Abstract
Most chloroplast genes in vascular plants are organized into polycistronic transcription units, which generate a complex pattern of mono-, di-, and polycistronic transcripts. In contrast, most Chlamydomonas reinhardtii chloroplast transcripts characterized to date have been monocistronic. This paper describes the atpA gene cluster in the C. reinhardtii chloroplast genome, which includes the atpA, psbI, cemA, and atpH genes, encoding the α-subunit of the coupling-factor-1 (CF1) ATP synthase, a small photosystem II polypeptide, a chloroplast envelope membrane protein, and subunit III of the CF0 ATP synthase, respectively. We show that promoters precede the atpA, psbI, and atpH genes, but not the cemA gene, and that cemA mRNA is present only as part of di-, tri-, or tetracistronic transcripts. Deletions introduced into the gene cluster reveal, first, that CF1-α can be translated from di- or polycistronic transcripts, and, second, that substantial reductions in mRNA quantity have minimal effects on protein synthesis rates. We suggest that posttranscriptional mRNA processing is common in C. reinhardtii chloroplasts, permitting the expression of multiple genes from a single promoter.
The chloroplast genome of Chlamydomonas reinhardtii shares many similarities with the genomes of vascular plants. These genomes are circular DNA molecules that range in size from 120 to 200 kb and have two unique regions separated by large, inverted repeats. Although gene content is highly conserved, the distribution of genes along the chloroplast chromosome varies widely between vascular plants and C. reinhardtii (Sugiura, 1992).
In vascular plant chloroplasts there is substantial evidence for extensive co-transcription of genes in polycistronic operons (Sugita and Sugiura, 1996), which results in complex mRNA accumulation patterns. A striking example is the psbB gene cluster, which groups three PSII genes and two Cyt b6/Cyt f complex genes into the transcription unit psbB-psbT-psbH-petB-petD (Barkan, 1988; Kohchi et al., 1988; Westhoff and Herrmann, 1988). Approximately 20 RNA species could be resolved for spinach and maize, each apparently resulting from processing of a primary transcript containing all five coding regions. In maize, both monocistronic and polycistronic petB and petD transcripts were shown to be engaged in translation (Barkan, 1988), but the monocistronic petD transcript was a substantially better template for translation than its precursor forms (Barkan et al., 1994).
The situation in C. reinhardtii, with most chloroplast mRNAs accumulating as monocistronic transcripts, appears to be very different from that in vascular plants. However, several instances of co-transcription of two or more genes have been documented (Rochaix, 1996). Although in most cases co-transcription was demonstrated by the accumulation of dicistronic mRNAs, the mode of petA-petD transcription suggests that the degree of co-transcription in C. reinhardtii chloroplasts may be greatly underestimated. Although only monocistronic transcripts for the petA and downstream petD genes accumulate in wild-type cells, deletion of the petD promoter still allowed the accumulation of wild-type levels of monocistronic petD mRNA as well as a small amount of a petA-petD co-transcript (Sturm et al., 1994). Apparently, in the absence of a functional petD promoter, the petD gene can be transcribed from the upstream petA promoter. In this case, a lack of transcription termination downstream of petA combines with efficient 5′ processing of petD mRNA to generate mature petD transcripts (Sakamoto et al., 1994). It is likely that both promoters are used in wild-type strains. Other monocistronic transcripts in C. reinhardtii chloroplasts may be generated similarly by a combination of co-transcription and processing.
The atpA gene encodes the α-subunit of the chloroplast ATP synthase (Dron et al., 1982b; Hallick, 1984; Leu et al., 1992). Both a monocistronic atpA transcript of 2.2 kb and a possible precursor form of slightly larger size have been detected in C. reinhardtii (Dron et al., 1982b). The relative amounts of the monocistronic atpA transcript and the putative precursor were found to vary in the nuclear mutants ncc1 (Drapier et al., 1992) and crp3 (Levy et al., 1997), suggesting that transcript maturation and/or stabilization in this region is complex and governed by at least two nuclear factors. Here we present a transcriptional analysis of the atpA gene cluster, the most complex analyzed to date in C. reinhardtii. We show that multiple promoters and mRNA-processing events together result in the accumulation of multiple, overlapping transcripts, reminiscent of transcription patterns typical in vascular plant chloroplasts.
MATERIALS AND METHODS
Strains and Growth Conditions
The wild-type Chlamydomonas reinhardtii strain used as a recipient for the creation of ΔatpA was P17, which was obtained by transformation of the atpB deletion strain CC373 to prototropy with a wild-type atpB gene (Stern et al., 1991). Cells were grown in Tris-acetate-phosphate medium (Harris, 1989), pH 7.2, at 25°C with 5.9 μmol photons m−2 s−1 of continuous illumination.
Plasmid Constructs
The nomenclature used for C. reinhardtii ctDNA restriction fragments is described by Harris (1989). Plasmid pΔatpA carries a 2004-bp deletion from the left end of R7 to a HindIII site immediately downstream of psbI. It was constructed by subcloning the 4.1-kb BamHI-EcoRI fragment from R15 and the 1.5-kb HindIII-EcoRI fragment from R7 into the BamHI and EcoRI sites of pUC19 (Yanisch-Perron et al., 1985) after the EcoRI site of the 4.1-kb fragment and the HindIII site of the 1.5-kb fragment were filled in with the Klenow fragment of DNA polymerase.
Plasmid pΔ1 carries a 632-bp deletion from a PacI site immediately downstream of the atpA stop codon to the same HindIII site, and was constructed as follows. The 4.1-kb BamHI-EcoRI fragment from R15 was subcloned into the BamHI and EcoRI sites of pBluescript (Promega) to generate pR15-1. The cloned R7 fragment was digested with PacI, repaired with the Klenow fragment, digested with EcoRI, and then purified. Separately, R7 was digested with HindIII, repaired with the Klenow fragment, and digested with BamHI, which is located in the multiple cloning site next to the EcoRI site of R7. These two purified fragments were ligated into the EcoRI and BamHI sites of pUC19 to generate pR7-1, which carries the deletion between the PacI and HindIII sites. To add upstream sequences to facilitate homologous recombination, the 2.9-kb EcoRI fragment of pR7-1 was inserted into the EcoRI site of pR15-1, yielding pΔ1.
Plasmid pΔ2 carries a 266-bp deletion from a HpaI site immediately downstream of the psbI initiation codon to the same HindIII site and was constructed as follows. R7 was digested to completion with EcoRI and partially with HpaI, and the 1.65-kb EcoRI-HpaI fragment was purified. Separately, the R7 fragment was digested with HindIII, repaired with the Klenow fragment, and digested with BamHI, which is located in the multiple cloning site adjacent to the EcoRI site of R7. These two fragments were inserted into the EcoRI and BamHI sites of pUC19 to generate pΔ3-1. Finally, to add upstream sequences to facilitate homologous recombination, the 3.2-kb EcoRI fragment of pΔ3-1 was inserted into the EcoRI site of pR15-1, yielding pΔ2.
Plasmid pΔ3 carries a 313-bp deletion between two HpaI sites, the first located 60 bp downstream of the atpA stop codon, and the second immediately downstream of the psbI initiation codon, and was constructed as follows. Plasmid pR7, carrying the R7 fragment in pUC19, was digested with HpaI and re-ligated to generate pEcoRI12-1. To facilitate homologous recombination, the 3.2-kb EcoRI fragment from pEcoRI12-1 was inserted into the EcoRI site of pR15-1, yielding pΔ3.
To construct the psbI-uidA fusion gene, a 331-bp ApoI-ScaI promoter test fragment was cloned into pBluescript, excised with XhoI and SmaI, and used to replace the petD promoter-5′ UTR fragment of plasmid pDG2 (Sakamoto et al., 1993). The resultant plasmid was a possible transcriptional fusion of psbI and uidA, flanked by the 3′ UTR of rbcL. The atpH-uidA promoter fusion was constructed by first subcloning a 460-bp EcoRI-RsaI fragment of R8 containing 370 bp of the 5′ noncoding region and 90 bp of the atpH coding region into EcoRI-SmaI-digested pBluescript, to create pHG1. A 2-kb BamHI-SacI fragment of pBI221 (Clontech, Palo Alto, CA) containing the uidA-coding region was inserted into the BamHI and SacI sites of pHG1 to obtain pHG2, generating a translational fusion of the atpH amino terminal to the entire uidA coding region. The EcoRI-SacI fragment of pHG2 was inserted into EcoRI-EcoRV-digested pBluescript after blunting the SacI site, yielding pHG3. A 440-bp HindIII-SacI fragment containing the rbcL 3′ UTR was subcloned from pUC-atpX-aad (Goldschmidt-Clermont, 1991) into the HindIII and SacI sites of pHG3, yielding pHG4. Then, the SacI-KpnI fragment of pHG4 was inserted into BamHI-KpnI-digested pUC19 after blunting the SacI and BamHI sites with the Klenow fragment of DNA polymerase, yielding pHG40. Finally, the 2.9-kb SalI fragment of pHG40, carrying the expression cassette, was inserted into the BglII site of pΔ26 (Stern et al., 1991) after partially filling in the SalI site with dTTP and dCTP and the BglII site with dATP and dGTP, yielding pHG5 and pHG5-R, respectively. Plasmid pHG5 carries the atpH-uidA-rbcL cassette in tandem with atpB, whereas pHG5-R has the cassette in the convergent orientation.
Plasmids 3′ rbcL(+) and 3′ rbcL(−) were constructed as follows. Plasmid pATPA-2, which was obtained from S. Ketchner (laboratory of F.-A.W.), contains a 4.37-kb HindIII-XbaI fragment beginning 1.25 kb upstream of the atpA start codon and ending within cemA. A 500-bp fragment containing the rbcL 3′ UTR and identical to that used in pUC-atpX-aad was excised from plasmid pFAR12 (Choquet et al., 1998) with SmaI and HindIII, and blunted with the Klenow fragment of DNA polymerase. Plasmid pATP-2 was linearized at an Eco47III site located approximately 120 bp downstream of the atpA mRNA 3′ end and 245 bp upstream of the cemA start codon, and the rbcL 3′ UTR fragment was inserted, yielding clones with the insert in both orientations.
Chloroplast Transformation
Strain ΔatpA was obtained by co-transformation of the wild-type strain P17 with pΔatpA and pCrBH4.8, which contains a version of the rrn16 gene conferring spectinomycin resistance, as described previously (Chen et al., 1993). ΔatpA was determined to be homoplasmic by DNA-filter hybridizations and by its inability to grow on medium lacking acetate. Strains Δ1, Δ2, and Δ3 were obtained by bombarding ΔatpA with the corresponding plasmids and selecting for growth on minimal medium. Homoplasmicity was verified by DNA-filter hybridizations. The psbI and atpH promoter test constructs were introduced into the atpB deletion-mutant strain CC373 (Shepherd et al., 1979), and transformants were selected on medium lacking acetate. Integration of the chimeric uidA genes described above was verified by PCR and DNA-filter hybridizations. Strains 3′ rbcL(+) and 3′ rbcL(−) were created by bombarding ΔatpA cells as described previously (Kuras and Wollman, 1994), and selecting for photosynthetic growth on minimal medium under bright light.
RNA Analysis
Total RNAs were extracted from 20-mL cultures at a density of approximately 2 × 106 cells mL−1 following a method described for Saccharomyces cerevisiae (Schmitt et al., 1990). Cells were pelleted and resuspended in 400 μL of 50 mm NaC2H3O2, pH 4.8, 10 mm EDTA, and 100 μm aurintricarboxylic acid. The cells were transferred to 1.5-mL microcentrifuge tubes, and SDS was added to a final concentration of 2%. Then, 1 volume of phenol equilibrated at pH 4.8 was added and the mixture was incubated for 4 min at 65°C and quick frozen in liquid N. After thawing, the aqueous phase was recovered by centrifugation and RNA was precipitated with ethanol. RNA samples were fractionated in formaldehyde-agarose gels, transferred to nylon membranes under vacuum, and hybridized with 32P-labeled probes made by random priming, as described previously (Drapier et al., 1992). Gels were analyzed using a phosphor imager (Molecular Dynamics, Sunnyvale, CA). Probes used were as follows, unless designated otherwise: for atpA, the 947-bp EcoRI-PstI fragment of R7; for psbI, the 215-bp RsaI-NsiI fragment of R7; for cemA, the 683-bp XbaI-EcoRI fragment of R7; for atpH, the 291-bp AflIII-ClaI fragment of R8; and for rbcL, the fragment R15-4 of Dron et al. (1982a).
The probe for S1 nuclease protection of the cemA and atpH 3′ ends was labeled by Klenow fill-in at the EcoRI site between R7 and R8, and then isolated from an agarose gel after digestion with BglI. S1 nuclease protection followed a published protocol (Ausubel et al., 1990) with slight modifications. Total RNA was co-precipitated with the radiolabeled probe and resuspended in 16 μL of formamide and 4 μL of 5× hybridization buffer containing 200 mm Pipes, pH 6.4, 2 m NaCl, and 5 mm EDTA. This mixture was denatured at 65°C for 10 min and annealed at 30°C overnight. S1 nuclease buffer (300 μL) containing 5 or 10 units of S1 nuclease μg−1 RNA was added to the annealed mixture and incubated for 1 h at 30°C. The products were collected by ethanol precipitation and analyzed by alkaline gel electrophoresis using a γ-32P-labeled 1-kb ladder (GIBCO-BRL) for molecular mass standards. Primer extension (Sturm et al., 1994) and RNase protection using uniformly labeled antisense RNA probes (Levy et al., 1997) were carried out as described previously. The probe for RNase protection of the atpA 3′ end was made from plasmid p22ApS, a 331-bp ApoI-ScaI fragment extending from 26 bp upstream of the atpA translation termination codon to 23 bp downstream of the psbI 5′ end. Run-on transcription assays were carried out as described by Gagne and Guertin (1992) with modifications (Stern and Kindle, 1993).
Protein Analysis
Pulse-labeling experiments were carried out as described previously (Drapier et al., 1992) in the presence of an inhibitor of cytoplasmic translation (6.6 μg mL−1 cycloheximide). Proteins of solubilized cells were separated in urea/SDS-polyacrylamide gels (Piccioni et al., 1981), and radioactive polypeptides were detected using a phosphor imager. For immunoblots, proteins from unlabeled, solubilized cells were separated as described above, transferred to nitrocellulose, incubated with specific polyclonal antibodies followed by 125I-protein A, as described previously (de Vitry et al., 1989). Detection and quantification of labeling were performed using a phosphor imager. Chloroplast F1F0 ATP synthase anti-α- and anti-β-subunit sera were kindly provided by C. Lemaire (Centre de Génétique Moléculaire, Gif-sur-Yvette, France). Antiserum against OEE2 was obtained in our laboratory.
RESULTS
Tetracistronic Transcription Unit in the atpA Region of the Chloroplast Genome
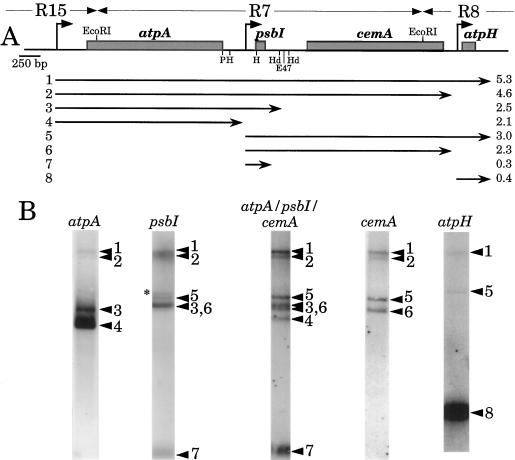
Three genes are found in the region immediately downstream of the atpA gene, as shown in Figure 1A: psbI, which encodes a small PSII subunit (Boudreau et al., 1994; Kunstner et al., 1995); cemA, which encodes a putative envelope membrane protein involved in C uptake (Rolland et al., 1997); and atpH, which encodes subunit III of the chloroplast ATP synthase (Lemaire and Wollman, 1989b). When total RNA was extracted from a wild-type strain and analyzed by RNA-filter hybridization with an intragenic atpA DNA fragment, two major transcripts of 2.1 and 2.5 kb were identified, as well as two less-abundant transcripts of 4.6 and 5.3 kb (Fig. 1B, atpA probe). The 2.1-kb band (transcript 4) has the size expected for monocistronic atpA mRNA, and represents approximately 90% of the total signal. Transcript 3 accounts for approximately 10% of the signal, and transcripts 1 and 2 accounts for less than 1% each.
Figure 1.
Transcription of the C. reinhardtii chloroplast atpA gene cluster. A, Map of the atpA region of the chloroplast genome. R15, R7, and R8 are EcoRI restriction fragments (Rochaix, 1980). Other restriction sites are: P, PacI; H, HpaI; Hd, HindIII; and E47, Eco47III. The three promoters are indicated by bent arrows. The extents of transcripts are shown as numbered arrows, with estimated sizes in kilobase pairs shown at the right. B, RNA accumulation in wild-type cells. Blots of total RNA were hybridized with the probes shown at the top of each lane as described in Methods. The atpA-psbI-cemA probe was the 513-bp ScaI-DraII fragment of R7. Not all blots are from the same gel, so the relative migration of some species varies slightly. The asterisk in the psbI lane indicates a transcript of unknown origin, which is not seen when a larger probe is used (third lane).
The sizes of the four atpA-containing transcripts strongly suggested that the four genes clustered in the atpA region were co-transcribed as a tetracistronic transcript. Support for the hypothesis of co-transcription was obtained by hybridizing similar RNA blots with probes for psbI, cemA, and atpH, as shown in Figure 1B; the deduced extents of the transcripts from this region are shown as numbered arrows in Figure 1A. The results show that atpA is present as part of tetra-, tri-, di-, and monocistronic transcripts 1 through 4. Hybridization with psbI revealed three additional mRNAs: transcript 5 (psbI + cemA + atpH), transcript 6 (psbI + cemA), and transcript 7 (monocistronic psbI). The atpH probe revealed one additional transcript, transcript 8, which corresponds to monocistronic atpH. The cemA probe failed to detect a monocistronic cemA transcript. This probe labeled only transcripts 1, 2, 5, and 6, each of which carries at least one other coding region upstream of cemA. Transcripts 1 through 8 accumulated in widely different amounts, the most abundant being the monocistronic atpA and atpH mRNAs.
Promoters in the atpA Gene Cluster
The transcripts shown in Figure 1 could be transcribed from a single atpA-proximal promoter, with RNA processing generating the remainder of the transcripts. Alternatively, functional promoters could also lie immediately upstream of psbI and atpH, where they might be required for expression of their respective genes or may be redundant with the atpA-proximal promoter.
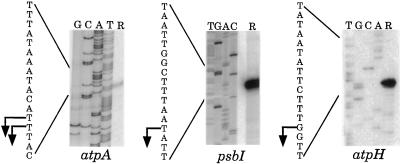
To localize promoters within this region, we first mapped mRNA 5′ ends upstream of atpA, psbI, and atpH by performing primer-extension experiments. As shown in Figure 2, the atpA 5′ end mapped to two consecutive thymidines located 390 and 391 nt upstream of the AUG initiation codon. These termini are 35 nt upstream of those estimated by an S1 nuclease protection assay (Dron et al., 1982b), and are within a putative promoter element (Dron et al., 1982b). Single 5′ ends were mapped for psbI and atpH. The atpH terminus is preceded by a sequence that matches the palindromic TATAAT(AT) consensus sequence previously observed in C. reinhardtii chloroplast promoters (Klein et al., 1992); this sequence starts at position −13 relative to the mature 5′ end. A- and T-rich sequences are also found surrounding or immediately upstream of the atpA and psbI 5′ ends, although neither matches the consensus. Given the A- and T-rich nature of ctDNA intergenic regions, these similarities may be fortuitous.
Figure 2.
5′-end mapping of atpA gene-cluster transcripts. Lanes R show primer-extension experiments with total RNA from wild-type cells, with the gene indicated under the panel. Relative to the translation-initiation codon, the primer for atpA annealed from +92 to +75, that for psbI annealed from +96 to +80, and that for atpH annealed from +27 to +9.
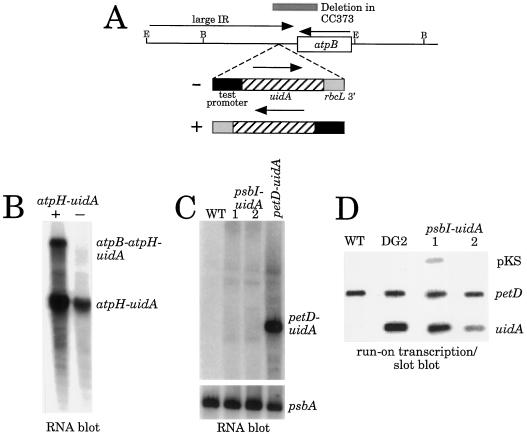
One reliable assay for promoter activity is to fuse putative promoters upstream of reporter genes, and to introduce these chimeric genes into chloroplasts by biolistic transformation. When DNA upstream of the atpA-coding region was fused to the bacterial aadA gene, spectinomycin-resistant transformants were recovered. These transformants accumulated chimeric atpA-aadA transcripts (Goldschmidt-Clermont, 1991), confirming that an active promoter lies upstream of atpA. To test for potential psbI and atpH promoters, upstream regions were fused to the Escherichia coli uidA gene, which encodes GUS, and the chimeric genes were introduced into C. reinhardtii chloroplasts. As shown in Figure 3A, the reporter genes were placed downstream of the atpB gene, in a vector previously used to express uidA fusion genes (Blowers et al., 1990, 1993; Sakamoto et al., 1993).
Figure 3.
Analysis of chimeric uidA promoter fusions. A, The site of insertions of chimeric genes into the chloroplast genome. RNA-filter hybridizations are shown for atpH test constructs (B) and psbI test constructs (C). Both blots were hybridized with a uidA-coding-region probe; a psbA-coding-region probe was used for normalization of the psbI blot. For atpH, + and − indicate strains carrying the fusions in opposite orientations (A). For psbI, 1 and 2 are independent transformants in the (−) orientation, and the petD-uidA lane contains total RNA isolated from the strain DG2, which is known to accumulate uidA mRNA in vivo (Sakamoto et al., 1993). D, Run-on transcription from psbI-uidA transformants are shown. Two micrograms of each of the three plasmids shown at the right was fixed to a nylon filter using a slot-blot apparatus. pKS, pBluescript. Four separate filters were hybridized with 32P-labeled transcripts from freeze-thaw-permeabilized cells of the strains shown across the top. Nonspecific hybridization can be seen for psbI-uidA lane1 (pKS) and for the wild type (uidA).
Figure 3B shows that when the atpH-uidA cassette was introduced either in tandem (+) or convergent (−) with the atpB gene, monocistronic uidA transcripts could be visualized by RNA-filter hybridizations; an additional co-transcript with atpB accumulated when the genes were in the tandem (+) orientation. Extension from a uidA primer revealed similar 5′ termini for monocistronic transcripts from both strains (data not shown); the sizes of the products were consistent with the mapping shown in Figure 2. These results strongly suggest that there is an atpH-specific promoter. Although we cannot completely rule out the possibility that uidA mRNA is produced exclusively by read through of atpB in the (+) orientation or from within the chloroplast genome's large, inverted repeat in the (−) orientation, followed by RNA processing directed by atpH 5′ sequences, we believe that this is very unlikely. The results of earlier studies (Stern and Kindle, 1993) suggested that in the (+) orientation, such read-through transcripts would either accumulate as dicistronic mRNAs or would be degraded by the atpB 3′ processing machinery, and that there is little transcription from within the chloroplast genome's large, inverted repeat.
Results with psbI were more equivocal. psbI-uidA transformants bearing the cassette convergent to the atpB gene did not accumulate a discrete uidA transcript, but did accumulate low levels of heterogeneous uidA-hybridizing transcripts (Fig. 3C). These transcripts could have arisen by a low level of read-through from the large, inverted repeat but, if so, they did not exhibit the discrete termini that might have been expected. To measure the transcription rate of the uidA-coding region in psbI-uidA transformants, cells were permeabilized by freeze-thaw cycles, and nascent RNAs were labeled with [32P]UTP. Strains used were a wild-type strain lacking a uidA gene, the strain DG2, which contains a petD-uidA reporter gene known to be expressed in vivo (Sakamoto et al., 1993), and two independent psbI-uidA transformants. The labeled RNAs were hybridized with filter-bound plasmid DNAs comprising vector only, the petD-coding region as a control, or the uidA gene. The results in Figure 3D clearly show that the uidA gene is transcribed in psbI-uidA transformants at a rate similar to that in DG2, and at a much higher rate than a promoterless uidA gene inserted into the same site (see fig. 7C in Sturm et al., 1994). Slight background hybridization to vector sequences was also observed.
From these results we infer that although the psbI insertion in the chimeric gene does confer promoter activity, it lacks RNA processing and/or stability elements that are required to form stable psbI transcripts in wild-type cells. Although the test fragment extends 309 bp upstream of the mature psbI 5′ end, including the entire intergenic region and 22 bp of the psbI 5′ UTR, additional sequences in the 5′ UTR or coding region may be required for transcript processing or stabilization. One candidate sequence is an imperfect, inverted repeat (ATAGTTAtTAAN5TAtTAACTAT) beginning at position −57 relative to the psbI initiation codon. Taken together, our data are most consistent with a model in which each coding region in the atpA gene cluster, with the exception of cemA, is preceded by a promoter element. Nonetheless, the data do not distinguish whether the mature 5′ termini are formed by transcription initiation or RNA processing.
Transcript 3′ Termini
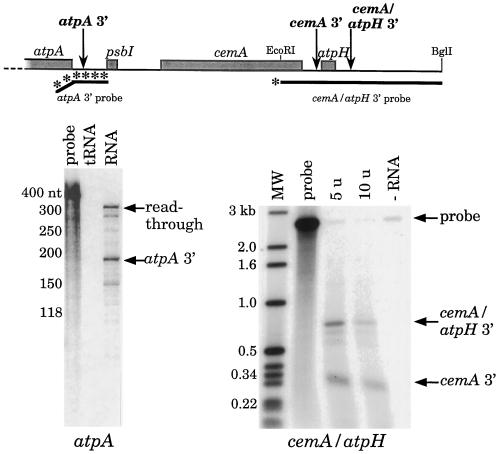
The RNA-filter-hybridization data suggested that there were four unique mRNA 3′ ends in the atpA gene cluster. It was of interest to map these ends to determine whether they coincided with obvious secondary structures, and also to determine whether any were coincident with 5′ ends of other transcripts within the cluster.
3′ ends were mapped to a resolution of ± 15 nt by RNase (atpA) or S1 nuclease (cemA and atpH) protection (Fig. 4). For atpA, two major protected products and several minor products were seen. The lower band corresponds to the 3′ end of the monocistronic atpA message (transcript 4), whereas the upper band represents full protection of the nonvector sequences in the probe, and thus corresponds to transcripts 1 through 3. The 5′ protection products from transcripts 5 through 7 would be 26 nt in length and thus not visible in this gel. The other minor bands are probably experimental artifacts, although we cannot rule out the possibility that they represent genuine in vivo 3′ termini. A single probe was used to map the 3′ ends of cemA and atpH, and two major protected bands were seen. The end of the smaller fragment, labeled cemA, maps downstream of cemA and presumably represents the 3′ ends of transcripts 2 and 6, whereas the larger band, labeled atpH, is consistent with a 3′ end downstream of atpH, and probably represents transcripts 1 and 5. (The DNA fragment fully protected by transcript 8 would not yield a labeled product.) The minor products just below the atpH band are probably artifacts, since they would map within the atpH-coding region. Taken together, our 3′ mapping data are consistent with the diagram in Figure 1.
Figure 4.
3′-End mapping of atpA gene cluster transcripts. Mapping was carried out by RNase (atpA) or S1 nuclease protection with the indicated number of units per microgram of RNA (cemA and atpH). Locations of probes and deduced 3′ ends are shown at the top of the figure and are described in Methods; the cemA 3′ end is close to or coincident with the atpH 5′ end. For atpA, the tRNA lane contained 10 μg of yeast tRNA instead of C. reinhardtii total RNA. For cemA and atpH, total RNA from strain ΔatpA was used; this strain accumulates increased levels of transcripts ending at cemA. Note that the probe for atpA was a uniformly labeled RNA (the bent part indicates pBluescript vector sequences), whereas that for cemA and atpH was an end-labeled DNA fragment. Electrophoresis was in a denaturing polyacrylamide gel for atpA, and in an alkaline agarose gel for cemA and atpH. The atpH 3′ end is marked on the gel as cemA/atpH 3′ because with this probe only cemA-atpH co-transcripts will be visible as protected fragments. MW, Molecular weight.
Regulatory Elements in the atpA-psbI-cemA Intergenic Regions
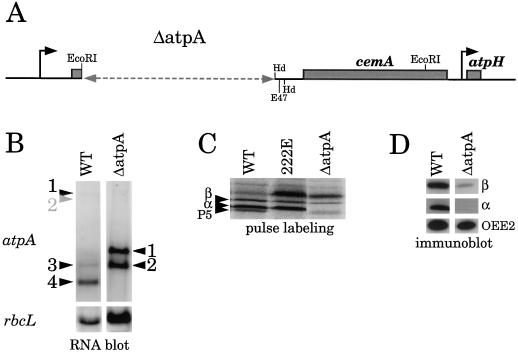
To further examine the role of the region between atpA and cemA in the expression of this gene cluster, we made three deletions in the psbI region. To facilitate the introduction of these deletions into the chloroplast genome, we first created a nonphotosynthetic recipient strain, ΔatpA. As shown in Figure 5A, ΔatpA lacks most of the atpA gene and also psbI. This strain was created by co-transformation with a plasmid conferring spectinomycin resistance in 16S rDNA (see Methods). Figure 5B shows RNA accumulation, and Figure 5, C and D, respectively, show protein synthesis and accumulation in ΔatpA. A 5′ atpA probe revealed two transcripts corresponding to the deleted version of transcripts 1 and 2 of wild-type cells. When protein synthesis was examined by pulse labeling, the α-subunit was undetectable, as expected, but β-subunit synthesis occurred at roughly wild-type levels. However, steady-state accumulation of the β-subunit was strongly reduced, consistent with a posttranslational degradation mechanism (Lemaire and Wollman, 1989a).
Figure 5.
Characterization of the ΔatpA deletion strain. A, The horizontal gray dashed line shows the extent of the deletion. B, atpA-hybridizing transcripts, numbered as in Figure 1, after hybridization of an RNA blot with a 480-bp DraII-EcoRI fragment of R15, the 5′ end of the atpA gene. For the wild type (WT), transcript 2 is not visible in this exposure and is therefore labeled in gray. C, Pulse-labeling with [14C]acetate for 5 min, as described in Methods; β and α indicate subunits of the ATP synthase, and P5 is a PSII subunit. Strain 222E does not synthesize P5 because of a nuclear mutation causing instability of psbB mRNA (Monod et al., 1992), and was used as a negative control for this protein. In strains 222E and ΔatpA, there is increased labeling of the Rubisco large subunit, which migrates just above the β-subunit. This is typically seen for nonphotosynthetic mutants of C. reinhardtii under these experimental conditions (see fig. 3 in Drapier et al., 1992). D, Immunoblot using the antisera indicated at the right. The diminished accumulation of the β-subunit in ΔatpA reflects a posttranslational instability of the protein.
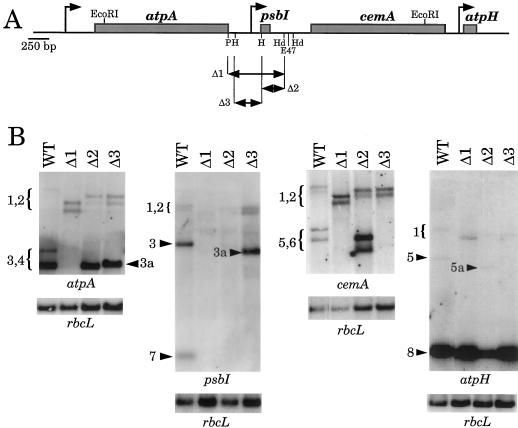
The three deletions mentioned above were designed to eliminate the atpA-psbI intergenic region (Δ3), the psbI-cemA intergenic region (Δ2), or both (Δ1). These deletions could remove intergenic 3′ and 5′ processing sites and/or transcription initiation sites. Figure 6A shows the extents of deletions in each of these constructs. Although atpA is essential for photosynthesis, psbI is dispensable (Kunstner et al., 1995). Therefore, we were able to obtain phototrophic transformants carrying the deletions in Δ1, Δ2, and Δ3 by selection on minimal medium after transformation of ΔatpA.
Figure 6.
RNA accumulation in the deletion strains Δ1, Δ2, and Δ3. A, Map of the atpA gene cluster as shown in Figure 1. The extents of the deletions in Δ1, Δ2, and Δ3 are shown. B, RNA accumulation, with strains shown at the top of each panel and probes shown at the bottom. Transcripts 1 and 2 accumulate to relatively low levels in wild-type (WT) cells, and are more easily visualized with the cemA probe. An rbcL probe was used as a loading control.
Figure 6B shows RNA-filter hybridization results with a series of probes from the atpA gene cluster and RNAs isolated from a wild-type strain and the deletion mutants. Using a probe from the atpA-coding region, two transcripts were detected for Δ1, and three each for Δ2 and Δ3. The two Δ1 transcripts are equivalent to RNAs 1 and 2 from the wild-type strain, although they are shorter as a result of the 355-nt deletion. Therefore, the Δ1 mutant appears to lack the cis elements necessary to generate the mono- and dicistronic RNAs terminating in this region (transcripts 3 and 4). The Δ2 and Δ3 mutants also accumulated deleted versions of RNAs 1 and 2, which are intermediate in size between those of Δ1 and wild-type cells. These results indicate that the formation of RNAs 1 and 2 does not require any sequences in the deleted regions, for example, for correct RNA folding. Furthermore, the accumulation of these transcripts is somewhat higher than in wild-type cells, consistent with the idea that when processing signals are present in the psbI region, RNAs 1 and 2 can serve as precursors for RNAs 3, 4, and others. However, the increase in RNAs 1 and 2 in Δ1 transformants is still less than would be expected if the normal levels of RNAs 3 and 4 were now present as longer transcripts. This indicates either that the deletions destabilize the longer RNAs or that regulatory mechanisms limit the accumulation of RNAs 1 and 2.
The atpA probe also identified major, shorter transcripts in the Δ2 and Δ3 transformants. The major transcript in Δ2 corresponds to the monocistronic atpA transcript, RNA 4, in wild-type cells. The accumulation of this transcript indicates that the site(s) of transcription termination and/or processing for the monocistronic atpA transcript is unaltered by the deletion downstream of psbI. The major atpA transcript in Δ3 also hybridized with a psbI coding-region probe (RNA 3a) and thus contains atpA and psbI 3′ sequences; we infer that it is a deleted version of transcript 3. Because this transcript terminates at the psbI 3′ processing site and no shorter transcript accumulates, we conclude that the processing site for monocistronic atpA lies within the region deleted in Δ3.
Figure 6B also shows results with three other probes, which confirm the identities of the transcripts described above. For example, a psbI probe detected nothing in Δ1 and Δ2, since these sequences had been deleted, but identified RNAs 1 and 2 in Δ3. Monocistronic psbI mRNA was seen only in the wild-type strain; we infer that Δ3 lacks sequences required for 5′-end formation of psbI mRNA. A cemA probe also identified full-length and internally deleted versions of RNAs 1 and 2. In addition, it hybridized with RNAs 5 and 6 from wild-type cells and internally deleted versions in Δ2. RNAs 5 and 6 did not accumulate in Δ1 or Δ3; we conclude that sequences required for transcription or 5′-end formation were deleted in these transformants. Finally, an atpH-specific probe detected transcripts 1, 5, and 8. As seen with the cemA probe, RNA 5 accumulated only in wild-type and Δ2 (RNA 5a) cells, presumably because signals for 5′-end formation have been deleted in the other strains. Accumulation of atpH RNA (RNA 8) was not dramatically affected, which was not surprising because the deletions lie far upstream of atpH.
The total content of atpA-containing transcripts in Δ2 and Δ3 was significantly reduced relative to those of wild-type cells when normalized to the control transcript rbcL. Based on phosphor imager quantification relative to rbcL in multiple experiments, these strains accumulated approximately 50 and 40% of the wild-type content, respectively, mostly as versions of RNA 3 or 4. In contrast, the amount of the longer RNAs 1 and 2 increased approximately 3-fold in Δ2 and 1.5-fold in Δ3 relative to wild-type RNAs 1 and 2. Moreover, the amounts of RNAs 5 and 6 increased approximately 4-fold in Δ2 relative to wild-type cells. In Δ1, atpA transcript accumulation was more severely affected; RNAs 1 and 2 represented approximately 15% of the total atpA transcripts accumulating in wild-type cells. These alterations in transcript accumulation reflect complex relationships between RNA processing and stability that may be influenced by transcript structure in the deletion strains, as well as by other, as yet undefined mechanisms.
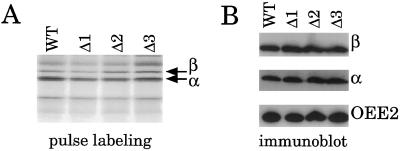
Translation of the ATPase α-Subunit
The atpA-containing mRNAs of Δ1, Δ2, and Δ3 varied from wild type both in terms of the polycistronic distribution of the atpA-coding region and in the total steady-state level of atpA transcripts. We wondered whether these variations might affect the synthesis and/or accumulation of the ATP synthase α-subunit. These were assessed by pulse labeling with [14C]acetate, and by immunoblotting, respectively. Figure 7A shows the results of an experiment in which wild-type, Δ1, Δ2, and Δ3 cells were pulse labeled for 5 min in the presence of cycloheximide to inhibit cytosolic translation. The rates of synthesis in both the α- and β-subunits were similar to those in wild-type cells, displaying the characteristic higher rate of synthesis for the α-subunit than for the β-subunit (Drapier et al., 1992). No differences were seen in protein accumulation as determined by immunoblot analysis (Fig. 7B). It is remarkable that Δ1, which accumulates only about 15% of the wild-type level of atpA-containing mRNAs, and only in the form of polycistronic transcripts, nevertheless displayed essentially wild-type rates of synthesis for the α-subunit.
Figure 7.
Protein synthesis and accumulation in strains Δ1, Δ2, and Δ3. The strains were pulse labeled for 5 min with [14C]acetate (A) or analyzed by immunoblotting (B). Labeling is as for Figure 5.
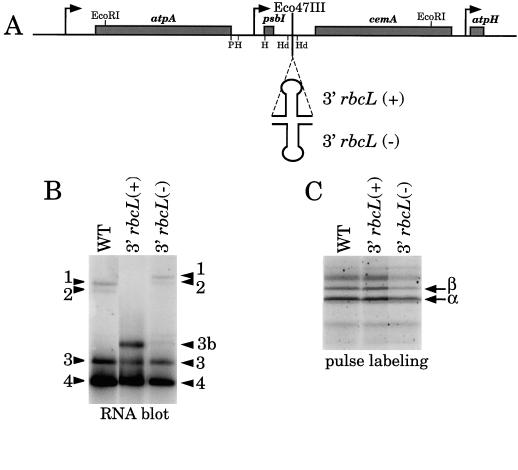
Interruption of the Cluster by Insertion of the rbcL 3′ UTR
One possible interpretation of the results shown in Figure 7A, namely that α-subunit synthesis was unaffected by the deletions in Δ1, Δ2, and Δ3, is that it is translated only from the larger polycistronic mRNAs, transcripts 1 and 2, even in wild-type cells. To determine whether the α-subunit could be synthesized from mono- or dicistronic atpA mRNA, we constructed an atpA gene cluster that was modified to prevent accumulation of the tri- and tetracistronic atpA transcripts. To do this, the 3′ UTR of rbcL was inserted between the psbI and cemA genes, as shown in Figure 8A. In the sense (+) orientation, this sequence was suggested to act as a transcriptional attenuator, since RNA downstream of the stem loop did not accumulate in vivo (Blowers et al., 1993). However, because the rbcL 3′ UTR is not an efficient transcription terminator in vivo (Rott et al., 1996), we think it more likely that RNA processing and destabilization of the downstream sequences are responsible for the transcript accumulation patterns. The rbcL sequence was inserted in both the (+) and (−) orientations at an Eco47III site approximately 120 bp downstream of the mapped 3′ end of psbI.
Figure 8.
Analysis of transformants with an insertion of the rbcL 3′ UTR downstream of psbI. A, Map of the atpA gene cluster showing the site of the rbcL 3′ UTR insertion. B, RNA-filter hybridization analysis using an atpA coding-region probe. Transcripts are numbered as in Figure 1. C, Separation of proteins after 5 min of pulse labeling with [14C]acetate. WT, Wild type.
The RNA-accumulation and protein-synthesis patterns of transformants homoplasmic for these modifications are shown in Figure 8, B and C, respectively. RNA-filter hybridization with an atpA probe revealed altered patterns of transcript accumulation relative to the wild-type strain for both 3′ rbcL(+) and 3′ rbcL(−). In the (+) strain, transcripts 1 and 2 were undetectable, and a new transcript (3b) accumulated. The size of transcript 3b is consistent with that expected for an RNA containing atpA and psbI, and terminating at the 3′ end of the rbcL insertion. For the (−) strain, the pattern was similar to that in the wild type. Transcripts 1 and 2 migrated more slowly because of the rbcL insertion, and two new transcripts with sizes consistent with 3′ ends near the antisense rbcL insertion accumulated to a low level. Other experiments have shown that the 3′ UTR of rbcL can act as an inefficient RNA 3′-end-formation element in this orientation (Rott et al., 1998).
To determine whether the lack of tri- and tetracistronic transcripts in the (+) transformants affected α-subunit synthesis, a pulse-labeling experiment was performed. As shown in Figure 8C, the α-subunit was synthesized at a rate equivalent to wild type, despite the fact that tri- and tetracistronic atpA transcripts did not accumulate to an appreciable level. We conclude that these longer RNAs are not obligate substrates for α-subunit synthesis.
DISCUSSION
Complex Gene Cluster in the C. reinhardtii Chloroplast Genome
We have described the organization and expression of a four-gene cluster encoding two components of the ATP synthase, a PSII polypeptide, and a putative envelope membrane protein involved in C uptake in chloroplasts. Although there are other closely spaced genes in C. reinhardtii chloroplasts, we define this set of four genes as a cluster based on the accumulation of abundant transcripts containing two, three, or four coding regions.
Although gene clusters are common in land-plant chloroplast genomes, there are only a handful of examples of genes that are known to be co-transcribed in C. reinhardtii. These include exon 2 of psaA with psbD (Choquet et al., 1988), atpE with the 3′ part of rps7 (Robertson et al., 1990), psbB with psbT (Johnson and Schmidt, 1993; Summer et al., 1997), psbF with psbL (Mor et al., 1995), rps9-ycf4-ycf3-rps18 (Boudreau et al., 1997), and petA with petD, as discussed in the introduction. Other possibly co-transcribed gene clusters in C. reinhardtii are psbF-psbL-petG-ORF56 (Fong and Surzycki, 1992) and ribosomal protein genes related to the E. coli S10 and spc operons (Harris et al., 1994). More exhaustive analysis of transcript patterns in C. reinhardtii chloroplasts may reveal additional examples of co-transcribed gene clusters.
Transcription Initiation and RNA-Processing Events in the atpA Gene Cluster
The atpA gene cluster has a partially redundant transcriptional organization, with independent promoters proximal to atpA, psbI, and atpH. Although tri- and tetracistronic atpH-containing transcripts accumulate in wild-type cells, the predominant atpH transcript is monocistronic. It is difficult to assess the contribution of the atpH promoter to atpH transcription because monocistronic atpH mRNA might be produced either primarily by transcription from the atpH promoter or by transcription from the atpA- or psbI-proximal promoter followed by RNA processing. In fact, the atpA-proximal promoter could suffice for transcription of the entire gene cluster. This situation is slightly more complex than that of the petA-petD region, where the downstream petD monocistronic transcript is presumably generated by the same processing event whether transcription is initiated at the petA or petD promoter (Sakamoto et al., 1994; Sturm et al., 1994). A similar situation may exist for psbB and the downstream psbH gene; psbH appears to contain a promoter able to provide wild-type levels of RNA when cut off from the psbB promoter (Summer et al., 1997). Rapid RNA processing may partially explain the relatively simple patterns of transcript accumulation in C. reinhardtii chloroplasts despite the fact that co-transcription of gene clusters is more common than originally thought. However, it is clear that redundant promoters exist in a number of gene clusters, and may afford a selective advantage by allowing more finely tuned responses to changing environmental conditions.
Our model for transcription of the atpA gene cluster invokes three sites of transcription initiation, up to three sites of transcript 5′ processing, and four sites of transcription termination or 3′ processing. The 5′ termini of all transcripts could be formed directly by transcription initiation or, alternatively, by 5′ processing. We favor processing as the mechanism for 5′-end formation because no primary transcripts have been detected in C. reinhardtii chloroplasts by capping with α32P-GTP and vaccinia virus guanylyltransferase, a method that works readily for chloroplast mRNAs of land plants (Sugita and Sugiura, 1996).
Sites of 3′-end formation were analyzed by transcript 3′-end mapping and deletion analysis. From data shown in Figure 4, we place the atpA 3′ end approximately 150 nt downstream of the UAA stop codon. There is a small, inverted repeat immediately upstream of this terminus (GcAUUUA… [7 nt]… UAAAUaC) that could form a weak stem-loop structure. In contrast, the 3′ end of psbI mRNA was mapped 75 nt downstream of the UAA stop codon, approximately 25 nt downstream of a strong, potential stem-loop structure (UAAUUUAGCUAAGAGAU-UGUUAccuUAACAAUCUCUUAGCUAAAUUA). Such structures are known to stabilize discrete chloroplast transcripts in C. reinhardtii (Stern et al., 1991; Blowers et al., 1993; Lee et al., 1996). In comparing the relative accumulation of transcripts 3 and 4, there is clearly no correlation with the theoretical stability of the stem-loop and transcript accumulation. This phenomenon has been previously noted for vascular-plant chloroplast mRNAs tested in vitro (Stern and Gruissem, 1987) and is also consistent with the orientation dependence of 3′ inverted repeats in stabilizing chimeric chloroplast mRNAs in C. reinhardtii (Blowers et al., 1993; Rott et al., 1998). These observations suggest that RNA-binding proteins or other RNA structures play a pivotal role in determining transcript abundance.
The 3′ end of cemA mRNA mapped approximately 180 nt downstream of the UAA stop codon, immediately downstream of an inverted repeat (AACCAAAGAAUAUaAUAUUCU-UUGGUU). The atpH 5′ end also maps in this region, to the second G in the inverted repeat sequence. This raises the possibility that the cemA 3′ end and the atpH 5′ end are formed by a common RNA-processing event, but the imprecision of the 3′ mapping must be taken into account and, thus, the ends may overlap slightly and be in competition for the processing machinery. Because the atpH 3′ end was mapped to a region for which we do not have the nucleotide sequence, it cannot be determined if there are obvious secondary structures close to it. Whether these 3′ termini are formed by transcription termination or RNA processing is unknown, but based on data for spinach (Stern and Gruissem, 1987) and C. reinhardtii (Stern and Kindle, 1993; Rott et al., 1996) chloroplasts, termination is unlikely to occur at a significant rate.
Therefore, many of the mRNAs from the atpA gene cluster may be produced by processing larger transcripts. Although a precursor-product relationship cannot be directly proven, we found that deleting the atpA 3′-processing site (strain Δ3) or the psbI 3′-processing site (strain Δ2) resulted in an increased accumulation of the larger atpA polycistronic transcripts (Fig. 6). The effect was even more pronounced when both processing sites were deleted (strain Δ1). This is consistent with the suggestion that RNA 1 is the primary transcript from which all of the smaller atpA transcripts are generated, analogous to the mechanism by which the many psbB operon transcripts are formed in land plants (Westhoff and Herrmann, 1988). However, we cannot rule out the possibility that a transcription-termination signal was deleted in the deletion strains, thereby increasing the frequency of transcriptional read through.
Monocistronic mRNAs accumulate for all genes except cemA, which is present only as di- or polycistronic mRNAs 1, 2, 5, and 6. Although there is no direct evidence that cemA is expressed at the protein level in C. reinhardtii, it is a highly conserved gene, with 10 entries in the database, including crop plants, Marchantia, black pine, Synechocystis PCC 6803, and Porphyra. The putative protein in C. reinhardtii would necessarily be translated from a nonmonocistronic message, which would be somewhat unusual for this organism. However, it is possible that cemA is inefficiently translated, and the lack of 5′ processing serves as a regulatory mechanism. In C. reinhardtii petD, subunit IV translation from a dicistronic petA-petD message may be inefficient or impossible (Sturm et al., 1994), and monocistronic petD seems to be the preferred form for translation in maize chloroplasts (Barkan et al., 1994). However, dicistronic mRNAs are the only mRNAs present for psbF-psbL (Mor et al., 1995), and monocistronic mRNAs could not be detected for ycf3 or ycf4, which are known to be expressed at the protein level (Boudreau et al., 1997). Therefore, translation of downstream open reading frames in dicistronic transcripts is clearly possible in C. reinhardtii chloroplasts.
The cemA and psbI Genes Are Dispensable for Photosynthesis
The three deletions introduced downstream of atpA in strains Δ1, Δ2, and Δ3 inactivated psbI but did not abolish phototrophic growth of the transformants. This observation confirms a previous report (Kunstner et al., 1995) that psbI is dispensable for photosynthesis. This report also showed that psbI inactivation nevertheless caused a partial loss of PSII activity. In agreement with this result, we found that Δ1, Δ2, and Δ3 each had substantially altered fluorescence-induction patterns, and accumulated approximately 20% of the wild-type level of PSII proteins (data not shown).
The function of cemA is still under investigation. The cemA gene product was originally reported as a chloroplast envelope membrane protein of 34 kD (Sasaki et al., 1993). However, no protein of this size was detected after in vivo labeling of C. reinhardtii chloroplast envelope membranes with 35S (Clemetson et al., 1992). A later report suggested that the cemA protein could bind heme (Willey and Gray, 1990), whereas a cemA (cotA) mutant of Synechocystis PCC 6803 had defects in CO2 transport (Katoh et al., 1996). C uptake was recently reported to be affected in C. reinhardtii ycf10 (cemA) deletion mutants (Rolland et al., 1997), and the strains were found to be photosynthetically competent but high-light sensitive. We have constructed a cemA-deletion strain (ΔAH) that harbors a deletion from the HpaI site immediately downstream of atpA to the EcoRI site in the carboxyl-terminal part of cemA (see Fig. 1). The ΔAH mutant was still capable of phototrophic growth (data not shown), in agreement with the results of Rolland et al. (1997) showing that cemA does not play an essential role in photosynthesis.
Changes in the Content of atpA Transcripts Do Not Affect α-Subunit Translation
In the strains described in the present study, the range of modifications in the pattern and number of atpA transcripts had no effect on the rate of α-subunit synthesis. The abundance of polycistronic messages beginning with atpA and having 3′ termini following cemA or atpH increased from 2- to 13-fold in Δ1, Δ2, and Δ3, whereas the deletion in Δ1 prevented monocistronic atpA mRNA accumulation altogether. In all cases, wild-type rates of α-subunit synthesis were observed. In contrast, the insertion in strain rbcL(+) (Fig. 8) caused a complete loss of tri- and tetracistronic atpA transcripts and, again, the rate of α-subunit synthesis was unaffected. Because the atpA-coding region lies the farthest upstream, these observations suggest that translation initiation at atpA is independent of the downstream coding regions.
The total amount of atpA-containing messages also varied among the wild-type strain and deletion mutants. The greatest effect was in Δ1, where only 15% of the wild-type amount remained, yet no effect on the rate of α-subunit synthesis was observed. The insensitivity of α-subunit synthesis rates to the number and type of atpA transcripts in C. reinhardtii has been noted previously. In the nuclear mutant ncc1, in which atpA transcript abundance declined by a factor of 10, there was only a moderate effect on the rate of α-subunit synthesis (Drapier et al., 1992). In addition, when C. reinhardtii cells were grown for 48 h in the presence of 5-fluorodeoxyuridine, an inhibitor of ctDNA replication, both the atpA gene-copy number and transcript level decreased by a factor of approximately 10, but the rate of α-subunit synthesis was unaffected (Hosler et al., 1989). Therefore, translation of the α-subunit is limited by some factor other than the availability of atpA transcripts. It follows that there is a large excess of atpA transcripts in wild-type cells, which are stable in spite of their translational inactivity. This is consistent with the 3-fold increase in the steady-state level in atpA transcripts reported for the F54 mutant, which is blocked at the level of α-subunit synthesis (Drapier et al., 1992). Together, these results would argue that in the case of atpA, ribosome association does not protect the transcript from degradation, but, instead, that their degradation may be a co-translational process. However, whether the translation-initiation complex has any role in RNA stability remains to be determined.
ACKNOWLEDGMENTS
D.B.S. performed part of this work as a recipient of a Guggenheim Fellowship and the Georges Morel Prize from the Institut National de Recherche Agronomique. We thank members of the Stern, Kindle, and Wollman laboratories for stimulating discussions.
Abbreviations:
- nt
nucleotides
- UTR
untranslated region
Footnotes
This work was supported by the Centre National de la Recherche Scientifique (grant no. UPR9072 to F.-A.W.) and by the National Science Foundation (grant no. MCB 9406550 to K.L.K. and D.B.S.).
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RG, Moore DD, Seidman JG, Smith JA, Struhl K (1990) Current Protocols in Molecular Biology. Green Publishing Associates and Wiley Interscience, New York
- Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic RNAs. EMBO J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Ellmore GS, Klein U, Bogorad L. Transcriptional analysis of endogenous and foreign genes in chloroplast transformants of Chlamydomonas. Plant Cell. 1990;2:1059–1070. doi: 10.1105/tpc.2.11.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blowers AD, Klein U, Ellmore GS, Bogorad L. Functional in vivo analyses of the 3′ flanking sequences of the Chlamydomonas chloroplast rbcL and psaB genes. Mol Gen Genet. 1993;238:339–349. doi: 10.1007/BF00291992. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Otis C, Turmel M. Conserved gene clusters in the highly rearranged chloroplast genomes of Chlamydomonas moewusii and Chlamydomonas reinhardtii. Plant Mol Biol. 1994;24:585–602. doi: 10.1007/BF00023556. [DOI] [PubMed] [Google Scholar]
- Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix J-D. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997;16:6095–6104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kindle K, Stern D. Initiation codon mutations in the Chlamydomonas chloroplast petD gene result in temperature-sensitive photosynthetic growth. EMBO J. 1993;12:3627–3635. doi: 10.1002/j.1460-2075.1993.tb06036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kück U, Bennoun P, Rochaix JD. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell. 1988;52:903–914. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman F-A (1998) Translation of cytochrome f is autoregulated through the 5′-untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA (in press) [DOI] [PMC free article] [PubMed]
- Clemetson JM, Boschetti A, Clemetson KJ. Chloroplast envelope proteins are encoded by the chloroplast genome of Chlamydomonas reinhardtii. J Biol Chem. 1992;267:19773–19779. [PubMed] [Google Scholar]
- de Vitry C, Olive J, Drapier D, Recouvreur M, Wollman F-A. Posttranslational events leading to the assembly of photosystem II protein complex: a study using photosynthesis mutants from Chlamydomonas reinhardtii. J Cell Biol. 1989;109:991–1006. doi: 10.1083/jcb.109.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier D, Girard-Bascou J, Wollman F-A. Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell. 1992;4:283–295. doi: 10.1105/tpc.4.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M, Rahire M, Rochaix J-D. Sequence of the chloroplast 16S rRNA gene and its surrounding regions of Chlamydomonas reinhardtii. Nucleic Acids Res. 1982a;10:7609–7620. doi: 10.1093/nar/10.23.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dron M, Rahire M, Rochaix JD. Sequence of the chloroplast DNA region of Chlamydomonas reinhardtii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J Mol Biol. 1982b;162:775–793. doi: 10.1016/0022-2836(82)90547-2. [DOI] [PubMed] [Google Scholar]
- Fong SE, Surzycki SJ. Organization and structure of plastome psbF, psbL, petG and ORF712 genes in Chlamydomonas reinhardtii. Curr Genet. 1992;21:527–530. doi: 10.1007/BF00351664. [DOI] [PubMed] [Google Scholar]
- Gagne G, Guertin M. The early genetic response to light in the green unicellular alga Chlamydomonas eugametos grown under light/dark cycles involves genes that represent direct responses to light and photosynthesis. Plant Mol Biol. 1992;18:429–445. doi: 10.1007/BF00040659. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19:4083–4090. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick RB. Identification and partial DNA sequence of the gene for the alpha-subunit of the ATP synthase complex of Chlamydomonas reinhardtii chloroplasts. FEBS Lett. 1984;177:374–376. doi: 10.1016/0014-5793(84)81298-3. [DOI] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham NW. Chloroplast ribosomes and protein synthesis. Microbiol Rev. 1994;58:700–754. doi: 10.1128/mr.58.4.700-754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler JP, Wurtz EA, Harris EH, Gillham NW, Boynton JE. Relationship between gene dosage and gene expression in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol. 1989;91:648–655. doi: 10.1104/pp.91.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Schmidt GW. The psbB gene cluster of the Chlamydomonas reinhardtii chloroplast sequence and transcriptional analyses of psbN and psbH. Plant Mol Biol. 1993;22:645–658. doi: 10.1007/BF00047405. [DOI] [PubMed] [Google Scholar]
- Katoh A, Lee KS, Fukuzawa H, Ohyama K, Ogawa T. cemA homologue essential to CO2 transport in the cyanobacterium Synechocystis PCC6803. Proc Natl Acad Sci USA. 1996;93:4006–4010. doi: 10.1073/pnas.93.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, De Camp JD, Bogorad L. Two types of chloroplast gene promoters in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1992;89:3453–3457. doi: 10.1073/pnas.89.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T, Yoshida T, Komano T, Ohyama K. Divergent messenger RNA transcription in the chloroplast psbB operon. EMBO J. 1988;7:885–892. doi: 10.1002/j.1460-2075.1988.tb02892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunstner P, Guardiola A, Takahashi Y, Rochaix JD. A mutant strain of Chlamydomonas reinhardtii lacking the chloroplast photosystem II psbI gene grows photoautotrophically. J Biol Chem. 1995;270:9651–9654. doi: 10.1074/jbc.270.16.9651. [DOI] [PubMed] [Google Scholar]
- Kuras R, Wollman F-A. The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J. 1994;13:1019–1027. doi: 10.1002/j.1460-2075.1994.tb06350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Bingham SE, Webber AN. Function of 3′ non-coding sequences and stop codon usage in expression of the chloroplast psaB gene in Chlamydomonas reinhardtii. Plant Mol Biol. 1996;31:337–354. doi: 10.1007/BF00021794. [DOI] [PubMed] [Google Scholar]
- Lemaire C, Wollman F-A. The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constituent subunits. J Biol Chem. 1989a;264:10228–10234. [PubMed] [Google Scholar]
- Lemaire C, Wollman F-A. The chloroplast ATP synthase in Chlamydomonas reinhardtii. II. Biochemical studies on its biogenesis using mutants defective in photophosphorylation. J Biol Chem. 1989b;264:10235–10242. [PubMed] [Google Scholar]
- Leu S, Schlesinger J, Michaels A, Shavit N. Complete DNA sequence of the Chlamydomonas reinhardtii chloroplast atpA gene. Plant Mol Biol. 1992;18:613–616. doi: 10.1007/BF00040681. [DOI] [PubMed] [Google Scholar]
- Levy H, Kindle KL, Stern DB. A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas chloroplasts. Plant Cell. 1997;9:825–836. doi: 10.1105/tpc.9.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod C, Goldschmidt-Clermont M, Rochaix J. Accumulation of chloroplast psbB RNA requires a nuclear factor in Chlamydomonas reinhardtii. Mol Gen Genet. 1992;231:449–459. doi: 10.1007/BF00292715. [DOI] [PubMed] [Google Scholar]
- Mor TS, Ohad I, Hirschberg J, Pakrasi HB. An unusual organization of the genes encoding cytochrome b-559 in Chlamydomonas reinhardtii: psbE and psbF genes are separately transcribed from different regions of the plastid chromosome. Mol Gen Genet. 1995;246:600–604. doi: 10.1007/BF00298966. [DOI] [PubMed] [Google Scholar]
- Piccioni RG, Bennoun P, Chua NH. A nuclear mutant of Chlamydomonas reinhardtii defective in photosynthetic photophosphorylation: characterization of the algal coupling factor ATPase. Eur J Biochem. 1981;117:93–102. doi: 10.1111/j.1432-1033.1981.tb06307.x. [DOI] [PubMed] [Google Scholar]
- Robertson D, Gillham NW, Boynton JE. Cotranscription of the wild-type chloroplast atpE gene encoding the CF1/CF0 epsilon subunit with the 3′ half of the rps7 gene in Chlamydomonas reinhardtii and characterization of frameshift mutations in atpE. Mol Gen Genet. 1990;221:155–163. doi: 10.1007/BF00261715. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D. Restriction fragments from Chlamydomonas chloroplast DNA. Methods Enzymol. 1980;65:785–795. doi: 10.1016/s0076-6879(80)65073-3. [DOI] [PubMed] [Google Scholar]
- Rochaix J-D. Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- Rolland N, Dorne AJ, Amoroso G, Sultemeyer DF, Joyard J, Rochaix JD. Disruption of the plastid ycf10 open reading frame affects uptake of inorganic carbon in the chloroplast of Chlamydomonas. EMBO J. 1997;16:6713–6726. doi: 10.1093/emboj/16.22.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott R, Drager RG, Stern DB, Schuster G. The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol Gen Genet. 1996;252:676–683. doi: 10.1007/BF02173973. [DOI] [PubMed] [Google Scholar]
- Rott R, Liveanu V, Drager RG, Stern DB, Schuster G. The sequence and structure of the 3′ untranslated regions of chloroplast transcripts are important determinants of mRNA accumulation and stability. Plant Mol Biol. 1998;36:307–314. doi: 10.1023/a:1005943701253. [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Kindle KL, Stern DB. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of β-glucuronidase translational fusions. Proc Natl Acad Sci USA. 1993;90:497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W, Sturm NR, Kindle KL, Stern DB. petD mRNA maturation in Chlamydomonas reinhardtii chloroplasts: the role of 5′ endonucleolytic processing. Mol Cell Biol. 1994;14:6180–6186. doi: 10.1128/mcb.14.9.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sekiguchi K, Nagano Y, Matsumo R. Chloroplast envelope protein encoded by chloroplast genome. FEBS Lett. 1993;316:93–98. doi: 10.1016/0014-5793(93)81743-j. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd HS, Boynton JE, Gillham NW. Mutations in nine chloroplast loci of Chlamydomonas affecting photosynthetic functions. Proc Natl Acad Sci USA. 1979;76:1353–1357. doi: 10.1073/pnas.76.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern DB, Kindle KL. 3′ end maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol Cell Biol. 1993;13:2277–2285. doi: 10.1128/mcb.13.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Radwanski ER, Kindle KL. A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991;3:285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm N, Kuras R, Buschlen S, Sakamoto W, Kindle KL, Stern DB, Wollman F-A. The petD gene is transcribed by functionally redundant promoters in Chlamydomonas reinhardtii chloroplasts. Mol Cell Biol. 1994;14:6171–6179. doi: 10.1128/mcb.14.9.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast genome. Plant Mol Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- Summer EJ, Schmid VHR, Bruns BU, Schmidt GW. Requirement for the H phosphoprotein in photosystem II of Chlamydomonas reinhardtii. Plant Physiol. 1997;113:1359–1368. doi: 10.1104/pp.113.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts: the psbB operon from spinach. Eur J Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- Willey DL, Gray JC. An open reading frame encoding a putative haem-binding polypeptide is cotranscribed with the pea chloroplast gene for apocytochrome f. Plant Mol Biol. 1990;15:347–356. doi: 10.1007/BF00036920. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]