Abstract
The Dwarf crayfish or Cambarellinae, is a morphologically singular subfamily of decapod crustaceans that contains only one genus, Cambarellus. Its intriguing distribution, along the river basins of the Gulf Coast of United States (Gulf Group) and into Central México (Mexican Group), has until now lacked of satisfactory explanation. This study provides a comprehensive sampling of most of the extant species of Cambarellus and sheds light on its evolutionary history, systematics and biogeography. We tested the impact of Gulf Group versus Mexican Group geography on rates of cladogenesis using a maximum likelihood framework, testing different models of birth/extinction of lineages. We propose a comprehensive phylogenetic hypothesis for the subfamily based on mitochondrial and nuclear loci (3,833 bp) using Bayesian and Maximum Likelihood methods. The phylogenetic structure found two phylogenetic groups associated to the two main geographic components (Gulf Group and Mexican Group) and is partially consistent with the historical structure of river basins. The previous hypothesis, which divided the genus into three subgenera based on genitalia morphology was only partially supported (P = 0.047), resulting in a paraphyletic subgenus Pandicambarus. We found at least two cases in which phylogenetic structure failed to recover monophyly of recognized species while detecting several cases of cryptic diversity, corresponding to lineages not assigned to any described species. Cladogenetic patterns in the entire subfamily are better explained by an allopatric model of speciation. Diversification analyses showed similar cladogenesis patterns between both groups and did not significantly differ from the constant rate models. While cladogenesis in the Gulf Group is coincident in time with changes in the sea levels, in the Mexican Group, cladogenesis is congruent with the formation of the Trans-Mexican Volcanic Belt. Our results show how similar allopatric divergence in freshwater organisms can be promoted through diverse vicariant factors.
Introduction
The freshwater crayfish subfamily Cambarellinae is comprised of the unique genus Cambarellus, with 17 recognized species and a disjunctive distribution across the freshwater streams of the Gulf Cost of the United States and North and Central México (Fig. 1) [1]. The subfamily is unique because of the exceptionally small body size of its species. They typically reach only 4 cm compared to most crayfish averaging a maximum body size of >5 cm; hence, the reference to the genus as the “Dwarf” crayfishes. Their distribution goes from the Swanee River in northern Florida, eastward through the southern Mississippi River watershed to southern Illinois and continues southwest to the Nueces River in Texas [2], [3]. In México, Cambarellus has a discontinuous distribution with three distant and isolated populations from the northern states of Chihuahua, Coahuila and Nuevo León and then along the Trans-Mexican Volcanic Belt (TMVB) [4], [5]. The genus contains species largely inhabiting lakes and lentic habitats. The evolutionary history of such a broad and disjunct distribution of species is unclear and our goal with this study is to shed some light on the biogeography of the Cambarellinae.
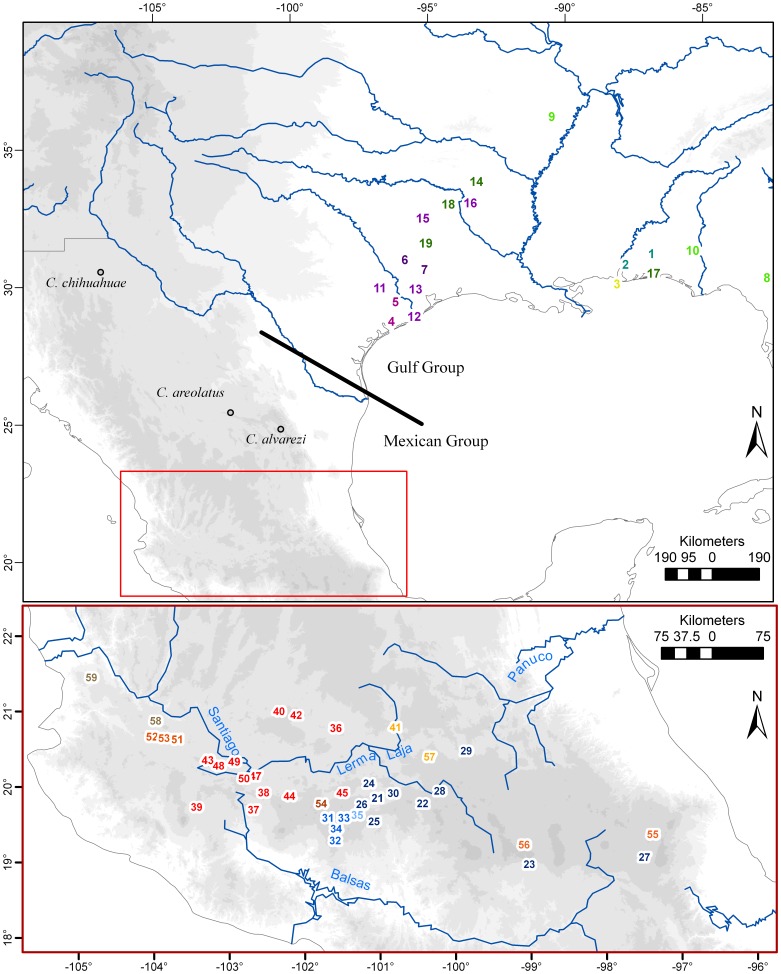
Figure 1. Map of localities sampled.
Map of localities sampled in this study, numbers are referred to in Table 1. Sample locations are colored to represent different clades recovered by phylogenetic analyses (see Fig. 3). Open circles correspond to the only locality records for the three species not included in the analyses as they were not found during sampling, or did not amplify during PCR reactions. Gray background refers to elevation (500–6000 m).
A series of apomorphic morphological characters define the subfamily and, therefore, monophyly has been accepted since its proposal. These include, as for other crayfish groups, genital morphology, which is particularly important, but also a small body size, specific branchial formula, movable and enlarged annulus ventralis (female genitalia) and the absence of the cephalic process in the first pair of pleopods (male genitalia) [1], [2], [6]. The morphological unity of these characters that define the subfamily contrasts with the wide morphological variation in other characters described for populations of several species [2], [5], [7]. This diversity within and among species makes designation and identification difficult, especially for widely distributed species [2], [5].
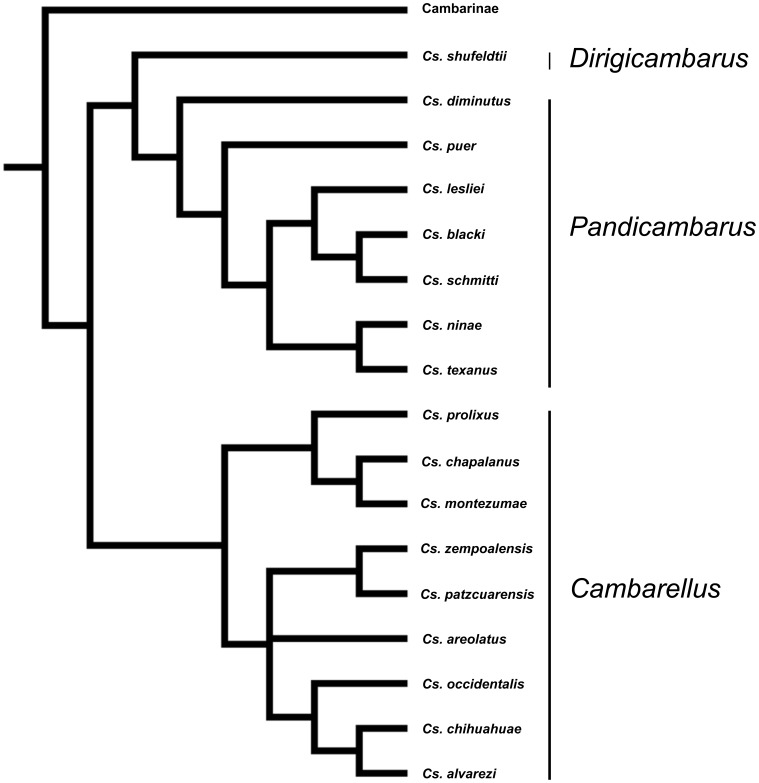
Despite the intriguing geographic distribution and species diversity in the Cambarellinae, the only phylogenetic hypothesis for species relationships in the group is based on phenotypic information and genital morphology [2]. With this hypothesis (Fig. 2) three subgenera were proposed; Pandicambarus (containing seven species), the monotypic Dirigicambarus, both comprised of species occurring north of the Rio Grande (the Gulf Group), and Cambarellus, containing species south of the Rio Grande (the Mexican Group) [2]. However, no apomorphic characters have been proposed to support these subgeneric classifications and no formal phylogenetic hypothesis has been evaluated using either molecular or morphological characters. Therefore, we propose to estimate a robust phylogenetic hypothesis for the group using an extensive molecular data set. We then use this phylogenetic framework to evaluate a coherent taxonomy for the group and to test biogeographic hypotheses regarding the origin and spread of the dwarf crayfish.
Figure 2. Morphologic hypothesis tested.
Phylogenetic hypothesis based on morphologic analysis of the monotypic subfamily Cambarellinae (genus Cambarellus), indicated are the subgenera previously proposed, mainly based on genital morphology (Fitzpatrick, 1983).
We also examine diversification patterns in the subfamily through the estimated phylogenetic history of the species within the subfamily. Phylogenetic diversity patterns are impacted by geographic features and geologic history due to their effects on allopatric speciation [8]. Given the contrasting geographical features (Fig. 1) coupled with their distinct geological histories occupied by the different groups in Cambarellinae, we will use reconstructed molecular phylogenies to serve as models of lineages through time (LTT), that will allow us to test the tempo and pattern of change across lineages [9], [10], [11]. In the present study, we used our molecular dataset on the subfamily Cambarellinae to infer the timing and mode of lineage accumulation (patterns of speciation minus extinction) which allows us to determine whether there have been contrasting patterns in rates of diversification between the two geographical components of this group; namely, those defined as the Gulf and Mexican Groups, as a result of contrasting biogeographic histories. Finally, we identify a geological timescale consistent with biogeographic factors and cladogenetic events in this group.
Materials and Methods
Sampling and Sequencing
No specific permits were required for the described field studies, as none of the studied species were included in any endangered list, at national or international levels at the time of sampling (comprising the years 2005 and 2006). Including field and museum localities, 59 geographic locations covering 14 of the 17 species were collected throughout the distributional range of the subfamily Cambarellinae (Fig. 1). Taxonomic identification was carried out using existing keys [12]. The two main ranges for the subfamily were covered, along the Neartic and the Transition zone of North America, from the Mississippi River basin to the TMVB in central México. Most of the species could be sampled, but those tissues from species with very restricted distribution ranges and/or being collected in a reduced number of times in wild were obtained from museum specimens (National Museum of Natural History, Smithsonian Institution) (Table 1). Detailed data about samples included are summarized in the Table S1.
Table 1. Sampling localities and Genbank accession numbers from individuals of Cambarellus used in this study.
| + | Species id fromthis study | Subgenus(Fitzpatrick,1983) | GeneBank accession numbers | ||||
| 16S | 12S | cox1 | 28S | H3 | |||
| 1 | Cambarellus blacki | Pandicambarus | JX127836 | JX127697 | JX127977 | JX127568 | JX127429 |
| 1 | Cambarellus blacki | Pandicambarus | JX127837 | JX127698 | JX127978 | JX127569 | JX127430 |
| 2 | Cambarellus diminutus * | Pandicambarus | JX127810 | JX127953 | JX127545 | JX127405 | |
| 3 | Cambarellus lesliei * | Pandicambarus | JX127809 | JX127952 | JX127544 | JX127404 | |
| 4 | Cambarellus ninae | Pandicambarus | JX127814 | JX127957 | JX127549 | JX127409 | |
| 5 | Cambarellus ninae ** | Pandicambarus | JX127833 | JX127694 | JX127974 | JX127565 | JX127426 |
| 6 | Cambarellus puer1233 | Pandicambarus | JX127822 | JX127686 | JX127965 | JX127557 | JX127417 |
| 7 | Cambarellus puer | Pandicambarus | JX127815 | JX127958 | JX127550 | JX127410 | |
| 8 | Cambarellus schmitti | Pandicambarus | JX127811 | JX127954 | JX127546 | JX127406 | |
| 9 | Cambarellus schmitti | Pandicambarus | JX127838- | JX127699- | JX127979- | JX127570- | JX127431- |
| JX127855 | JX127714 | JX127996 | JX127587 | JX127447 | |||
| 10 | Cambarellus schmitti | Pandicambarus | JX127856 | JX127715 | JX127997 | JX127448 | |
| 11 | Cambarellus texanus | Pandicambarus | JX127832 | ||||
| 12 | Cambarellus texanus *** | Pandicambarus | JX127834 | JX127695 | JX127975 | JX127566 | JX127427 |
| 13 | Cambarellus texanus | Pandicambarus | JX127819 - | JX127683 – | JX127962 – | JX127554 – | JX127414 – |
| JX127821 | JX127685 | JX127964 | JX127556 | JX127416 | |||
| 14 | Cambarellus shufeldtii | Dirigicambarus | JX127812 | JX127955 | JX127547 | JX127407 | |
| 15 | Cambarellus shufeldtii | Dirigicambarus | JX127816 | JX127959 | JX127551 | JX127411 | |
| 16 | Cambarellus shufeldtii | Dirigicambarus | JX127817 | JX127960 | JX127552 | JX127412 | |
| 17 | Cambarellus shufeldtii | Dirigicambarus | |||||
| 18 | Cambarellus shufeldtii | Dirigicambarus | JX127835 | JX127696 | JX127976 | JX127567 | JX127428 |
| 19 | Cambarellus shufeldtii | Dirigicambarus | JX127857 | JX127998 | JX127588 | JX127449 | |
| 20 | Cambarellus shufeldtii | Dirigicambarus | JX127818 | JX127961 | JX127553 | JX127413 | |
| 21 | Cambarellus zempoalensis | Cambarellus | JX127725 | JX127599 | JX127868 | JX127460 | JX127320 |
| 21 | Cambarellus zempoalensis | Cambarellus | JX127770 | JX127644 | JX127913 | JX127505 | JX127365 |
| 22 | Cambarellus zempoalensis | Cambarellus | JX127747 | JX127621 | JX127890 | JX127482 | JX127342 |
| 22 | Cambarellus zempoalensis | Cambarellus | JX127759 | JX127633 | JX127902 | JX12749 | JX127354 |
| 22 | Cambarellus zempoalensis | Cambarellus | JX127772 | JX127646 | JX127915 | JX127507 | JX127367 |
| 22 | Cambarellus zempoalensis | Cambarellus | JX127773 | JX127647 | JX127916 | JX127508 | JX127368 |
| 23 | Cambarellus zempoalensis | Cambarellus | JX127750 | JX127624 | JX127893 | JX127485 | JX127345 |
| 23 | Cambarellus zempoalensis | Cambarellus | JX127756 | JX127630 | JX127899 | JX127491 | JX127351 |
| 24 | Cambarellus zempoalensis | Cambarellus | JX127786 | JX127660 | JX127929 | JX127521 | JX127381 |
| 25 | Cambarellus zempoalensis | Cambarellus | JX127744 | JX127618 | JX127887 | JX127479, | JX127339 |
| 25 | Cambarellus zempoalensis | Cambarellus | JX127753 | JX127627 | JX127896 | JX127488 | JX127348 |
| 25 | Cambarellus zempoalensis | Cambarellus | JX127794 | JX127668 | JX127937 | JX127529 | JX127389 |
| 26 | Cambarellus zempoalensis | Cambarellus | JX127743 | JX127617 | JX127886 | JX127478 | JX127338 |
| 26 | Cambarellus zempoalensis | Cambarellus | JX127755 | JX127629 | JX127898 | JX127490 | JX127350 |
| 26 | Cambarellus zempoalensis | Cambarellus | JX127765 | JX127639 | JX127908 | JX127500 | JX127360 |
| 26 | Cambarellus zempoalensis | Cambarellus | JX127791 | JX127665 | JX127934 | JX127526 | JX127386 |
| 27 | Cambarellus zempoalensis | Cambarellus | JX127732 | JX127606 | JX127875 | JX127467 | JX127327 |
| 28 | Cambarellus zempoalensis | Cambarellus | JX127771 | JX127645, | JX127914 | JX127506 | JX127366 |
| 28 | Cambarellus zempoalensis | Cambarellus | JX127793 | JX127667 | JX127936 | JX127528 | JX127388 |
| 29 | Cambarellus zempoalensis | Cambarellus | JX127736 | JX127610 | JX127879 | JX127471 | JX127331 |
| 29 | Cambarellus zempoalensis | Cambarellus | JX127754 | JX127628 | JX127897 | JX127489 | JX127349 |
| 29 | Cambarellus zempoalensis | Cambarellus | JX127789 | JX127663 | JX127932 | JX127524 | JX127384 |
| 29 | Cambarellus zempoalensis | Cambarellus | JX127805 | JX127679 | JX127948 | JX127540 | JX127400 |
| 30 | Cambarellus zempoalensis | Cambarellus | JX127798 | JX127672 | JX127941 | JX127533 | JX127393 |
| 30 | Cambarellus zempoalensis | Cambarellus | JX127799 | JX127673 | JX127942 | JX127534 | JX127394 |
| 31 | Cambarellus patzcuarensis | Cambarellus | JX127728 | JX127602 | JX127871 | JX127463 | JX127323 |
| 31 | Cambarellus patzcuarensis | Cambarellus | JX127740 | JX127614 | JX127883 | JX127475 | JX127335 |
| 31 | Cambarellus patzcuarensis | Cambarellus | JX127751 | JX127625 | JX127894 | JX127486 | JX127346 |
| 31 | Cambarellus patzcuarensis | Cambarellus | JX127774 | JX127648 | JX127917 | JX127509 | JX127369 |
| 32 | Cambarellus patzcuarensis | Cambarellus | JX127802 | JX127676 | JX127945 | JX127537 | JX127397 |
| 32 | Cambarellus patzcuarensis | Cambarellus | JX127803 | JX127677 | JX127946 | JX127538 | JX127398 |
| 33 | Cambarellus patzcuarensis | Cambarellus | JX127741 | JX127615 | JX127884 | JX127476 | JX127336 |
| 33 | Cambarellus patzcuarensis | Cambarellus | JX127775 | JX127649 | JX127918 | JX127510 | JX127370 |
| 34 | Cambarellus patzcuarensis | Cambarellus | JX127779 | JX127653 | JX127922 | JX127514 | JX127374 |
| 35 | Cambarellus sp. (cladeIII) | Cambarellus | JX127738 | JX127612 | JX127881 | JX127473 | JX127333 |
| 35 | Cambarellus sp. (cladeIII) | Cambarellus | JX127752 | JX127626 | JX127895 | JX127487 | JX127347 |
| 36 | Cambarellus chapalanus | Cambarellus | JX127726 | JX127600 | JX127869 | JX127461 | JX127321 |
| 36 | Cambarellus chapalanus | Cambarellus | JX127760 | JX127634 | JX127903 | JX127495 | JX127355 |
| 37 | Cambarellus chapalanus | Cambarellus | JX127733 | JX127607 | JX127876 | JX127468 | JX127328 |
| 37 | Cambarellus chapalanus | Cambarellus | JX127734 | JX127608 | JX127877 | JX127469 | JX127329 |
| 37 | Cambarellus chapalanus | Cambarellus | JX127737 | JX127611 | JX127880 | JX127472 | JX127332 |
| 37 | Cambarellus chapalanus | Cambarellus | JX127764 | JX127638 | JX127907 | JX127499 | JX127359 |
| 37 | Cambarellus chapalanus | Cambarellus | JX127830 | JX127693 | JX127972 | JX127564 | JX127425 |
| 38 | Cambarellus chapalanus | Cambarellus | JX127795 | JX127669 | JX127938 | JX127530 | JX127390 |
| 38 | Cambarellus chapalanus | Cambarellus | JX127796 | JX127670 | JX127939 | JX127531 | JX127391 |
| 38 | Cambarellus chapalanus | Cambarellus | JX127800 | JX127674 | JX127943 | JX127535 | JX127395 |
| 39 | Cambarellus chapalanus | Cambarellus | JX127742 | JX127616 | JX127885 | JX127477 | JX127337 |
| 39 | Cambarellus chapalanus | Cambarellus | JX127766 | JX127640 | JX127909 | JX127501 | JX127361 |
| 40 | Cambarellus chapalanus | Cambarellus | JX127783 | JX127657 | JX127926 | JX127518 | JX127378 |
| 41 | Cambarellus chapalanus | Cambarellus | JX127748 | JX127622 | JX127891 | JX127483 | JX127343 |
| 41 | Cambarellus chapalanus | Cambarellus | JX127749 | JX127623 | JX127892 | JX127484 | JX127344 |
| 41 | Cambarellus chapalanus | Cambarellus | JX127768 | JX127642 | JX127911 | JX127503 | JX127363 |
| 41 | Cambarellus chapalanus | Cambarellus | JX127792 | JX127666 | JX127935 | JX127527 | JX127387 |
| 41 | Cambarellus chapalanus | Cambarellus | JX127797 | JX127671 | JX127940 | JX127532 | JX127392 |
| 42 | Cambarellus chapalanus | Cambarellus | JX127781 | JX127655 | JX127924 | JX127516 | JX127376 |
| 43 | Cambarellus prolixus | Cambarellus | JX127729 | JX127603 | JX127872 | JX127464 | JX127324 |
| 43 | Cambarellus prolixus | Cambarellus | JX127761 | JX127635 | JX127904 | JX127496 | JX127356 |
| 43 | Cambarellus prolixus | Cambarellus | JX127762 | JX127636 | JX127905 | JX127497 | JX127357 |
| 44 | Cambarellus chapalanus | Cambarellus | JX127735 | JX127609 | JX127878 | JX127470 | JX127330 |
| 44 | Cambarellus chapalanus | Cambarellus | JX127769 | JX127643 | JX127912 | JX127504 | JX127364 |
| 45 | Cambarellus chapalanus | Cambarellus | JX127801 | JX127675 | JX127944 | JX127536 | JX127396 |
| 46 | Cambarellus chapalanus | Cambarellus | JX127739 | JX127613 | JX127882 | JX127474 | JX127334 |
| 46 | Cambarellus chapalanus | Cambarellus | JX127758 | JX127632 | JX127901 | JX127493 | JX127353 |
| 47 | Cambarellus chapalanus | Cambarellus | JX127745 | JX127619 | JX127888 | JX127480 | JX127340 |
| 47 | Cambarellus chapalanus | Cambarellus | JX127746 | JX127620 | JX127889 | JX127481 | JX127341 |
| 48 | Cambarellus prolixus | Cambarellus | JX127806 | JX127680 | JX127949 | JX127541 | JX127401 |
| 48 | Cambarellus prolixus | Cambarellus | JX127807 | JX127681 | JX127950 | JX127542 | JX127402 |
| 49 | Cambarellus prolixus | Cambarellus | JX127808 | JX127682 | JX127951 | JX127543 | JX127403 |
| 50 | Cambarellus chapalanus | Cambarellus | JX127804 | JX127678 | JX127947 | JX127539 | JX127399 |
| 51 | Cambarellus sp. (clade V) | Cambarellus | JX127780 | JX127654 | JX127923 | JX127515 | JX127375 |
| 52 | Cambarellus sp. (clade V) | Cambarellus | JX127782 | JX127656 | JX127925 | JX127517 | JX127377 |
| 53 | Cambarellus sp. (clade V) | Cambarellus | JX127787 | JX127661 | JX127930 | JX127522, | JX127382 |
| 53 | Cambarellus sp. (clade V) | Cambarellus | JX127788 | JX127662 | JX127931 | JX127523 | JX127383 |
| 54 | Cambarellus sp. (clade VI) | Cambarellus | JX127730 | JX127604 | JX127873 | JX127465 | JX127325 |
| 54 | Cambarellus sp. (clade VI) | Cambarellus | JX127757 | JX127631 | JX127900 | JX127492 | JX127352 |
| 54 | Cambarellus sp. (clade VI) | Cambarellus | JX127790 | JX127664 | JX127933 | JX127525 | JX127385 |
| 55 | Cambarellus montezumae | Cambarellus | JX127731 | JX127605 | JX127874 | JX127466 | JX127326 |
| 55 | Cambarellus montezumae | Cambarellus | JX127763 | JX127637 | JX127906 | JX127498 | JX127358 |
| 56 | Cambarellus montezumae | Cambarellus | JX127776 | JX127650 | JX127919 | JX127511 | JX127371 |
| 56 | Cambarellus montezumae | Cambarellus | JX127777 | JX127651 | JX127920 | JX127512 | JX127372 |
| 56 | Cambarellus montezumae | Cambarellus | JX127778 | JX127652 | JX127921 | JX127513 | JX127373 |
| 57 | Cambarellus sp. (clade VIII) | Cambarellus | JX127727 | JX127601 | JX127870 | JX127462 | JX127322 |
| 57 | Cambarellus sp. (clade VIII) | Cambarellus | JX127767 | JX127641 | JX127910 | JX127502 | JX127362 |
| 58 | Cambarellus occidentalis | Cambarellus | JX127784 | JX127658 | JX127927 | JX127519 | JX127379 |
| 58 | Cambarellus occidentalis | Cambarellus | JX127785 | JX127659 | JX127928 | JX127520 | JX127380 |
| 59 | Cambarellus occidentalis | Cambarellus | JX127813 | JX127956 | JX127548 | JX127408 | |
| Procambarus toltecae | JX127823 | JX127687 | JX127966 | JX127558 | JX127418 | ||
| Procambarus acutus1 | JX127824 | JX127688 | JX127967 | JX127559 | JX127419 | ||
| Procambarus acutus2 | JX127827 | JX127970 | JX127562 | JX127422 | |||
| Procambarus llamasi1 | JX127825 | JX127689 | JX127968 | JX127560 | JX127420 | ||
| Procambarus llamasi2 | JX127826 | JX127690 | JX127969 | JX127561 | JX127421 | ||
| Procambarus clarkii | JX127829 | JX127692 | JX127971 | JX127563 | JX127424 | ||
| Procambarus bouvieri | JX127828 | JX127691 | JX127423 | ||||
| Orconectes deanae | JX127859 | JX127717 | JX128000 | JX127590 | JX127451 | ||
| Orconectes ronaldi | JX127865 | JX127722 | JX128005 | JX127596 | JX127457 | ||
| Orconectes virilis1 | JX127866 | JX127723 | JX128006 | JX127597 | JX127458 | ||
| Orconectes virilis2 | JX127860 | JX127591 | JX127452 | ||||
| Cambarus brachydactylus ++ | DQ411732 | DQ411729 | DQ411783 | DQ411802 | |||
| Cambarus maculatus | JX127864 | JX127721 | JX128004 | JX127595 | JX127456 | ||
| Cambarus pyronotus | JX127862 | JX127719 | JX128002 | JX127593 | JX127454 | ||
| Cambarus striatus | JX127861 | JX127718 | JX128001 | JX127592 | JX127453 | ||
| Fallicambarus byersi | JX127863 | JX127720 | JX128003 | JX127594 | JX127455 | ||
| Fallicambarus caesius | JX127867 | JX127724 | JX128007 | JX127598 | JX127459 | ||
| Fallicambarus fodiens | JX127858 | JX127716 | JX127999 | JX127589 | JX127450 | ||
Locality number, as depicted in Figure 1.
Type specimens or type localities.
Morphologically identified as C. shufeldtii.
Morphologically identified as C. puer.
Sequence from the study of Buhay et al. 2007, tissue originally from the Carnegie Museum of Natural History.
Populations termed as ‘C. sp’ are new proposed taxa, according to phylogenetic structure (see Figure 3).
Populations from clade I are included in the lineage of C. zempoalensis, species which has to be re-examined by incorporing C. montezumae lermensis in the analysis.
The central goal of this work is to estimate a robust phylogenetic hypothesis for relationships among the species within the subfamily to test taxonomic hypotheses, biogeographic hypotheses, and speciation hypotheses. As phylogenies are most accurately estimated using broad taxonomic sampling as well as extensive character sampling, we attempted to sample all species within the subfamily (but are missing three of them) and collected sequence data from five different gene regions (three mitochondrial and two nuclear). We sequenced the mitochondrial genes 16S rDNA (16S), 12S rDNA (12S) and Cytochrome Oxidase subunit I (COI). These genes have good phylogenetic signal in crustaceans [13] and are considered optimal choices to characterize the genetic variation in crustacean groups. Nuclear genes sequenced were 28S rDNA large ribosomal unit (28S) and Histone 3 (H3) gene, which also have some variation among species and are particularly good at discerning deeper nodes [13].
PCR amplifications using gene specific primers (Table 2) were carried out in 25 µL reactions containing: 1× PCR buffer, 0.5 µM of each primer, 0.2 mM of each dNTP, 1.5 mM MgCl2, 1 U Taq polymerase (Biotools), and about 10–50 ng of template DNA. The cycling profile for PCR amplifications was 3 min at 94°C (1 cycle), 30 s at 94°C, 30 s at the primer-specific melting temperature and 60 s at 72°C (30 cycles), followed by a final extension of 4 min at 72°C. PCR products were visualized in 1.0% agarose gels (1×TBE) and stained with SYBR-Safe (Invitrogen). Fragments were sequenced on an ABI 3730XL DNA Analyzer. Sequences of the different gene fragments were aligned using MUSCLE [14]. In the case of the COI gene, recommendations to detect the occurrence of possible nmtDNA were carried out for each sequence. These included the identification of stop codons, repeated sequencing of samples, nonsynonymous substitution and unusual levels of genetic divergence in samples from the same population [15], [16].
Table 2. Primer and PCR conditions used in this study to amplify different gene regions.
| Generegion | primers | sequence | Tm(°C) | Reference |
| COI | COIAR | GTTGTTATAAAATTHACTGARCCT | 48.5 | This study |
| COIBF | GCYTCTGCKATTGCYCATGCAGG | 48.5 | This study | |
| COIBR | TGCRTAAATTATACCYAAAGTACC | 48.5 | This study | |
| COICF | ACCTGCATTTGGRATAGTATCTC | 48.5 | This study | |
| COICR | GAAWYTTYAATCACTTCTGATTTA | 48.5 | This study | |
| COIDF | CTGGRATTGTTCATTGATTTCCT | 48.5 | This study | |
| ORCO1F | AACGCAACGATGATTTTTTTCTAC | 48.5 | [75] | |
| ORCO1R | GGAATYTCAGMGTAAGTRTG | 48.5 | [75] | |
| 16S | 1471 | CCTGTTTANCAAAAACAT | 46 | [76] |
| 16S-1472 | AGATAGAAACCAACCTGG | 46 | [76] | |
| 12S | 12sf | GAAACCAGGATTAGATACCC | 53 | [77] |
| 12sr | TTTCCCGCGAGCGACGGGCG | 53 | [77] | |
| 28S | 28s-rD1a | CCCSCGTAATTTAAGCATATTA | 52 | [78], [79] |
| 28s-rD3b | CCYTGAACGGTTTCACGTACT | 52 | [78], [79] | |
| 28s-rD3a | AGTACGTGAAACCGTTCAGG | 52 | [78], [79] | |
| 28s-rD4b | CCTTGGTCCGTGTTTCAAGAC | 52 | [78], [79] | |
| 28sA | GACCCGTCTTGAAGCACG | 52 | [78], [79] | |
| 28S B | TCGGAAGGAACCAGCTAC | 52 | [78], [79] | |
| H3 | H3 AF | ATGGCTCGTACCAAGCAGACVGC | 57 | [80] |
| H3 AR | ATATCCTTRGGCATRATRGTGAC | 57 | [80] |
Phylogenetic Analyses
Partition homogeneity tests were carried out on the concatenated matrix using PAUP v. 4.0b10 [17]. We examined homogeneity across partitions by gene and by codon position for protein-translated fragments (Table 3). We estimated phylogenies using Maximum Likelihood (ML) and Bayesian Inference (BI) approaches. Additionally, we used 15 species of the family Cambaridae as outgroups: Cambarus maculatus, C. striatus, C. pyronotus, C. brachidactylus, Orconectes ronaldi, O. virilis, O. deanae, Fallicambarus caesius, F. fodiens, F. byersi, Procambarus bouvieri, P. clarkii, P. llamasi, P. acutus and P. toltecae (Table 1).
Table 3. Substitution model and phylogenetic performance of each gene fragment.
| Gene | Size (pb) | Substitution model/gamma parameter/Invariable sites | Variable sites | PI | %PI | |
| AICc | BIC | |||||
| 16S | 501 | HKY+G; 0.232 | HKY+G; 0.230 | 199 | 121 | 24.1 |
| 12S | 358 | K80+G; 0.219 | TVM+G; 0.213 | 143 | 80 | 22.3 |
| COI | 1527 | HKY+G; 0.321 | HKY+G; 0.321 | 530 | 502 | 32.8 |
| 28S | 992 | TIM3+G; 0.031 | TIM3+G; 0.031 | 39 | 28 | 2.8 |
| H3 | 322 | JC; – | HKY+I; 0.834 | 31 | 24 | 7.4 |
| All | 3700 | GTR+G; 0.256 | GTR+G; 0.254 | 1431 | 847 | 22.8 |
In order to identify the most appropriate evolutionary model of nucleotide substitution (Table 2), we considered the Akaike corrected information criterion (AICc) [18], and the Bayesian Information Criterion (BIC) [19] as estimated using the program jModeltest [20]. A phylogenetic tree was constructed under ML using PHYML 3.0 [21] and AICc-selected parameters for the concatenated matrix. The tree search was started with an initial BIONJ tree estimation followed by a Subtree Pruning and Regrafting (SPR) topological moves algorithm. We assessed confidence in branches using 1000 nonparametric bootstrap [22] replicates under the best-fit evolutionary model.
Bayesian inference of phylogeny was implemented in MrBayes v. 3.1.2 [23], following the BIC-selected parameters and applying a Monte Carlo Markov Chain (MCMC) search procedure for 10 million generations. Sequences were partitioned by codon position for COI and by gene for the rest of fragments, using the parameters found by BIC as priors and unlinking the run parameters. Convergence between the different run parameters in paired simultaneous runs (4 chains by run), trees were sampled every 100 generations and run length was adjusted considering an adequate sampling based on average standard deviation of split frequencies being <0.01 [24]. We examined the results and determined the burn-in period as the set of trees saved prior to log likelihood stabilization and convergence as estimated using Tracer 1.4.1 [25], eventually the first 10% trees. Tracer was also used to check for convergence between chain runs and optimal values of run parameters. Confidence in nodes was assessed from the posterior probabilities along the MCMC run. Highly supported nodes are termed herein as those with a value of 95% or more in posterior probabilities and bootstrap values.
We tested our resulting topology against the phylogenetic hypotheses put forth by Fitzpatrick [2]; namely, the three subgenera are monophyletic and show the following relationships ((Dirigicambarus, Pandicambarus),Cambarellus). Topology constrained ML scores were estimated for each hypothesis in PAUP*. Congruence with alternative hypotheses was evaluated in a ML framework applying the Shimodaira-Hasegawa (SH; [26]) test and the Approximate Unbiased (AU) test [27] with 50,000 RELL bootstrap replicates as implemented in TreeFinder [28]. We also tested these hypotheses using a Bayesian approach by identifying the alternative hypothesis within the set of Bayesian tree topologies and testing for significant differences. To do so, we filtered the post-burnin Bayesian topologies included in the set of trees with the constraint topology in PAUP* [17].
Divergence Dating
In order to propose an accurate time frame for phylogenetic divergence processes, we estimated mean node ages and their 95% highest posterior densities (HPDs) using Bayesian relaxed molecular clock methods [29] as implemented in BEAST ver. 1.6.1 [30]. In this method, tests of evolutionary hypotheses are not conditioned on a single tree topology, which allows for simultaneous evaluation of topology and divergence times while incorporating uncertainty in both. A uniform Yule tree prior was specified, as appropriate for hierarchical rather than reticulate relationships, and a subsampling of one representative of every lineage was included to avoid over-representation of certain individual lineages with more sampling. We applied the optimal model of data partitioning and DNA substitution identified by BIC for each gene (COI, 16S, 12S, 28S and H3) and for codon positions for COI. An uncorrelated relaxed lognormal molecular clock was applied to model rate variation across branches, and pertinence of a relaxed estimation was checked after verifying that the distribution of the coefficient of variation was >1. The dating analysis was performed with the total matrix, but calibration of the molecular clock was done using COI and 16S mutation rates only, as information on rates of mutation of these two fragments is widely described in multiple groups and for which there is extensive fossil calibrated divergence time data in crustaceans [31], [32]. As a representation of these substitution rates, we considered the range to include extreme values reported, which extends between 0.23–1.1% per million years (PMY) for 16S [33], [34] and 0.7–1.3% PMY for COI [35], [36], [37]. These sets were introduced as uniform prior distributions, as no evidence justifies a specific distribution of rates in our data, avoiding the introduction any additional bias to the rate values assumed. Considering the geographic distribution of the genus, a geological calibration was also included as identified with the uplifting of the TMVB, which began around 12 MYA [38]. This age was set as a maximum for MRCA of the Mexican species. Additionally, fossil calibration was included in one point as the minimum age to account from the oldest fossil from the genus Procambarus [a Procambarus primaevus, 52.6–53.4 MYA, [39]]. Monophyly was not enforced for any node. Analyses were run for 20 million generations with a sampling frequency of 2000 generations. Tracer was used to determine the appropriate burn-in by monitoring run parameters by ensuring all effective sample sizes (ESS) were larger than 200 and independent runs converged. Two million generations were discarded before recording parameters and four independent runs were performed to ensure values were converging on similar estimates.
Diversification Patterns
The two main components of the subfamily occupy two regions highly contrasting in topography and biogeographic history. Thus, a second objective in this study was to describe the patterns of cladogenesis involved in the evolutionary history of Cambarellinae and to test the hypothesis that the different biogeographic histories from the two different geographic ranges of the subfamily (i.e., the Mexican and Gulf Groups), could lead to contrasting cladogenetic patterns evidenced by possible diversification shifts. Shifts in birth and death rates can leave distinctive signatures in phylogenies, resulting in departures from linearity in semi-log LTT plots [9], [11]. We compared diversification rates from the reconstructed phylogeny of the entire subfamily and of the two main clades (Mexican Group vs. Gulf Group) to different null models of diversification by using the Birth-Death Likelihood method (BDL). This temporal method was used to test different hypothesis of cladogenesis rate shifts [40]. BDL uses maximum likelihood estimates of speciation rate parameters and a likelihood score per tree, and test different rate-variable models against null models of rate-constancy under the Akaike Information Criterion (AIC) [18]. To provide an indication of the diversification rates in each case, we generated a logarithm LTT plot using the LASER package version 2.2 [41]. The LTT plot was generated from the Maximum Clade Credibility tree from BEAST, after pruning the terminals not included in each clade tested using TreeEdit v1.0a10 [42] and rooting the basal age to the one observed from the dating analysis. To test for significant departures from the null hypothesis of rate-constancy, observed ΔAICRC from our data was compared to those from the different rate diversification models using BDL as implemented in the LASER package version 2.2 [41]. The test statistic for diversification rate-constancy is calculated as: ΔAICRC = ΔAICRC−ΔAICRV, where AICRC is the Akaike Information Criterion score for the best fitting constant-rate diversification model, and AICRv is the AIC for the best fitting variable-rate diversification model. Thus, a positive value for ΔAICRC indicates that a rate-variable model best approximates the data. We tested five different models, of which two are rate-constant and three are rate-variable: 1) the constant-rate birth model (Yule) [the Yule process; [43]] with one parameter λ and μ set to zero; 2) the constant-rate birth-death model with two parameters λ and μ (BD); 3) a pure birth rate-variable model (yule2rate) where the speciation rate λ1 shifts to rate λ2 at time ts, with three parameters (λ1, λ2, ts); density-dependent speciation models with two variants, 4) exponential (DDX) and 5) logistic (DDL). Significance of the change in AIC scores was tested by generating a distribution of scores. This was done through simulation of 9000 trees using yuleSim in LASER, for the entire Cambarellinae subfamily and each geographic group, reflecting our sampling size in each case and having the same speciation rate as under the pure-Birth model.
Results
Phylogeny
We sequenced three mitochondrial (16S (519 bps), 12S (365 bps) and COI (1527 bps)) and two nuclear (28S 1100 bps and H3 322 bps) gene fragments resulting in 3833 characters (2411 mitochondrial and 1422 nuclear) and giving a series of substitution models (Table 3). These new data have been deposited in GenBank (Table 1). COI-like sequences were found in seven cases, identified by the occurrence of one or several stop-codons along the sequence and an unusual sequence divergence, which affected position in the tree and divergence regarding the other sequences coming from the same population. These sequences were removed from data sets and not considered for any analysis. As previously reported [15], when working with COI sequences in crayfish these sequences have to be specially checked to ensure they are mitochondrial.
The most variable fragment was 12S, followed by COI and 16S (variable sites: COI = 530/1527, 16S = 199/519 12S = 143/365; besides this, COI showed the highest proportion of parsimony informative (PI) sites: COI = 419, 16S = 121, 12S = 80) (Table 3). As expected, nuclear fragments were the most conservative (for the mitochondrial set, variable sites = 1187, PI = 783; for the nuclear set, variable sites = 244, PI = 64). The complete combined data set contained 1431 variable sites (∼37%), and 847 PI (∼22%).
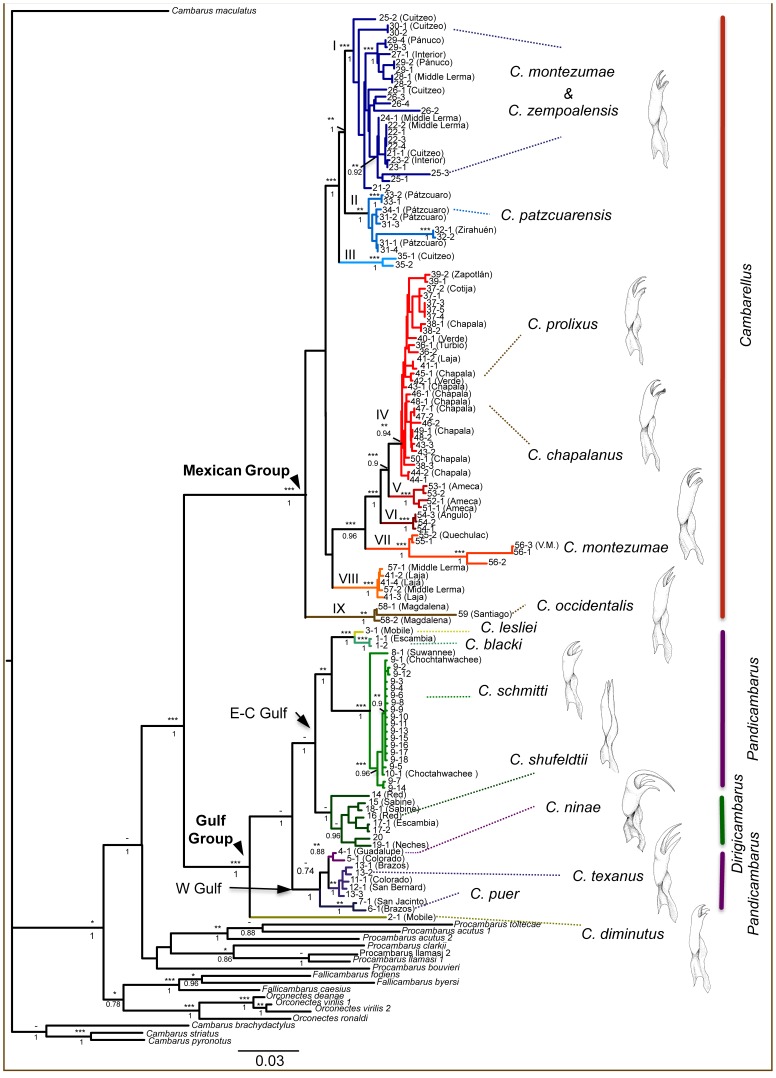
The topologies recovered by mitochondrial and nuclear analyses based on ML and BI methods were similar (Figure 3), although some discrepancies can be found in some terminal taxa arrangements and between genera-outgroup relationships, principally concerning the relative positions of Cambaridae genera representatives. Both topologies show Cambarellus as a monophyletic clade (Figure 3). Within Cambarellus we found two divergent clades which correspond to the two distinct geographic ranges of the genus based on a highly supported node by ML and BI analyses (more than 95% of nodal support values). The first lineage included the species from the Mexican Group, coincident with the TMVB in México. The second lineage included the Gulf Group, containing the species distributed in USA. Only results from the combined analyses of mitochondrial and nuclear information are shown, as nuclear evidence did not have enough phylogenetic signal to distinguish relationships within each geographic group (Mexican and Gulf Groups). As shown in different studies, mitochondrial and nuclear information could resolve different portions of the phylogeny (i.e., shallow vs. deep levels of tree, [44], [45] ) and that was one of the major reasons for combining these data types in this study. The hypothesis explaining this is that long-branch attraction might be more common among deeper nodes, and that slow-evolving nuclear DNA might help to resolve such issues [46], [47].
Figure 3. Phylogenetic tree of Cambarellus genus.
Phylogenetic tree of Cambarellus based on three mitochondrial and two nuclear genes. Bootstrap support from ML (above) and Posterior Probabilities from Bayesian Inference (bellow) are indicated on each node. ***Stands for 95 or more, **for 85–94 and *for 75–84 support values from ML analyses. Drawings correspond to male genital morphology, which is the base for traditional taxonomy of subgenus and species in the group. Individual 5–1 was morphologically identified as C. shufeldtii, but is considered here as C. ninae based on the phylogenetic position in tree.
Topology tests rejected the null hypothesis of an equally good explanation for all the constrained and the unconstrained topologies. The topology obtained in this study showed a significantly better Likelihood score (L = −27483.1) than the monophyletic grouping of Pandicambarus subgenus. Our phylogenetic estimate resulted in a monophyletic subgenus Cambarellus and Dirigicambarus, but Dirigicambarus was nested within the paraphyletic Pandicambarus (Fig. 3). We tested the monophyly of the Pandicambarus by forcing this alternative topology and we can reject this hypothesis by the results of SH and AU tests (likelihood values for the alternative hypothesis/p values for SH and AU - 27565.1/0.043, 0.047). Except for the division within Pandicambarus, Fitzpatrick’s notion of relationships among the subgenera is supported by our resulting topology, except for the non-monophyletic Pandicambarus as Pandicambarus and Dirigicambarus are nested together as a sister clade that is then sister to Cambarellus as proposed by Fitzpatrick. Bayesian inference also failed to support the monophyly of Pandicambarus failing to find a monophyletic Pandicambarus in 9900 trees resulting from the MCMC search.
Species were generally well recovered as monophyletic groups for most of those included in the Gulf Group, but a different situation is depicted for the Mexican Group (Figure 3). The clades highly supported by phylogenetic analyses have a geographic concordance, supporting the hypothesis that geographic events could have been important factors influencing cladogenesis in the genus, especially those regarding geographic features of the TMVB. Phylogenetic structuring between all Mexican taxa did not support the monophyly of some of the species currently recognized, as the highly supported clades showed representatives of multiple named species, suggesting that some of the named species did not form monophyletic assemblages.
Low 16S divergences can be observed between taxa. Divergences obtained between those contained in the Gulf Group were higher than those from the Mexican Group. The mean sequence divergence considering the likelihood model within the former was DHKY = 4.13%, and that within the latter was DHKY = 1.18% (Table 4).
Table 4. Uncorrected (below diagonal) and ML 16S rDNA distances (above diagonal) between phylogenetic groups of Cambarellus.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | Within Group unco | Within Group HKY | |
| C. zempoalensis Lerma-Cuitzeo | 0.33 | 0.98 | 1.17 | 0.89 | 2.33 | 1.57 | 2.58 | 1.25 | 12.11 | 11.25 | 10.9 | 10.04 | 11.63 | 11.38 | 13.01 | 15.91 | 18.47 | 18.98 | 23.74 | 0.16 | 0.16 | |
| C. patzcuarensis | 0.32 | 1.13 | 1.41 | 1.13 | 2.7 | 1.93 | 2.96 | 1.58 | 11.72 | 10.88 | 11.15 | 10.51 | 11.82 | 11.55 | 12.76 | 15.56 | 17.94 | 18.41 | 22.73 | 0.13 | 0.13 | |
| C. sp. LaMintzita | 0.93 | 1.07 | 1.53 | 1.36 | 2.9 | 1.97 | 3.13 | 1.75 | 12.67 | 11.8 | 11.24 | 10.63 | 12.15 | 11.9 | 14.31 | 17.12 | 19.31 | 18.71 | 24.64 | 1.24 | 1.3 | |
| C. chapalanus Chapala | 1.09 | 1.29 | 1.42 | 0.77 | 1.91 | 1.31 | 2.04 | 1.01 | 12.17 | 11.31 | 10.79 | 9.43 | 10.89 | 10.62 | 12.79 | 15.56 | 18.84 | 17.12 | 26.92 | 0.64 | 0.68 | |
| C. sp. Ameca | 0.85 | 1.07 | 1.28 | 0.74 | 2.01 | 1.2 | 2.24 | 0.83 | 13.8 | 12.81 | 12.23 | 10.64 | 12.24 | 11.66 | 14.64 | 17.41 | 20.2 | 19.11 | 30.08 | 0.11 | 0.11 | |
| C. sp. Zacapu | 2.08 | 2.37 | 2.57 | 1.74 | 1.85 | 1.87 | 1.14 | 1.81 | 12.44 | 11.55 | 11.7 | 10.94 | 12.29 | 11.37 | 14.13 | 17.29 | 20.6 | 18.79 | 29.79 | 0.58 | 0.6 | |
| C. montezumae Xochimilco | 1.43 | 1.72 | 1.8 | 1.21 | 1.13 | 1.7 | 2.15 | 0.95 | 12.24 | 11.35 | 11.54 | 9.5 | 10.91 | 10.56 | 12.79 | 15.33 | 18.9 | 18.07 | 29.03 | 0.31 | 0.32 | |
| C. sp. Vegil | 2.31 | 2.6 | 2.77 | 1.86 | 2.06 | 1.09 | 1.97 | 1.82 | 12.25 | 11.36 | 11.02 | 10.22 | 11.41 | 10.41 | 13.59 | 15.98 | 18.79 | 17.73 | 29.43 | 0.00 | 0.00 | |
| C. occidentalis | 1.16 | 1.45 | 1.61 | 0.95 | 0.8 | 1.67 | 0.9 | 1.69 | 12.56 | 11.67 | 12.07 | 9.57 | 10.95 | 10.55 | 13.24 | 15.1 | 19.11 | 17.88 | 29.03 | 0.14 | 0.14 | |
| C. lesliei | 8.27 | 8.11 | 8.57 | 8.3 | 9.09 | 8.45 | 8.35 | 8.3 | 8.48 | 0.41 | 3.26 | 2.76 | 3.09 | 3.16 | 4.89 | 9.64 | 12.25 | 13.1 | 20.47 | 0.00 | 0.00 | |
| C. ninae | 7.85 | 7.68 | 8.15 | 7.87 | 8.63 | 8.01 | 7.91 | 7.86 | 8.04 | 0.41 | 2.75 | 2.26 | 2.65 | 2.64 | 4.32 | 8.89 | 11.53 | 12.51 | 19.73 | 0.00 | 0.00 | |
| C. schmitti | 7.73 | 7.85 | 7.94 | 7.68 | 8.44 | 8.1 | 8.05 | 7.66 | 8.27 | 2.85 | 2.44 | 3.89 | 4.62 | 4.66 | 6.34 | 10.94 | 14.47 | 15.49 | 20.77 | 0.00 | 0.00 | |
| C. shufeldtii La Fayette | 7.21 | 7.43 | 7.54 | 6.9 | 7.58 | 7.72 | 6.96 | 7.25 | 6.97 | 2.44 | 2.04 | 3.26 | 1.82 | 1.82 | 4.45 | 7.99 | 13.31 | 14.55 | 20.55 | 0.00 | 0.00 | |
| C. shufeldtii | 8.03 | 8.12 | 8.31 | 7.69 | 8.39 | 8.42 | 7.72 | 7.9 | 7.73 | 2.74 | 2.39 | 3.84 | 1.68 | 1.51 | 4.54 | 8.41 | 13.06 | 13.54 | 22.04 | 1.58 | 1.69 | |
| C. texanus | 7.91 | 7.99 | 8.19 | 7.55 | 8.11 | 7.99 | 7.53 | 7.41 | 7.5 | 2.79 | 2.36 | 3.85 | 1.67 | 1.42 | 4.8 | 7.42 | 12.11 | 13.25 | 22.08 | 0.85 | 0.88 | |
| C. puer | 8.51 | 8.41 | 9.09 | 8.42 | 9.27 | 9.06 | 8.45 | 8.71 | 8.62 | 4.01 | 3.6 | 4.89 | 3.62 | 3.76 | 3.92 | 7.02 | 13.75 | 15.46 | 20.96 | 0.80 | 0.72 | |
| C. diminutus | 9.96 | 9.84 | 10.43 | 9.82 | 10.6 | 10.56 | 9.75 | 9.92 | 9.62 | 6.92 | 6.5 | 7.52 | 5.91 | 6.25 | 5.64 | 5.37 | 16.3 | 15.91 | 25.11 | 0.00 | 0.00 | |
| Procambarus | 10.78 | 10.61 | 11.15 | 10.92 | 11.42 | 11.57 | 10.95 | 10.89 | 10.99 | 8.48 | 8.11 | 9.42 | 8.87 | 8.86 | 8.39 | 9.05 | 10.04 | 14.51 | 22.17 | 7.31 | 10.84 | |
| Orconectes | 11.1 | 10.92 | 11.09 | 10.5 | 11.27 | 11.16 | 10.8 | 10.81 | 10.76 | 8.89 | 8.67 | 9.95 | 9.55 | 9.16 | 9.06 | 9.92 | 10.17 | 9.36 | 20.52 | 0.00 | 0.00 | |
| Cambarus | 12.78 | 12.51 | 13.06 | 13.6 | 14.55 | 14.38 | 14.13 | 14.26 | 14.14 | 11.65 | 11.43 | 11.82 | 11.69 | 12.29 | 12.22 | 11.85 | 13.14 | 11.95 | 11.67 | 0.00 | 0.00 |
The Mexican Group is composed of several clades highly supported by ML and BI analyses (95–100% support, termed with roman numerals in Figure 1), which also show geographic concordance. Some geographic overlapping between clades was observed, mainly along the Lerma Basin. The Clade I included populations from the Cuitzeo and Middle-Lerma basins, morphologically assigned to C. montezumae. C. zempoalensis from type locale was placed inside this clade as well. Cambarellus patzcuarensis from the basins of Pátzcuaro and Zirahuén were contained in Clade II and sister clade to Clade I. The third and more divergent clade (Clade III) consisted of a population from La Mintzita, geographically close to the Cuitzeo basin.
Clade IV consisted of populations from the basin of Chapala and its tributaries (Duero River), as well as its neighboring basins, Cotija and Zapotlán. This group included two species, C. chapalanus and C. prolixus, both found in Lake Chapala and associated with different habitat conditions. Also included here were populations from up-stream tributaries of the Santiago River, which originates as an outflow of the Chapala Lake. Clade V contained populations from the river Ameca basin. Clade VI contained the population from Zacapu Lagoon. The Clade VII included two populations from the eastern-limits of the distribution of the genus in the TMVB, the populations of Xochimilco (type locality for C. montezumae) from the Valley of México basin and the crater lake Quechulac. The Clade VIII was composed of two populations from the northern margin of the Middle-Lerma basin and the Clade IX by populations from the basins of the Santiago and Magdalena rivers, in the west part of TMVB.
Gulf Group relationships depict a phylogenetic structuring corresponding to geographic ranges. C. diminutus corresponds to the most divergent lineage, while two clades were recovered with high ML and BI support corresponding to a west-east pattern. The first clade contained most of the species from the Central and East Gulf Coast (CEG), except C. diminutus, and included four recognized species. Populations of C. shufeldtii from the Mississippi river basin form a monophyletic group, while C. blacki, C. lesliei, and C. schmitti are grouped together in a sister clade to the latter, geographically covering the eastern extreme distribution range of the genus in the Gulf Group from the Mobile Bay, Alabama to the Swuanee River, Florida. A similar grouping is observed in the second clade of the Gulf Group, containing populations from the West Gulf Coast (WG), mainly in the south-west part of Texas, where C. puer was recovered as a sister lineage to the clade grouping C. texanus and C. ninae.
Diversification Patterns and Dating
Log-likelihood scores with the molecular clock enforced and not enforced were −13.893 and −13.767, respectively. As the LRT rejected the null hypothesis of a global molecular clock (χ2 252, P = 0.001), the sequences analyzed did not evolve at a homogenous rate along all branches and we proceeded to use a relaxed molecular clock (Fig. 4) as a result.
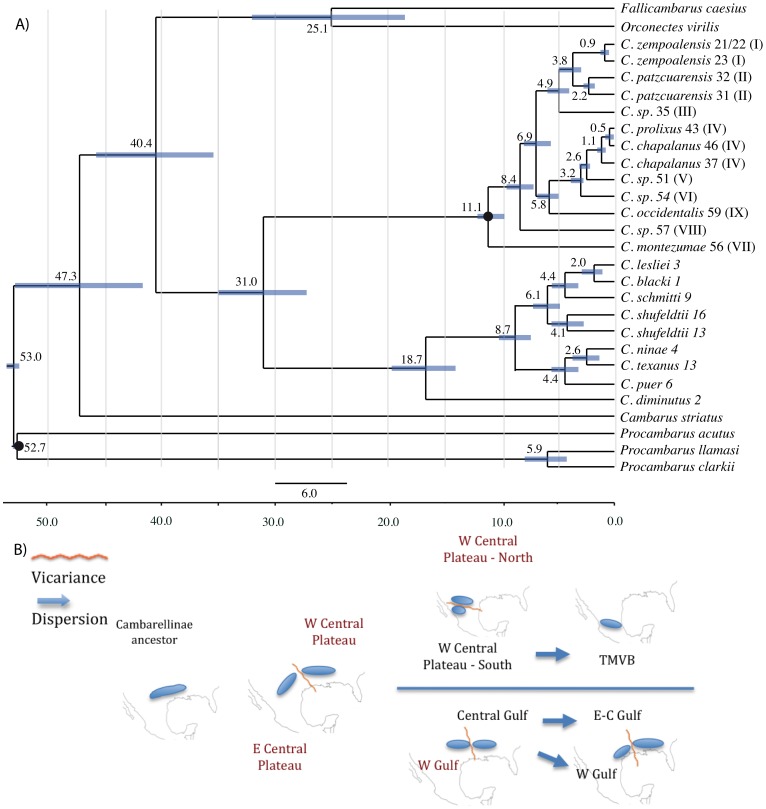
Figure 4. Molecular dating of cladogenetic events.
Dates and major biogeographic events inferred during cladogenesis of the Cambarelline subfamily. A) Ultrametric tree resulting from the dating analysis. Mean ages are indicated in each node (MYA), and 95% HDP intervals are shown as blue bars. Black dots indicate node used for calibration (oldest fossil recorded for Procambarus). Numbers correspond to localities and roman numerals to clades from phylogenetic tree (Figure 3). B) Major cladogenetic events inferred from phylogenetic structure and dating. Red names refer to extinct lineages.
Ages from the dating analysis were recovered with consistency through repetitions (Figure 4). The crown age for the tree was 53 Myr (95% highest posterior density [HPD] interval for node heights/ages: 52.6–53.7 Myr), which corresponds to the separation of the genus Procambarus from the rest of the groups. We estimated an approximate age of 31.0 Myr (27.4–34.9 Myr 95% HPD) for the TMRCA of clade containing the Cambarellinae. MRCA for the terminals included in the two lineages of the Gulf Group is approximately 16.7 Myr (13.9–19.7 Myr 95% HPD). MRCA of the Mexican Group was dated around 11.1 (9.8–11.9 Myr 95% HPD). We propose some major biogeographic events inferred from the phylogenetic structure, which depicted different vicariant and dispersion events along the evolutionary history of Cambarellinae (depicted in Figure 4.).
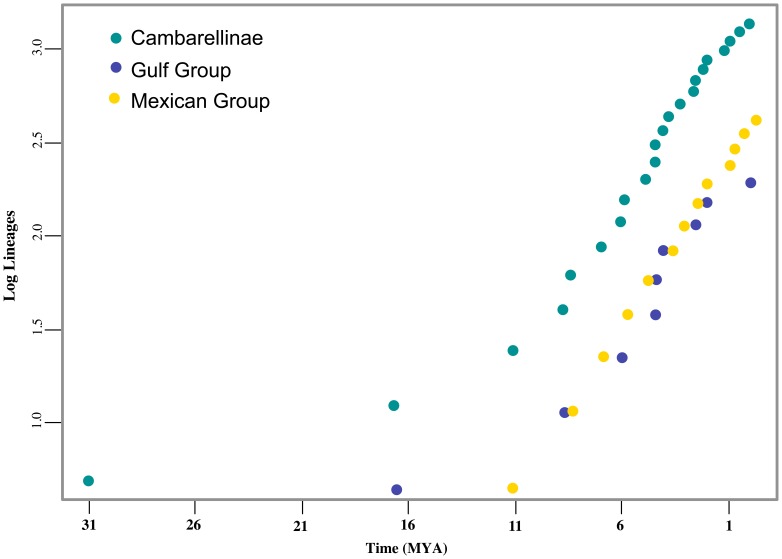
The LTT plots track the temporal accumulation of lineages in a clade and indicate that the subfamily Cambarellinae did not significantly deviate from a constant model of diversification during its evolutionary history, as evidenced in the LTT analyses for the entire subfamily (including both, Gulf and Mexican Groups, see Fig. 5). LTTs rate-constancy models received better AIC scores, and they were not significantly different from the best rate-variable model for all analyses (Table 5). The pure birth speciation rate model was identified as having the lowest AIC value amongst the other models tested for the subfamily together and the two groups separately. Although the Mexican Group showed the highest diversification rate (under pureBirth model r = 0.174), it is still a low value as compared to recognized shifts in diversification in other animal groups ranging from 0.4 to 0.8 speciation events per million years [48], [49].
Figure 5. Diversification patterns through time.
LTT plot for the Cambarellinae subfamily (green), the Mexican Group (yellow) and the Gulf Group (blue).
Table 5. Results of the Birth-Death Likelihood analysis based on fitting different diversification models to Cambarellinae and its containing groups (Gulf and Mexican Groups).
| Group | pureBirth | BD | DDL | DDX | yule2rate | |
| Cambarellinae | Parameters | r1 = 0.122 | r1 = 0.053 | r1 = 0.122 | r1 = 0.064 | r1 = 0.043 |
| a = 0.706 | k = 476707.6 | x = −0.297 | r2 = 0.152 | |||
| Ln(L) | 16.600 | 15.815 | 16.601 | 15.969 | 14.668 | |
| AIC | −35.201 | −35.631 | −37.201 | −35.939 | −35.336 | |
| ΔAIC | 0 | −0.430 | −2 | −0.738 | −0.135 | |
| Gulf | r1 = 0.106 | r1 = 0.088 | r1 = 0.197 | r1 = 0.127 | r1 = 0.153 | |
| a = 0.229 | k = 12.126 | x = −0.117 | r2 = 0.059 | |||
| st = 4.116 | ||||||
| Ln(L) | 12.101 | 12.089 | 11.783 | 12.083 | 11.395 | |
| AIC | −26.202 | −28.179 | −27.567 | −28.166 | −28.791 | |
| ΔAIC | 0 | −1.977 | −1.365 | −1.964 | −2.589 | |
| Mexican | r1 = 0.174 | r1 = 0.174 | r1 = 0.276 | r1 = 0.296 | r1 = 0.210 | |
| a = 0.0 | k = 21.007 | x = 0.283 | r2 = 0.120 | |||
| st = 2.225 | ||||||
| Ln(L) | 10.201 | 10.201 | 9.894 | 10.049 | 9.834 | |
| AIC | −22.402 | −24.402 | −23.789 | −24.096 | −25.669 | |
| ΔAIC | 0 | −2 | −1.387 | −1.694 | −3.267 |
r = net diversification rate (speciation events per million years);
a = extinction fraction;
st = time of rate shift (MYA);
k = carrying capacity prameter;
x = rate change parameter;
Ln(L) = Log-Likelihood;
AIC = Akaike information criterion;
ΔAIC = change in AIC relative to pureBirth.
Quick inspection of the LTT plots shows some differences between the cladogenesis of the entire subfamily and that of the Gulf and Mexican Groups alone (Figure 5). However, according to the BDL analysis, the diversification rate-constancy statistic ΔAICRc was found to be similar between them, being −0.135 for the entire subfamily, −1.38 for the Mexican Group and −1.36 for the Gulf Group, indicating that the data are a better fit to the constant rather than variable rate model of diversification in all cases. Goodness-of-fit tests indicated that the mean Bayes LTT from the entire subfamily was not significantly different from expectations under any of the rate constancy models (AIC pureBirth and BD = 35.20 and 35.63, respectively). The values from the BDL analysis of the Mexican and the Gulf Groups were not significantly different than the critical values found under the different simulated constant rate models (for AIC pureBirth = 22.40 and BD = 24.40 for the Mexican Group and AIC pureBirth = 26.20 and BD = 28.17 for the Gulf Group). These results are consistent with a lack of evidence about episodes of shifts in diversification rates along the evolutionary history of Cambarellinae or its two groups separately.
Discussion
Phylogenetic Relationships
Our results are consistent with the monophyly of the Cambarellinae subfamily, previously proposed from morphology and a set of apomorphic characters [2]. The combination of mitochondrial and nuclear markers provide sufficient information to resolve the relationships between highly supported clades, namely the Gulf (Pandicambarus/Dirigicambarus) and Mexican (Cambarellus) Groups and included clades (Figure 3). Less resolution is observed at the deeper nodes of the Mexican Group, where several clades were not supported by all analyses. It is possible, as commonly argued for polytomies, that such patterns could be related to an acceleration of speciation rates in a short period of time [50]. Species sampling in this study is not complete, as three species are still to be added to the phylogenetic analysis. These correspond to C. alvarezi, C. areolatus and C. chihuahuae from North of Mexico and have almost no collection records. Populations from the aforementioned species are currently under serious threat or possibly extinct, as we did not find any specimens in our attempts to collect them. Their rarity is possibly due to extreme habitat alteration or drought, a situation reported as critical for freshwater fauna in some of the localities from where they have been recorded [51], [52]. Their future inclusion, if possible (mostly through museum collections or captive populations), could provide valuable insight into the phylogenetic relationships within the subfamily, especially between the Mexican and Gulf Groups defined here.
Several differences can be found between the phylogenetic relationships emerging from this work and the previous hypothesis [2]. First, relationships between species in the Gulf Group are not congruent with several assumptions made from morphology, especially regarding the phylogenetic meaning of genitalia variation. Although species are generally well recovered as monophyletic, their relationships are not congruent. As evidenced by topology tests carried out in this study, sister relationships proposed by genital morphology between the two subgenera from the Gulf Group (Pandicambarus and Dirigicambarus) is not supported. Instead, Dirigicambarus (composed by C. shufeldtii) is recovered as a sister taxon of a clade containing C. lesliei and C. schmitti. This would leave the subgenus Pandicambarus as paraphyletic, ultimately questioning also its phylogenetic validity. Maintaining of the subgenus Dirigicambarus for C. shufeldtii could be also questioned, as no phylogenetic evidence supports it, pointing out that genital distinctiveness in this species could be the result of drift events or selective processes along its history. Besides its proposition as a member of a separate subgenus, C. shufeldtii has been recognized as a derived rather than a plesiomorphic representative [2], an assumption supported in this study. Therefore, we recommend that the subgenus Dirigicambarus be disregarded and that the genus Cambarellus should contain only two subgenera, namely Cambarellus and Pandicambarus that correspond to the Mexican and Gulf clades, respectively (resulting in Cambarellus shufeldtii being considered a member of the subgenus Pandicambarus). Our phylogenetic results support the hypothesis of C. diminutus as having plesiomorphic character states for the Gulf Group. Its unique morphological traits (outlined in [2]) are in agreement with this hypothesis.
Taxonomic Implications
Numerous species concepts have been proposed that emphasize different features for delimiting species. Sometimes, this has led to contrasting conclusions regarding species limits and the number of species in many groups. A ‘unified species concept’ was advocated that emphasizes the common element found in many species concepts, which is that species are separately evolving lineages [53]. This unified concept also allows the use of diverse lines of evidence to test species boundaries [e.g., monophyly at one or multiple DNA loci, morphological diagnosability, ecological distinctiveness, etc. [53], [54] and is the species concept we follow in this study.
There were two cases in which the inferred topology did not recover species’ monophyly in the Gulf Group. The first one shown by one individual morphologically assigned to C. shufeldtii (Locality 5, Colorado Basin), which grouped with individuals of C. ninae and the other by one individual morphologically assigned to C. puer (Locality 12, San Bernard Basin), grouped with individuals of C. texanus. The most plausible explanation for this could be the finding of introgression of C. shufeltii, supported by the overlapping ranges of these species in east Texas. As a common consequence, introgression between species with smaller ranges could be favored when they share similar regions with widely distributed species like C. shufeldtii and C. puer (e.g., [55]). The aforementioned hypothesis needs to be supported with faster-evolving nuclear markers, which allow the differentiation between species, and could be approached in the near future.
For the Mexican Group, the phylogenetic structure shows a geographic correspondence. This observation supports the hypothesis that cladogenesis in the group has been influenced by geological history. This geographic correspondence could explain why instead of recovering species, cladogenetic structure recovered different hydrological units as monophyletic. This is the case for the widely distributed C. montezumae, which is not recovered as monophyletic, as several populations morphologically assigned to this species were located in different clades in the Mexican Group. In fact, several populations morphologically assigned to C. montezumae form a paraphyletic group, as C. zempoalensis is recovered inside this group. Another example concerns C. prolixus, included inside the wider genetic variation of C. chapalanus. However, the striking morphological distinctiveness of C. prolixus suggests a very recent processes of divergence between this species and C. chapalanus which may be missed by the genetic markers used here [see [56] for discussion on the relative importance of genetic markers versus selected morphological differences in species studies]. Based on an unified species criterion, we found support for all described species in the Mexican Group from TMVB, which match to the terminal clades in tree (Figure 3) plus C. prolixus, which possesses contrasting morphological and ecological features. These clades correspond to six described species: 1) C. zempoalensis, corresponding to the population from Zempoala. Temporarily, we consider this species as valid but this needs to be confirmed with an analysis including the ‘lermensis form’ (in the terms of Villalobos’ proposal) [57]. This is because when considering the range of this clade, it probably includes the aforementioned form, from the upper Lerma Basin. As such, C. montezumae lermensis would be raised to species rank and C. zempoalensis would stand as a junior synonym; all populations found in clade I would temporarily correspond to C. zempoalensis, until confirmation of the above mentioned issue regarding its synonymy with C. montezumae lermensis; 2) C. patzcuarensis, for those populations from the Patzcuaro basin; 3) C. chapalanus, from the basin of Chapala and adjacent basins; 4) C. prolixus, from certain habitat conditions at Chapala Lake; 5) C. montezumae, from the Valley of Mexico and adjacent basins and 6) C. occidentalis, from the lower part of the Río de Santiago basin, at the western extreme of the distribution in México. In addition, we found several monophyletic clades, and in congruence to the same criterion, we propose they correspond to no recognized species, those from the terminal clades in tree (Figure 3): 1) clade III, for the population from La Mintzita spring; 2) clade V, for populations from Ameca basin; 3) clade VI, for populations from the Zacapu Lagoon and 4) clade VIII, for certain populations from the northern side of the Middle Lerma Basin (populations of La Laja basin and Vegil, see Table S1).
Rates of Cladogenesis and Contrasting Cladogenetic Forces
Unlike the cladogenetic structure, the rate at which cladogenesis took place in Cambarellus does not seem to be affected by geologic events. Even when most of the cladogenetic events in the Mexican Group are probably the result of vicariance corresponding to geological features as the formation of the TMVB, a geologic region that has been proposed to affect cladogenesis in different freshwater groups [58], [59], this study has found no effect of geologic events on speeding or reducing cladogenesis rates. Although different in nature, and affected by contrasting geographic ranges, vicariant events in both groups lead to similar cladogenetic trajectories, demonstrating the impact of climatic and geologic forces on allopatric speciation.
All these lines of geological evidence indicate that the historical geographic range of the hypothesized ancestral species of Cambarellinae in México and the Southeast of the United States have changed dramatically over time. Additionally, some other effects could have played a roll in speciation in both groups. Although the continental ice sheets during the Pleistocene glacial periods in North America never extended into the study area, these glaciations had some profound indirect effects in freshwater faunas in México and are hypothesized to have permitted dispersal by stream captures, local inland or estuarine flooding, and interconnecting drainages due to lowered sea levels during the late Neogene [60].
Biogeography
Our results support that MRCA for the Cambarellinae existed in the Eocene, ∼40.4 MYA (35.2–45.7 MYA). A singular biogeographic event inferred from this study comes from the separation of the two major clades, which could be related to the Eocene-Oligocene boundary, a transition documented to strongly affect terrestrial, marine and freshwater dwellers, as evidenced by significant extinctions and taxonomic turnovers in a wide range of groups [61], [62]. In this case, the formation of the Rio Grande Rift could have vicariant effects on the ancestors of both groups.
We postulate that historical vicariant events are related to change in geographical barriers and climate in both groups while dispersal events of some species are responsible for occupying the current wider range, with their current absence related to extinction periods. While these biogeographic events are present in both groups, contrasting vicariance and dispersal impacts on distributions are not unusual for freshwater crayfishes [63]. Here we explain some possible alternatives, inferred from congruence in timing of cladogenetic events. Species cladogenesis in the Gulf and Mexican Groups are best explained by an allopatric mechanism of speciation because no overlap is observed between sister taxa in Cambarellus [8]. Estimation of divergence times provides a temporal scenario of these events, allowing for a relationship of earth history with hypothesized vicariant mechanisms proposed to promote allopatric speciation. We postulate that divergence patterns in these groups are contrasting in several ways. First, date estimates agree with a more ancient diversification in the Gulf Group than in the Mexican Group, even when possible events related to species diversification from Northern Mexico could not be inferred. It is possible that the latter predate the ones observed for the Mexican Group. Second, while the Gulf Group cladogenesis could be more related to climatic oscillations, orogenic impacts could have been more important for diversification of the extant species in the Mexican Group. The absence of Cambarellus from the Río Grande basin could be explained by a generalized extinction of its populations related to the high desiccation rate since the Tertiary [64]. The ultimate evidence of that would be the presence of C. chihuahuae from the Guzman basin in the Southern part of the Río Grande Rift. This high extinction rate could explain the current disjunct geographic pattern between the Mexican and Gulf Groups. It seems reasonable to consider that as a consequence of the extinction rate along the former contact zone between the groups. It would not be surprising to find relict populations from both groups if further sampling efforts in this region could take place, which could modify their known range and find regions containing both lineages.
Proposed Vicariant Events Promoting Speciation
The first diversification event in the Gulf Group was dated to Early Miocene, ∼16.7 MYA (13.9–19.7 MYA), and corresponded to the separation of the C. diminutus lineage. It is possible that extinction events could explain the observation of a unique well differentiated branch leading to C. diminutus, although a wider genetic variation not yet sampled from this lineage could be possible, which would be consistent with the wide morphological variation previously observed [2]. Orogenic activity dating to this period corresponds at the SE of United States with the formation of the Edwards Plateau, and the Miocene increased activity along the Balcones Fault. The next cladogenesis recorded for the Gulf Group is congruent with Late Miocene times, approximately 8.7 MYA (7.3–10.3 MYA), originating the Western (WG) and Eastern Gulf Coast (EG) species groups. These speciation events are consistent with sea levels along the gulf coast driven by climatic oscillations since the Middle Miocene. These were characterized by a dramatic rise in sea level between 80 and 100 m above the present day sea level [65], [66]. As a consequence, a marine incursion took place along the coast of the Gulf of Mexico, which could be important in the split of West and Central-East distribution ranges and keep them separated long enough to induce a strong speciation event.
In the Late Miocene there was a sharp drop of 80–100 m below present sea levels, extending Gulf of Mexico tributaries further south. This southward extension of Gulf Coastal rivers created connections between tributaries that were isolated during periods of higher sea level. The Late Miocene drop in sea level correlates with the estimated age of the first speciation events among the extant species of Cambarellus from the Gulf Group. Later, the Pliocene (2.5–5.5 MYA) was characterized by a 50–80 m rise above current sea level, but this incursion lasted for only a short time, approximately one million years [66]. Sea levels dropped in the Late Pliocene, and during the Pleistocene there were at least three major fluctuations in sea level, none rising higher than 10–20 m above the current level [66], [67]. All these events could affect the most recent speciation events and possible inter-basin connection between the Gulf species of Cambarellus could allow for dispersal of some of the today widely distributed taxa along the coastal drainages.
As a lentic-habitat dweller is the widespread way of life for the subfamily, it is reasonable to think that this could be the same situation for the ancestors of the different groups. In this case, formation of Paleolakes during the Middle Miocene (∼10.8 MYA) along the Northern Central Plateau of México and South-East United States could be important features driving early cladogenesis in the Mexican Group.
The pattern of distribution observed in Cambarellus agrees with those proposed for other freshwater organisms, like the Plateau Track and western Mountain Track [64]. To explain similar distributions in fish genera such as Ictalurus, Moxostoma, and Micropterus, these patterns suggest former hydrographic exchanges across the present arid plateau. Based on faunal composition and the finding of sister taxa between those regions like Tampichthys/Codoma and Algansea/Agosia sister pairs [68], possible connections between drainages of the South Western Gulf Slope (Nueces, Colorado and Guadalupe rivers) and those from the northern Río Grande tributaries have been suggested [69]. These connections could explain the presence of the Northern Central Plateau species (C. alvarezi, C. areolatus and C. chihuahuae), especially joined to lake habitats. Extensive lakes associated with the past Río Grande inflow have been documented to cover much of north-western Chihuahua and southern New Mexico in Pleistocene times, like the Lake Cabeza de Vaca [70].
The reduction in volume of lacustrine habitats in the Central Plateau by climatic events may have resulted in a high rate of late Cenozoic extinction [64], and the patchy distribution pattern of Cambarellus in this region. This high rate of desiccation, now increased by human activities [51], could have eroded diversity in this region, driving to extinction most of the Cambarellus populations in the Northern Central Plateau of México and could also explain the current absence of Cambarellus from the rivers south of the West Gulf Coast drainages. Partial extirpation from a formerly continuous range due to increasing dry rate during the Tertiary, has also been seen in different fish groups with a similar range, like Goodeidae and Cyprinodontidae [71]. Additionally and continuing southward, former connections between Northern and Western Central Plateau rivers could explain the presence of C. occidentalis in the Lower Santiago basin and from there a connection to the rest of the TMVB could be inferred.
Along the TMVB, the Lerma-Santiago river system is the main drainage. Previous connections between the Lerma River and northeastern and western drainages have been suggested for Goodeidae and Cyprinid fish [71]. Similar to what has been postulated for freshwater fish groups like the families Atherinidae, Goodeidae Cyprinidea, diversification in Cambarellus along the TMVB could have been related to an ancient and successive fragmentation of the Lerma-Santiago drainage across extensive lacustrine systems from the Miocene to Pleistocene [71], [72].
Separation of the main clades of Cambarellus in the TMVB is dated along the late Miocene and Pliocene (10.8–4.6 MYA), a period of high geological activity in México [73]. Formation of the TMVB advanced in a West-East direction [74], and this could influence the separation of clades from the main groups of Cambarellus. This formation could have begun before the presence of Cambarellus in TMVB, given its absence on the Pacific basins south to the Zapotlán basin, at its western margin. Major diversification of the genus took place in an interval of time of less than 9 MYA.
This study found evidence consistent with a long and complex evolutionary history of the Cambarellinae. The group’s distribution has been modified extensively by geologic and climatic factors. Although there appear to be contrasting causes for cladogenesis between the two groups, they have similar diversification rates. In addition, these results showed that genital and morphological changes widely used in the subfamily in particular and in crayfish in general, should be compared with other kinds of evidence in order to make more robust use of morphological differences for evolutionary inferences.
Supporting Information
Voucher numbers and localities of the individuals analyzed. Proposed new taxa are indicated as Cambarellus sp., and correspond to the terminal clades indicated with roman numerals in phylogeny (see Figure 3).
(DOCX)
Acknowledgments
We wish to acknowledge the relevant support in lab work from Rebecca Scholl (BYU). Museum samples and access to specimens were kindly provided by Rafael Lemaitre and Karen Reed from US NMNH and Gonzalo Giribet from MCZ. Valuable help during field trips and with editing was provided by Patricia Ornelas García. We thank Daniel P. Johnson and Paul E. Moler for collection help and for providing some Cambarellus samples from the US Gulf Coast. Also, some important aspects of the manuscript were improved from fruitful discussions with Patricia Ornelas and Mario García and from the comments kindly made by Joe Castresana and two anonymous reviewers. Several samples from México were provided by Fernando Álvarez, José Luis Villalobos, Omar Domínguez and Rodolfo Pérez. Very useful suggestions on analyses were provided by Regina Cunha and Carina Cunha.
Funding Statement
Carlos Pedraza-Lara was partially supported by a predoctoral grant provided by CSIC, Spain. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Hobbs HHJ (1974) A checklist of the North and Middle American crayfishes (Decapoda: Astacidae and Cambaridae). Smithsonian Contributions to Zoology 166: iii+161, 161–294.
- 2.Fitzpatrick Jr JF (1983) A Revision of the Dwarf Crawfishes (Cambaridae, Cambarellinae). Journal of Crustacean Biology: 266–277.
- 3. Hobbs HHJ (1950) A new crayfish of the genus Cambarellus from Texas (Decapoda, Astacidae). Proceedings of the Biological Society of Washington 63: 89–96. [Google Scholar]
- 4. Hobbs H (1989) An Illustrated Checklist of the American Crayfishes (Decapoda, Astacidae, Cambaridae, Parastacidae). Smithsonian Contributions to Zoology 480: 1–236. [Google Scholar]
- 5.Villalobos A (1955) Cambarinos de la fauna Mexicana (Crustacea:Decapoda). Universidad Nacional Autónoma de México. 290 p.
- 6.Laguarda A (1961) Contribución al estudio comparativo de la fórmula branquial en la Familia Astacidae (Crustacea:Decapoda): Universidad Nacional Autónoma de México. 1–74 p.
- 7.Rojas Y, Alvarez F, Villalobos JL (2002) Morphological variation in the crayfish Cambarellus (Cambarellus) montezumae (Decapoda:Cambaridae). In: Escobar E, Álvarez F, editors. Modern approaches to the study of Crustacea. New York: Kluwer Academic/Plenum Publishers. 311–317.
- 8. Barraclough T, Vogler A (2000) Detecting the geographical pattern of speciation from species level phylogenies. Am Nat 155: 419–434. [DOI] [PubMed] [Google Scholar]
- 9. Harvey P, May R, Nee S (1994) Phylogenies without fossils. Evolution 48: 523–529. [DOI] [PubMed] [Google Scholar]
- 10. Nee S (2006) Birth-Death Models In Macroevolution. Annual Review of Ecology, Evolution and Systematics 37: 1–17. [Google Scholar]
- 11. Rabosky D (2007) Likelihood methods for detecting temporal shifts in diversification rates. Evolution 60: 1152–1164. [PubMed] [Google Scholar]
- 12. Hobbs HHJ (1974) Synopsis of the families and genera of crayfishes (Crustacea:Decapoda). Smithsonian Contributions to Zoology 164: 1–32. [Google Scholar]
- 13.Toon A, Finley M, Staples J, Crandall KA (2009) Decapod Phylogenetics and Molecular Evolution. In: Martin JW, Crandall KA, Felder DL, editors. Decapod Crustacean Phylogenetics. Boca Raton, FL: Taylor and Francis Group, LLC.
- 14. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buhay JE (2009) “COI-like” Sequences are Becoming Problematic in Molecular Systematic and DNA Barcoding Studies. Journal of Crustacean Biology 29: 96–110. [Google Scholar]
- 16. Song H, Buhay JE, Whiting MF, Crandall KA (2008) Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proceedings of the National Academy of Sciences 105: 13486–13491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swofford DL (1998) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sunderland, Massachussetts: Sinauer Associates.
- 18. Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- 19.Schwarz G (1978) Estimating the dimensions of a model. Ann Stat 6 461–464.
- 20. Posada D (2008) j Model Test: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253. [DOI] [PubMed] [Google Scholar]
- 21. Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology 52: 696. [DOI] [PubMed] [Google Scholar]
- 22. Felsenstein J (1985) Confidence-Limits on Phylogenies - an Approach Using the Bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 23. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Ronquist F (2005) Bayesian analysis of molecular evolution using MrBayes. Statistical methods in molecular evolution New York: Springer: 183–232.
- 25.Rambaut A, Drummond AJ (2007) Tracer v1.4, Available: http://beast.bio.ed.ac.uk/Tracer.Accesed 2011 Jun 3.
- 26. Shimodaira H, Hasegawa M (1999) Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution 16: 1114–1116. [Google Scholar]
- 27. Shimodaira H (2002) An approximately unbiased test of phylogenetic tree selection. Systematic Biology 51: 492. [DOI] [PubMed] [Google Scholar]
- 28.Jobb G (2008) TREEFINDER. Version of October 2008. Munich, Germany.
- 29. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology 4: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drummond A, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Breinholt J, Perez-Losada M, Crandall KA (2009) The timing of the diversification of the Freshwater Crayfishes. In: Boca Raton, FL: Taylor and Francis Group Martin JW, Crandall KA, Felder DL, editors. Decapod Crustacean Phylogenetics. LLC: 343–356. [Google Scholar]
- 32. Porter M, Pérez-Losada M, Crandall K (2005) Model-based multi-locus estimation of decapod phylogeny and divergence times. Molecular Phylogenetics and Evolution 37: 355–369. [DOI] [PubMed] [Google Scholar]
- 33.Cunningham C, Blackstone N, Buss L (1992) Evolution of king crabs from hermit crab ancestors. [DOI] [PubMed]
- 34. Stillman J, Reeb C (2001) Molecular phylogeny of eastern Pacific porcelain crabs, genera Petrolisthes and Pachycheles, based on the mtDNA 16S rDNA sequence: phylogeographic and systematic implications. Molecular Phylogenetics and Evolution 19: 236–245. [DOI] [PubMed] [Google Scholar]
- 35.Cook BD, Pringle CM, Hughes JM (2008) Phylogeography of an Island Endemic, the Puerto Rican Freshwater Crab (Epilobocera sinuatifrons). J Hered: esm126. [DOI] [PubMed]
- 36. Knowlton N, Weigt LA (1998) New dates and new rates for divergence across the Isthmus of Panama. Proceedings of the Royal Society of London Series B-Biological Sciences 265: 2257–2263. [Google Scholar]
- 37. Knowlton N, Weigt LA, Solorzano LA, Mills DK, Bermingham E (1993) Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the Isthmus of Panama. Science 260: 1629. [DOI] [PubMed] [Google Scholar]
- 38. Ferrari L, Lopez-Martinez M, Aguirre-Diaz G, Carrasco-Nunez G (1999) Space-time patterns of Cenozoic arc volcanism in central Mexico: From the Sierra Madre Occidental to the Mexican Volcanic Belt. Geology 27: 303–306. [Google Scholar]
- 39. Feldmann R, Grande L, Birkhimer C, Hannibal J, McCoy D (1981) Decapod fauna of the Green River formation (Eocene) of Wyoming. Journal of Paleontology 55: 788–799. [Google Scholar]
- 40. Rabosky DL (2007) Likelihood methods for detecting temporal shifts in diversification rates. Evolution 60: 1152–1164. [PubMed] [Google Scholar]
- 41.Rabosky D (2006) LASER: a maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. [PMC free article] [PubMed]
- 42.Rambaut A, Charleston M (2002) TreeEdit: phylogenetic tree editor, ver. 1.0 alpha 10. Department of Zoology, University of Oxford, Oxford.
- 43. Yule GU (1925) A Mathematical Theory of Evolution, Based on the Conclusions of Dr. J. C. Willis, F.R.S. Philosophical Transactions of the Royal Society of London Series B, Containing Papers of a Biological Character 213: 21–87. [Google Scholar]
- 44. Pereira SL, Baker AJ, Wajntal A (2002) Combined nuclear and mitochondrial DNA sequences resolve generic relationships within the Cracidae (Galliformes, Aves). Systematic Biology 51: 946–958. [DOI] [PubMed] [Google Scholar]
- 45. San Mauro D, Gower DJ, Massingham T, Wilkinson M, Zardoya R, et al. (2009) Experimental design in caecilian systematics: Phylogenetic information of mitochondrial genomes and nuclear rag1. Systematic Biology 58: 425–438. [DOI] [PubMed] [Google Scholar]
- 46. Fisher-Reid MC, Wiens J (2011) What are the consequences of combining nuclear and mitochondrial data for phylogenetic analysis? Lessons from Plethodon salamanders and 13 other vertebrate clades. BMC Evolutionary Biology 11: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wiens JJ, Kuczynski CA, Smith SA, Mulcahy DG, Sites Jr JW, et al. (2008) Branch lengths, support, and congruence: testing the phylogenomic approach with 20 nuclear loci in snakes. Systematic Biology 57: 420–431. [DOI] [PubMed] [Google Scholar]
- 48. Kozak KH, Weisrock DW, Larson A (2006) Rapid lineage accumulation in a non-adaptive radiation: phylogenetic analysis of diversification rates in eastern North American woodland salamanders (Plethodontidae: Plethodon). Proceedings of the Royal Society B: Biological Sciences 273: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ricklefs RE, Losos JB, Townsend TM (2007) Evolutionary diversification of clades of squamate reptiles. Journal of Evolutionary Biology 20: 1751–1762. [DOI] [PubMed] [Google Scholar]
- 50. Slowinski JB (2001) Molecular Polytomies. Molecular Phylogenetics and Evolution 19: 114–120. [DOI] [PubMed] [Google Scholar]
- 51. Contreras-Balderas S, Lozano-Vilano M (1996) Extinction of most Sandía and Potosí valleys(Nuevo León, México) endemic pupfishes, crayfishes and snails. Ichthyological exploration of freshwaters Munchen 7: 33–40. [Google Scholar]
- 52. Rodríguez-Almaraz G, Campos E (1994) Distribution and status of the crayfishes (Cambaridae) of Nuevo León, México. Journal of Crustacean Biology 14: 729–735. [Google Scholar]
- 53.Sites Jr JW, Marshall JC (2004) Operational criteria for delimiting species. Annual Review of Ecology, Evolution, and Systematics: 199–227.
- 54. De Queiroz K (2007) Species concepts and species delimitation. Systematic Biology 56: 879–886. [DOI] [PubMed] [Google Scholar]
- 55. Perry W, Feder J, Dwyer G, Lodge D (2001) Hybrid zone dynamics and species replacement between Orconectes crayfishes in a northern Wisconsin lake. Evolution 55: 1153–1166. [DOI] [PubMed] [Google Scholar]
- 56. Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends in Ecology & Evolution 15: 290–295. [DOI] [PubMed] [Google Scholar]
- 57. Villalobos A (1943) Estudios de los cambarinos mexicanos, I: Observaciones sobre Cambarellus montezumae (Saussure) y algunas de sus formas con descripcion de una subspecie nueva. Annales del Instituto de Biologia 14(2) 587–611: 587–611. [Google Scholar]
- 58. Mateos M, Sanjur OI, Vrijenhoek RC (2002) Historical biogeography of the livebearing fish genus Poeciliopsis (Poeciliidae : Cyprinodontiformes). Evolution 56: 972–984. [DOI] [PubMed] [Google Scholar]
- 59. Ornelas-García CP, Dominguez-Dominguez O, Doadrio I (2008) Evolutionary history of the fish genus Astyanax Baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol 8: 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conner J, Suttkus R (1986) Zoogeography of freshwater fishes of the western Gulf Slope of North America. The zoogeography of North American freshwater fishes: New York, John Wiley and Sons: 413–456.
- 61.McKinney M, McNamara K, Carter B, Donovan S (1992) Evolution of Paleogene echinoids: a global and regional view. Eocene-Oligocene climatic and biotic evolution: 349–367.
- 62.Thomas E (1992) Middle Eocene–late Oligocene bathyal benthic foraminifera (Weddell Sea): Faunal changes and implications for ocean circulation. Eocene-Oligocene climatic and biotic evolution Princeton Univ Press, Princeton, NJ: 245–271.
- 63. Crandall KA, Templeton AR (1999) The Zoogeography and Centers of Origin of the Crayfish Subgenus Procericambarus (Decapoda: Cambaridae). Evolution 53: 123–134. [DOI] [PubMed] [Google Scholar]
- 64.Miller RR, Smith ML (1986) Origin and geography of the fishes of Central Mexico. In: Hocutt CH, Wiley EO, editors. The Zoogeography of North American freshwater fishes. New York, USA: Wiley-Intersciences publication.
- 65. Haq BU, Hardenbol J, Vail PR (1987) Chronology of Fluctuating Sea Levels since the Triassic. Science 235: 1156–1167. [DOI] [PubMed] [Google Scholar]
- 66. Riggs S (1984) Paleoceanographic model of Neogene phosphorite deposition, US Atlantic continental margin. Science 223: 123. [DOI] [PubMed] [Google Scholar]
- 67.Swift C, Gilbert C, Bortone S, Burgess G, Yerger R (1986) Zoogeography of the freshwater fishes of the southeastern United States: Savannah River to Lake Pontchartrain. In: Hocutt CH, Wiley EO, editors. Zoogeography of North American freshwater fishes. New York: John Wiley and Sons. 213–255.
- 68. Schönhuth S, Doadrio I, Dominguez-Dominguez O, Hillis D, Mayden R (2008) Molecular evolution of southern North American Cyprinidae (Actinopterygii), with the description of the new genus Tampichthys from central Mexico. Molecular Phylogenetics and Evolution 47: 729–756. [DOI] [PubMed] [Google Scholar]
- 69.Smith M, Miller R (1986) The evolution of the Rio Grande Basin as inferred from its fish fauna, p. 457–485. In: Hocutt C, Wiley E, editors. The Zoogeography of North American Freshwater Fishes. New York: John Wiley and Sons.
- 70. Burrows R (1910) Geology of northern Mexico. Boletín de la Sociedad Geológica Mexicana 7: 85–103. [Google Scholar]
- 71. Doadrio I, Domínguez O (2004) Phylogenetic relationship within the fish family Goodeidae based on cytochrome b sequence data. Molecular Phylogenetics and Evolution 31: 416–430. [DOI] [PubMed] [Google Scholar]
- 72. Barbour CD (1973) A biogeographical history of Chirostoma (Pisces:Atherinidae): a species flock from the Mexican Plateau. Copeia 1973: 553–556. [Google Scholar]
- 73. Ferrari L, Conticelli S, Vaggelli G, Petrone CM, Manetti P (2000) Late Miocene volcanism and intra-arc tectonics during the early development of the Trans-Mexican Volcanic Belt. Tectonophysics 318: 161–185. [Google Scholar]
- 74. Ferrari L (2000) Avances en el conocimiento de la Faja Volcánica Transmexicana durante la última década. Boletín de la Sociedad Geológica Mexicana 53: 84–92. [Google Scholar]
- 75. Taylor CA, Hardman M (2002) Phylogenetics of the crayfish subgenus Crockerinus, genus Orconectes (Decapoda: Cambaridae), based on cytochrome oxidase I. Journal of Crustacean Biology. 22: 874–881. [Google Scholar]
- 76. Crandall KA, Fitzpatrick Jr JF (1996) Crayfish molecular systematics: using a combination of procedures to estimate phylogeny. Systematic Biology 45: 1. [Google Scholar]
- 77. Mokady O, Loya Y, Achituv Y, Geffen E, Graur D, et al. (1999) Speciation Versus Phenotypic Plasticity in Coral Inhabiting Barnacles: Darwin’s Observations in an Ecological Context. Journal of Molecular Evolution 49: 367–375. [DOI] [PubMed] [Google Scholar]
- 78. Whiting MF (2002) Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zoologica Scripta 31: 93–104. [Google Scholar]
- 79. Whiting MF, Carpenter JC, Wheeler QD, Wheeler WC (1997) The Strepsiptera Problem: Phylogeny of the Holometabolous Insect Orders Inferred from 18S and 28S Ribosomal DNA Sequences and Morphology. Syst Biol 46: 1–68. [DOI] [PubMed] [Google Scholar]
- 80. Colgan DJ, McLauchlan A, Wilson GDF, Livingston SP, Edgecombe GD, et al. (1998) Histone H3 and U2 snRNA DNA sequences and arthropod molecular evolution. Aust J Zool 46: 419–437. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Voucher numbers and localities of the individuals analyzed. Proposed new taxa are indicated as Cambarellus sp., and correspond to the terminal clades indicated with roman numerals in phylogeny (see Figure 3).
(DOCX)