin conjugates after incubation with recombinant human APC/C with or without APC15, the ubiquitylation mixture and CDH1 as indicated. (d) Western blot showing recombinant CDC20 and CDC20–ubiquitin conjugates after incubation with recombinant human APC/C with our without APC15, the ubiquitylation mixture and CDH1 as indicated. Reactions in c and d included both UBCH10 and UBE2S.
Next we generated recombinant APC/C lacking APC15 (Fig. 6a). To exclude that an APC15 related protein from the insect host cells (from the moth Spodoptera frugiperda) could substitute for human APC15 we analyzed our sample by mass spectrometry and searched for peptides derived from the sequence of S. frugiperda APC15, which we had identified by BLAST searches (Supplementary Fig. 1a). No peptides from S. frugiperda APC15 could be detected (data not shown). Human APC/C can therefore be assembled in the complete absence of APC15. Nevertheless, recombinant APC/C lacking APC15 incubated with CDH1 was similarly active in supporting the formation of cyclin-B1 and CDC20 ubiquitin conjugates as wild-type APC/C (Fig. 6c and d). Similar observations were made in ubiquitylation assays using a ubiquitin fusion to the cyclin-B1 fragment as a substrate to specifically assay E2 enzyme UBE2S instead of UBCH10, and in assays using CDC20 instead of CDH1 (Supplementary Fig. 4c).
APC15 mediates CDC20 auto-ubiquitylation by APC/CMCC
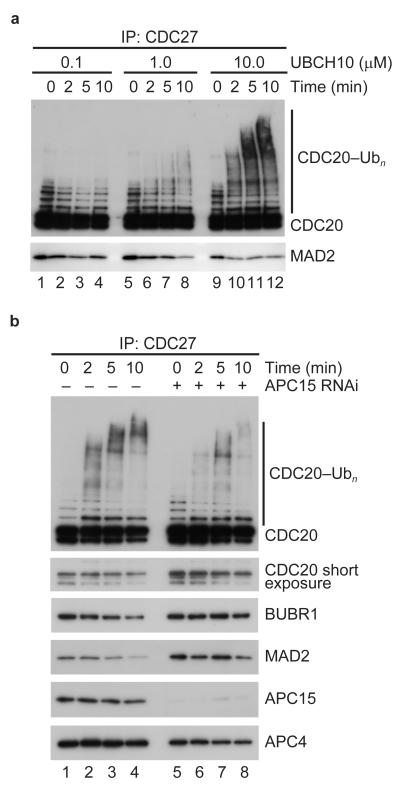
Although our data cannot exclude that APC15 has an effect on kinetic properties of ubiquitylation reactions which cannot be revealed by our assays, our results show unequivocally that APC15 is not essential for the ubiquitylation activity of APC/CCDH1 and APC/CCDC20. We therefore tested if APC15 might be required for the ubiquitylation of CDC20 when this protein is part of APC/CMCC. This reaction may depend on different mechanisms than substrate ubiquitylation mediated by APC/CCDH1 and APC/CCDC20, similarly to how Cdc20 ubiquitylation by APC/CCdh1 and Cdc20 auto-ubiquitylation have been shown to differ in yeast51. Because we were not able so far to reconstitute APC/CMCC using a fully recombinant system, we used in these assays APC/CMCC which had been isolated from APC15 depleted cells. We confirmed the finding by Reddy et al.27 that incubation of isolated APC/CMCC with ubiquitin, E1 and UBCH10 resulted in ubiquitylation of CDC20 and in dissociation of MAD2 from APC/CMCC (Fig. 7a). In contrast, we observed that CDC20 ubiquitylation was reduced when we used APC/CMCC from which APC15 had been depleted (Fig. 7b, lanes 5 to 8). This reduction was observed in seven out of seven independent experiments (Supplementary Fig. 5). Furthermore, the reduction in CDC20 ubiquitylation was observed despite the fact that more CDC20 was present in the APC/CMCC samples isolated from APC15 depleted cells than in samples containing wild-type APC/CMCC (Fig 7b, compare the APC4 immunoblot signals with the CDC20 signals in the shorter immunoblot exposure), and despite the fact that the depletion of APC15 by RNAi might not have been complete. These results indicate that APC15 is required for the efficient ubiquitylation of CDC20 when this protein is part of MCC, but not when CDC20 is a substrate of APC/CCDH1.
Figure 7.
APC15 is required for CDC20 auto-ubiquitylation by APC/CMCC. (a) Western blot showing CDC20 auto-ubiquitylation by APC/CMCC immobilized on CDC27-antibody beads incubated with increasing concentrations of UBCH10. After indicated time points, CDC27-antibody beads were washed and bound protein was eluted using antigenic peptides. (b) Western blot showing CDC20 auto-ubiquitylation by wild-type or APC15 depleted APC/CMCC immobilized on CDC27-antibody beads incubated with 10μM UBCH10. After indicated time points, CDC27-antibody beads were washed and bound protein was eluted using antigenic peptides.
Discussion
Because APC/C activity is essential for sister chromatid separation and mitotic exit APC/C has to be relieved from its inhibition by MCC once spindle assembly has been completed. Here we confirm and extend the finding that C11ORF51/APC15 is an APC/C subunit required for this event38-40. Importantly, we show for the first time that APC15 is not only essential for rapid MCC disassembly but also for efficient CDC20 auto-ubiquitylation by APC/CMCC and for proteasomal degradation in cells with an active SAC. This correlation supports the view that CDC20 ubiquitylation and degradation facilitate rapid MCC disassembly and APC/C activation27,29. Our results are more difficult to explain by the proposal that CDC20 degradation maintains the checkpoint30,40. We therefore favor the idea that CDC20 is constantly auto-ubiquitylated by APC/CMCC and degraded in cells with an active checkpoint, leading to continuous MCC disassembly.
According to this hypothesis, APC/C inhibition can only be maintained as long as newly synthesized CDC20 is assembled into MCCs. Once MCC production has ceased, newly synthesized CDC20 would rapidly replace MCC on APC/C, leading to APC/C activation. As first proposed by Varetti et al.29, checkpoint inhibition of the APC/C may therefore not be achieved by stable association of MCC with APC/C, but by a dynamic equilibrium of MCC assembly and disassembly which would lead to continuous turnover of MCC on APC/C.
This hypothesis has the potential to explain why CDC20 is continuously synthesized and degraded during prometaphase30 and how APC/C is activated rapidly and by default in metaphase, despite the fact that MAD2 binds to CDC20 tightly during prometaphase. The hypothesis also provides a rationale for a number of previous observations, for example why inhibition of the proteasome leads to accumulation of MCC on APC/C11,32,48-50. Interestingly, the MCC turnover hypothesis also implies that MCC’s association with the APC/C is not only required for APC/C inhibition, as was previously assumed to be the case, but may also have the opposite function, namely to overcome APC/C’s inhibition by MCC.
What may be the role of APC15 in this process? As a subunit of the APC/C, APC15 could simply be required for the ubiquitylation activity of the APC/C. However, we found that recombinant forms of human APC/CCDH1, whose generation we report here for the first time, can ubiquitylate both cyclin-B1 and CDC20 similarly well in the presence or absence of APC15, implying that APC/C’s ubiquitylation activity per se does not strictly depend on APC15. On the contrary, the ability of APC/CMCC to auto-ubiquitylate CDC20 is reduced after depletion of APC15, indicating that APC15 has a specific role in this process that is dispensable for substrate ubiquitylation by APC/CCDH1 and APC/CCDC20. The location of APC15 in the platform domain may provide a rationale for this phenomenon as our results indicate that APC15 is located in the vicinity of MCC’s binding site. APC15 could therefore be required for proper positioning of MCC on the APC/C, an effect that could be required for CDC20 auto-ubiquitylation. Alternatively, APC15 could have effects on the catalytic core of the APC/C, and these effects could be required for CDC20 auto-ubiquitylation but dispensable for conventional substrate ubiquitylation.
Interestingly, APC15 is distantly related to Mnd2, an APC/C subunit that had previously only been identified in budding yeast and other fungi. This finding confirms the notion that APC/C is highly conserved among eukaryotes, a phenomenon that is presumably reflecting the early appearance of APC/C during evolution and the importance of APC/C’s function and its tight regulation. Unlike APC15, Mnd2 was so far only known to have a role in meiosis I, where Mnd2 functions as an inhibitor of the meiosis specific APC/C coactivator Ama1 (refs. 52,53). However, because Ama1 is related in structure and function to CDC20, it is conceivable that Mnd2 and APC15 share the ability to control some property of APC/C coactivator proteins, a phenomenon that in the case of Mnd2 may lead to inactivation of Ama1, and in the case of APC15 to disassembly of MCC. It will, therefore, be interesting to test if Mnd2 inhibits Ama1 by a mechanism that depends on Ama1 auto-ubiquitylation by APC/CAma1, similarly to how the presence of APC15 enables CDC20 auto-ubiquitylation by APC/CMCC. Consistent with this possibility, and supporting the notion that APC15 and Mnd2 perform related functions, Foster and Morgan reported after submission of this manuscript that Mnd2 is also required for MCC-dependent Cdc20 auto-ubiquitylation and SAC inactivation in mitotic yeast cells54.
Materials and Methods
Cell culture
HeLa cells were grown in DMEM including 10% FBS (Invitrogen), 2 mM glutamine and 100 μg ml−1 penicillin/streptomycin (both from Sigma) and plated on 6-well plates containing 22 mm diameter coverslips for immunofluorescence microscopy (IFM), or on 15 cm tissue culture dishes. Cells were arrested in S phase via addition of 2.5 mM thymidine for 24 hours. To obtain mitotic cells with an active checkpoint, cells were incubated with 100 nM nocodazole (Sigma) for 14 hours and collected via mitotic shake-off. For proteasome inhibition 10 μM MG132 (Sigma) was used. Protein synthesis was inhibited by adding 20 μg ml−1 cycloheximide (Sigma) as described30.
RNA interference
HeLa cells were transfected with Lipofectamine RNAiMAX (Invitrogen) and 40 nM siRNA oligonucleotides for 48 hours before analysis by immunoblotting or IFM (C11_2: 5′-UCGCGGAGAAAGACAACAA-3′, C11_3: 5′-GGACAUGGAAGGCAACGAA-3′, C11_4: 5′-GGAUCGACCCUGUGUGGAA-3′; Ambion; MAD2: 5′-UUACUCGAGUGCAGAAAUA-3′, p31(comet): 5′-GGACACUAGUACCGCGAGU-3′; Dharmacon). C11_4 siRNA was used unless otherwise indicated. GL2 luciferase was used as control siRNA55. APC15 and firefly luciferase esiRNAs (obtained as a kind gift from Dr. Frank Buchholz) were used in Figure 2a and b.
Antibodies and immunoprecipitation
Polyclonal APC15 antibodies were generated by Gramsch Laboratories against a synthetic peptide (DEMNDYNESPDDGEV). APC2, CDC27, APC4, APC16, MAD2, BUBR1, CDC20, GFP antibodies were described before7,11,41. In addition the following commercial antibodies were used: CDC20 (1:200, sc-13162, Santa Cruz Biotechnology), cyclin-B1 (1:1,000, sc-245, monoclonal GNS1, Santa Cruz Biotechnology), α-tubulin (1:10,000, B-512, Sigma; 1:100, F2168, Sigma), BUB1 (1:500, MBL International), pH3Ser10 (1:1,000, 06-570, Millipore), anti-mouse IgG-HRP and anti-rabbit IgG-HRP (1:5,000, GE Healthcare), anti-mouse and anti-rabbit Alexa Fluor 568 and 647 (1:1,000, Invitrogen). APC/C was affinity-purified from HeLa cells using CDC27 antibody-crosslinked beads (Affi-prep protein A beads, BioRad). Cells were lysed for 30 min at 4°C in lysis buffer (CytoBuster supplemented with 20 mM β-glycerophosphate, 10 mM Na-pyrophosphate, 10 mM NaF, 1 mM Na3VO4, 1 μM okadaic acid, 1X complete protease inhibitor cocktail (Roche)). Beads were washed three times in washing buffer (20 mM Tris pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Tween-20). APC/C was recovered by elution with 2 bead volumes of 1 mg ml−1 antigenic peptide dissolved in elution buffer (20 mM Tris pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 0.1% (w/v) octyl-β-D-glucopyranoside) or 100 mM glycine-HCl at pH 2.2.
CDC20 auto-ubiquitylation by APC/CMCC and in vitro MCC disassembly
In vitro MCC dissociation and CDC20 auto-ubiquitylation was adopted from Reddy et al.27 with the following changes. APC/C purified from prometaphase arrested HeLa cells was immobilized on CDC27-antibody beads and incubated with 10 μM UbcH10, 0.5 μM E1, 1.5 mg ml−1 ubiquitin (Sigma), 30 mM Tris pH 7.5, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, ATP regenerating system (7.5 mM creatine phosphate, 1 mM ATP, 1 mM MgCl2), 10 mM dithiothreitol and 1 mg ml−1 BSA. The reactions were incubated at 23°C with 1,000 rpm shaking for indicated time points. After incubation with the ubiquitylation mixture, the CDC27-antibody beads were washed and bound protein was recovered by elution with antigenic peptide.
Time-lapse microscopy
HeLa cells stably expressing histone H2B-mCherry and mouse TUBB-LAP were used for live cell imaging analysis (histone H2B-mCherry plasmid is a gift from Dr. Jan Ellenberg). Cells were reverse transfected with 100 nM esiRNA using oligofectamine reagent (Invitrogen). The cells were cultured in μ-Slide 8 well (ibidi GmbH) and filmed 48 hours after transfection on a DeltaVision sectioning microscope system equipped with an IX71 microscope (Olympus) and a CoolSnap HQ CCD camera (Photometrics). Time-lapse images were taken at 5 min intervals as 13 sections with 1 μm z-steps, deconvolved, and maximally projected. Cell cycle stages were manually annotated to measure the duration of mitosis (from NEBD until anaphase onset) and mitotic exit (from anaphase until loss of microtubule bridges).
Immunofluorescence microscopy
Cells were fixed in 4% (w/v) paraformaldehyde in PBS, permeabilized in 0.1% (v/v) Triton X-100 in PBS, blocked with 3% (w/v) BSA in 0.01%TX-100/PBS and incubated with antibodies in blocking solution as described in figure legends. Images were taken on a Zeiss Axioplan 2 microscope with a 20x Plan-Neofluar objective lens to determine mitotic indices or on a Zeiss LSM700 confocal microscope with a 63x Plan-Apochromat objective lens at 0.38 μm z-distance for determining sub-mitotic stages.
Data quantification and analysis
Western blot signals were quantified using ImageJ (rsb.info.nih.gov/ij). Quantification of Western blots, live cell imaging and IFM was processed with Microsoft Excel 2007 and GraphPad Prism 5.
Electron microscopy and antibody labeling
Purified apo-APC/C was incubated with submolar concentrations of α-APC15-antibody and subjected to GraFix56. APC/C-antibody complexes were adsorbed to a thin film of carbon, transferred to an electron microscopic grid covered with a perforated carbon film, stained with 2% (w/v) uranyl formate and air dried at room temperature. Images were recorded at a magnification of 88,000x on a 4k × 4k CCD camera (TVIPS GmbH) using two-fold pixel binning (1.8 Å per pixel) in a Philips CM200 FEG electron microscope (Philips/FEI) operated at 160 kV acceleration voltage. Antibody-labeled APC/C complexes were analyzed as described11.
Expression and purification of recombinant human APC/C
Recombinant APC/C was expressed in insect cells (High Five SFM-adapted, Invitrogen) using the Bac to Bac baculovirus system (Invitrogen). Full-length open reading frames of individual human APC/C subunits (APC1, APC2, APC3/CDC27, APC5, APC7, APC8/CDC23, APC10/DOC1, APC11, APC13, APC15, APC16, and CDC26) were PCR amplified from cDNAs obtained (Open Biosystems) and cloned into pFastBac-1. A construct encoding APC6/CDC16 bearing a C-terminal fusion to a TEV cleavage site (GSENLYFQ) followed by two copies of StrepTagII (GSWSHPQFEKGSWSHPQFEK) was produced by cloning a PCR product generated with primers including the fusion sequences. APC4 was cloned into a pFastBac-1 derivative encoding an N-terminal GST tag followed by a TEV protease cleavage site. All clones were verified by DNA sequencing. Bacmids encoding each subunit were generated by transposition of DH10Bac E. coli (Invitrogen). Baculoviruses were produced by transfection of SF9 cells with FuGENE HD (Promega). P3 amplifications were used for all protein production. To express APC/C, High Five cells (Invitrogen) at a density of 1×106 ml−1 were co-infected with the 14 baculoviruses encoding the individual APC/C subunits, omitting the virus encoding APC15 as indicated. Cells were incubated post-infection for 24 hours at 27°C and two days at 20°C. APC/C was prepared from infected cells by a two-step affinity purification procedure, first utilizing the strep tag fused to APC6 and then the GST tag fused to APC4. All steps were conducted at 4°C. Cells were collected by centrifugation at 1,000 × g for 10 minutes and lysed by sonication in APC/C buffer (20 mM Tris pH 7.6, 150 mM NaCl, 1 mM DTT) containing complete EDTA-free protease inhibitors (Roche). Lysates were cleared by centrifugation at 15,000 rpm in an SS-34 rotor for 30 minutes, and the supernatant was incubated with streptactin-agarose (IBA) for 1 hour with agitation. Beads were transferred to a gravity flow column and washed with 10 column volumes of APC/C buffer. APC/C was eluted with 6 column volumes of buffer containing 2.5 mM desthiobiotin, passed over a glutathione-sepharose column by gravity flow, washed with 5 column volumes of APC/C buffer and eluted in APC/C buffer containing 10 mM reduced glutathione. The presence of all APC/C subunits was verified by mass spectrometry.
Recombinant APC/C ubiquitylation assays
Recombinant APC/C was assayed for ubiquitylation activity using a modification of a previously described method57. Immediately after preparation on ice, 6.5 μl of a “ubiquitylation mix” (190 nM E1, 190 nM UbcH10, 190 nM Ube2s, 1.5 mg ml−1 ubiquitin, 0.8 mg ml−1 BSA, 85 mM creatine phosphate, 12 mM ATP, 13 mM MgCl2, 5 mM Tris pH 7.5, 9 mM NaCl) was added to a 7 μl mixture of 80 nM recombinant APC/C and 2 μM substrate with or without 2 μM 3xMyc-His6-CDH1, as indicated in figures. Reactions were incubated at 25°C with 800 rpm shaking in a Thermomixer R (Eppendorf) for the indicated times and stopped by adding an equal volume of 2X Laemmli buffer without DTT. To distinguish between ubiquitin chain initiation and elongation, reactions were carried out with a fluorescently-labeled N-terminal fragment of cyclin-B1 or a fusion of ubiquitin to a fragment of cyclin-B1, UbcH10 or Ube2S and either CDH1 or CDC20 (Supplementary Note). These reactions were carried out at room temperature with 9 nM APC/C, 120 nM substrate, 1 μM coactivator in 15 mM Tris, 100mM NaCl, 5 mM MgATP, 0.25 mg ml−1 BSA, 190 nM E1, 190 nM of either E2, and initiated by addition of 125 μM ubiquitin. Reactions were separated by SDS-PAGE and FlAsH fluorescence was detected with a Storm 860 imager (Molecular Dynamics). To monitor 3xMyc-His6-CDC20 assays, samples were separated by SDS-PAGE on 12% polyacrylamide gels and Western blotted with an anti-CDC20 antibody (ab64877 Abcam).
Supplementary Material
Acknowledgements
Research in the laboratory of J.-M.P is supported by Boehringer Ingelheim, the Vienna Science and Technology Fund (WWTF LS09-13), the Laura Bassi Center for Optimized Structural Studies (FFG 822736) and the Austrian Science Fund (FWF special research program SFB F34 ‘Chromosome Dynamics’, grant W1221 ‘DK: Structure and Interaction of Biological Macromolecules’ and Wittgenstein award Z196-B20). Research in the laboratories of J.-M.P. and A.A.H. is supported by the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 241548 (MitoSys). Research in the laboratory of B.A.S. is supported by ALSAC/St. Jude and the Howard Hughes Medical Institute. Research in the laboratory of H.Stark is supported by the German Research Foundation (DFG) under grant agreement SFB860. Research in the laboratory of K.M. is supported by European Community’s Seventh Framework Programme under grant agreement no. 262067 (PRIME-XS). Y.T was supported by JSPS (Postdoctoral Fellowship for Research Abroad). N.G.B. is supported as a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
Accession numbers.
Electron Microscopy Database: recombinant human APC/C, EMD-2204.
References
- 1.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci. 2011;366:3595–604. doi: 10.1098/rstb.2011.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–8. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–7. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- 4.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–83. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hwang LH, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–4. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–7. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 7.Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–10. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 8.Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–69. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–43. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 10.Dube P, et al. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell. 2005;20:867–79. doi: 10.1016/j.molcel.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buschhorn BA, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2000;97:8973–8. doi: 10.1073/pnas.97.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leverson JD, et al. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–25. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, et al. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell. 2001;12:3839–51. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 17.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–65. [PMC free article] [PubMed] [Google Scholar]
- 18.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol Cell. 2005;18:543–53. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 19.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–8. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–13. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 21.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 23.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–25. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 25.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–43. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 26.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–5. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 28.Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107:5351–6. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–20. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–20. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao YF, et al. Cdk1-phosphorylated CUEDC2 promotes spindle checkpoint inactivation and chromosomal instability. Nat Cell Biol. 2011;13:924–33. doi: 10.1038/ncb2287. [DOI] [PubMed] [Google Scholar]
- 32.Jia L, et al. Defining pathways of spindle checkpoint silencing: functional redundancy between Cdc20 ubiquitination and p31(comet) Mol Biol Cell. 2011;22:4227–35. doi: 10.1091/mbc.E11-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teichner A, et al. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci U S A. 2011;108:3187–92. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–16. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miniowitz-Shemtov S, et al. Role of phosphorylation of Cdc20 in p31(comet)-stimulated disassembly of the mitotic checkpoint complex. Proc Natl Acad Sci U S A. 2012;109:8056–60. doi: 10.1073/pnas.1204081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia G, et al. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. Embo J. 2004;23:3133–43. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, et al. p31comet blocks Mad2 activation through structural mimicry. Cell. 2007;131:744–55. doi: 10.1016/j.cell.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–12. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 39.Hubner NC, et al. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–54. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–43. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–9. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kops GJ, et al. APC16 is a conserved subunit of the anaphase-promoting complex/cyclosome. J Cell Sci. 2010;123:1623–33. doi: 10.1242/jcs.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 44.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–15. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zachariae W, et al. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science. 1998;279:1216–9. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- 46.Hall MC, Torres MP, Schroeder GK, Borchers CH. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J Biol Chem. 2003;278:16698–705. doi: 10.1074/jbc.M213109200. [DOI] [PubMed] [Google Scholar]
- 47.Schreiber A, et al. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–32. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 48.Visconti R, Palazzo L, Grieco D. Requirement for proteolysis in spindle assembly checkpoint silencing. Cell Cycle. 2010;9:564–9. doi: 10.4161/cc.9.3.10581. [DOI] [PubMed] [Google Scholar]
- 49.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma HT, Poon RY. Orderly inactivation of the key checkpoint protein mitotic arrest deficient 2 (MAD2) during mitotic progression. J Biol Chem. 2011;286:13052–9. doi: 10.1074/jbc.M110.201897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foe IT, et al. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr Biol. 2011;21:1870–7. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oelschlaegel T, et al. The yeast APC/C subunit Mnd2 prevents premature sister chromatid separation triggered by the meiosis-specific APC/C-Ama1. Cell. 2005;120:773–88. doi: 10.1016/j.cell.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 53.Penkner AM, Prinz S, Ferscha S, Klein F. Mnd2, an essential antagonist of the anaphase-promoting complex during meiotic prophase. Cell. 2005;120:789–801. doi: 10.1016/j.cell.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 54.Foster SA, Morgan DO. The APC/C Subunit Mnd2/Apc15 Promotes Cdc20 Autoubiquitination and Spindle Assembly Checkpoint Inactivation. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 56.Kastner B, et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat Methods. 2008;5:53–5. doi: 10.1038/nmeth1139. [DOI] [PubMed] [Google Scholar]
- 57.Williamson A, Jin L, Rape M. Preparation of synchronized human cell extracts to study ubiquitination and degradation. Methods Mol Biol. 2009;545:301–12. doi: 10.1007/978-1-60327-993-2_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.