Abstract
microRNAs (miRNAs) are a class of negative regulators that take part in many processes such as growth and development, stress responses, and metabolism in plants. Recently, miRNAs were shown to function in plant nutrient metabolism. Moreover, several miRNAs were identified in the response to nitrogen (N) deficiency. To investigate the functions of other miRNAs in N deficiency, deep sequencing technology was used to detect the expression of small RNAs under N-sufficient and -deficient conditions. The results showed that members from the same miRNA families displayed differential expression in response to N deficiency. Upon N starvation, the expression of miR169, miR171, miR395, miR397, miR398, miR399, miR408, miR827, and miR857 was repressed, whereas those of miR160, miR780, miR826, miR842, and miR846 were induced. miR826, a newly identified N-starvation-induced miRNA, was found to target the AOP2 gene. Among these N-starvation-responsive miRNAs, several were involved in cross-talk among responses to different nutrient (N, P, S, Cu) deficiencies. miR160, miR167, and miR171 could be responsible for the development of Arabidopsis root systems under N-starvation conditions. In addition, twenty novel miRNAs were identified and nine of them were significantly responsive to N-starvation. This study represents comprehensive expression profiling of N-starvation-responsive miRNAs and advances our understanding of the regulation of N homeostasis mediated by miRNAs.
Introduction
Nitrogen (N) is one of the most important macronutrients essential for plant growth and development. N accounts for 1.5–2.0% of plant dry matter and is required for the synthesis of proteins, nucleotide acids, chlorophylls, and so on [1]. To grow and develop normally, plants must obtain sufficient N from the soil via their roots. However, there is not always sufficient N in the soil to meet the N requirements of plants, because soil N levels are affected by many factors such as soil erosion, rainwater leaching, and microbial consumption. Plants have already evolved a number of ways to conserve and mobilize internal N and to increase acquisition of external N. To adapt to N-limited conditions, plants must sense changes in internal and external N concentrations. The sensing and signal transduction networks that control plant responses to N deficiency are not well characterized.
Recent studies have revealed that plant small RNAs play a pivotal role in regulating gene expression at post-transcription levels. microRNAs (miRNAs), a class of non-coding small RNAs, originate from stem-loop structures of primary transcripts. They are processed into precursor RNA and then mature miRNAs 20–21 nt in length via the activities of Dicer-like proteins in the nucleus [2]. HYL1 (HYPONASTIC LEAVES 1) and SE1 (SERRATE 1) ensure the accurate processing of miRNAs [3]. The processed miRNAs are further methylated at the 3′ terminal by HEN1 (HUA ENHANCER 1) [4] and exported from the nucleus into the cytoplasm where they are incorporated into the RNA-induced silencing complex (RISC). In the RISC, the miRNAs recognize their targets through complementary base pairing and cleave target transcripts or/and repress target translation [5]–[7].
Plant miRNAs have been found in diverse plant species, including monocotyledons (e.g. Oryza sativa, Zea mays, Sorghum bicolor), dicotyledons (e.g. Arabidopsis thaliana, Medicago truncatula, Populus trichocarpa), mosses [8], and unicellular algae [9]. Plant miRNAs are involved in the regulation of plant growth and development, biotic and abiotic stresses, metabolic pathways and so on. Recently, plant miRNAs were shown to function in sensing nutrient stresses. For example, miR395 is induced by sulfate starvation and regulates sulfate accumulation and allocation by targeting APSs (ATP Sulfurylase) and SULTR2;1 (SULFATE TRANSPORTER 2;1), respectively [10]; miR399 is up-regulated by phosphate limitation and controls phosphate transport and redistribution by repressing PHO2 (PHOSPHATE 2) [11]. In addition, by deep sequencing small RNAs, miR778, miR827, miR828, and miR2111 were also identified as being responsive to phosphate starvation [12], [13]. The homeostasis of another essential nutrient, copper, is regulated by miR398, which directs the degradation of Copper/zinc Superoxide Dismutase mRNA when copper is limited [14]. miR397, miR408, and miR857 also mediate the regulation of copper homeostasis by targeting several Laccase genes [15]. miR169 plays an important role in regulating nodule development in M. truncatula and its over-expression leads to decreased expression of the MtHAP2-1 (HAPLESS 2-1) gene and a deficient N-fixation phenotype [16]. Meanwhile, over-expression of miR169 impairs the N-uptake system, leading to low N accumulation in Arabidopsis [17].
To date, however, there have been few studies on miRNA(s) involved in the N-deficiency response. It is unknown whether some miRNAs are specifically responsive to N limitation in Arabidopsis. Because of the vast dynamic range of miRNA expression, it is difficult to discover miRNAs expressed at low frequencies under normal growth conditions. Deep sequencing technologies can rapidly detect known and novel miRNAs with very high sensitivity [12], [13], [18], [19]. The aims of this study were to analyze the expression profiles of small RNAs in response to N deficiency and to identify novel miRNAs specifically induced by N deprivation. Therefore, Solexa sequencing technology was used to detect the small RNAs population in Arabidopsis grown under N-sufficient and N-deficient conditions. Our data provide insight into the authenticity of previously reported Arabidopsis miRNAs and identify some new miRNAs associated with N-starvation stress responses. It suggested that members from the same miRNA family showed differential accumulation in response to N starvation. Three conserved miRNAs were found to play roles in root system development under N starvation. Moreover, the finding that N starvation affects the expression of other nutrient stress-related miRNAs is intriguing, and adds new insight about how miRNAs regulate nutrient balance in plants.
Results and Discussion
Summary of small RNA profiles in response to N deficiency
To investigate the response of miRNAs to N deficiency, two small RNA libraries were constructed from N-sufficient (+N) and N-deficient (−N) seedlings by Solexa high-throughput sequencing technology. After adaptor trimming, the unique sequences were selected from the raw data and then mapped to the Arabidopsis genome. A total of ∼11 million raw reads were obtained from each library, and the clean reads after trimming adaptors accounted for ∼95%. Approximately 10 million sequence reads (89% and 91% of trimmed sequence reads in +N and −N libraries, respectively), corresponding to ∼1.5 million sequence signatures, mapped perfectly to the genome (The Arabidopsis Information Resource 8). The perfectly matched sequences were used for further analysis.
Most of the RNA sequences were within the range of 19 to 24 nt, which accounted for 90% of total clean reads. The correlation between the length of small RNAs and the proportion of total sequence reads was examined (Figure S1A). Two distinctive distributions were observed in +N and −N libraries. The library from the N-deficient samples showed fewer 24 nt sequences and more 20 nt sequences compared with those in the library from the N-sufficient samples. Subsequently, common and specific sequences were identified by comparing the two libraries. The +N-specific, −N-specific, and common sequences between the two libraries were 45.6, 38.95, and 15.99% respectively, for unique small RNAs, and 9.45, 7.98, and 82.57%, respectively, for total small RNAs (Figure S1B). The small RNAs were classified into rRNA, snRNA, snoRNA, tRNA, and other RNA according to their origin, and the proportion of each class was calculated (Table 1). A significant increase in small RNAs derived from tRNA was observed under N-deficient conditions; this trend was also observed under phosphate deficiency [12].
Table 1. Summary of small RNA sequencing data.
| class | +N | −N | ||
| Unique sRNA | Total sRNA | Unique sRNA | Total sRNA | |
| rRNA | 18852(0.87%) | 307668(3.12%) | 16787(0.86%) | 281621(2.73%) |
| snRNA | 1116(0.05%) | 2188(0.02%) | 1907(0.10%) | 5497(0.05%) |
| snoRNA | 1668(0.08%) | 3297(0.03%) | 2209(0.11%) | 6236(0.06%) |
| tRNA | 6099(0.28%) | 301115(3.11%) | 6404(0.33%) | 1007667(9.80%) |
| other | 2131218(98.7%) | 9067082(93.7%) | 1915407(98.6%) | 8984342(87.4%) |
| total | 2158953 | 9681350 | 1942714 | 10285363 |
The small RNA sequences were used to analyze previously characterized miRNAs in Arabidopsis (miRBase release 14.0, www.miRbase.org). All sequences that matched perfectly to known miRNAs from miRbase were used for statistical analyses. A total of 186 miRNAs from the combined RNA sequence reads were identified. These miRNAs belonged to 98 miRNA families and represented 89% of all known unique Arabidopsis miRNA sequences and 83% of all known miRNA families (Table S1).
Expression profiling of known miRNAs in response to N starvation
The small RNA sequences from the N-deficient and N-sufficient samples were analyzed for the presence of previously characterized miRNAs in Arabidopsis. The miR156 family was the most abundant (∼1,400,000 reads), although many miRNAs were expressed at low frequencies (read count fewer than 10).
Deep sequencing can distinguish and measure miRNA sequences with even one nucleotide change, which means that different members from the same miRNA family can be distinguished. Based on the sequencing results, different miRNA family members were expressed at vastly different frequencies. For example, miR158a and miR158b were represented by 331232 and 3383 reads respectively under N-sufficiency conditions, but 115256 and 2850 reads respectively under N-deficiency conditions (Table S2). Similar trends were observed for other miRNA families, such as miR156, miR169, and miR172.
Deep sequencing can also accurately quantify miRNA abundance. Among the miRNAs retrieved, the differentially expressed miRNAs with greater than 3-fold relative change in sequence count were identified (Figure 1). These miRNAs were defined as N-starvation-responsive miRNAs in the present study.
Figure 1. Differentially expressed miRNAs in response to N deficiency.
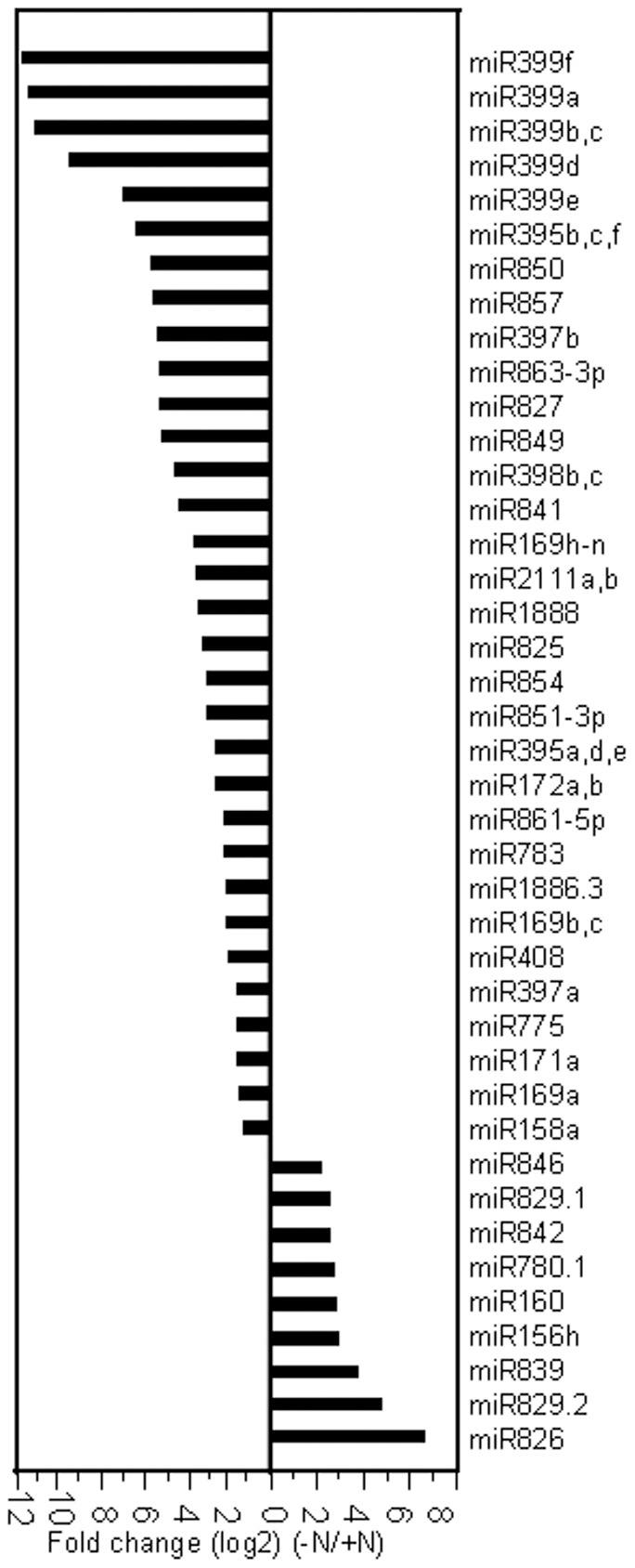
The significantly differentially expressed miRNAs with greater than 3-fold relative change were shown.
Homologous miRNA members conversely respond to N starvation
Most conserved plant miRNA families contain more than two members. Recent studies revealed that ancient plant miRNAs originated from the inverted duplication of target gene sequences and some miRNA members are distributed in clusters. However, it is unclear whether different species of the same family have different functions. Over-expression of different miRNA species has resulted in very similar phenotypes, implying that all miRNA members can effectively suppress their target genes. Deep sequencing technology provides an approach to detect expression of different mature miRNAs. miR169 is a conserved plant miRNA that is found in diverse plant species. In M. truncatula, miR169 plays a role in regulating nodule development [16]. In Arabidopsis, N starvation decreased the expression of miR169. Plants overexpressing miR169 accumulated less N and were more sensitive to N starvation [17]. The Arabidopsis miR169 gene family contains 14 members, represented by four different mature miRNA species; miR169a, miR169bc, miR169d–g, and miR169h–n (Figure 2A). Unlike most miRNA families whose members show similar expression patterns, the different species of the miR169 family showed differential responses to N starvation (Table S2). The read count of the miR169d–g mature sequence increased under N deficiency, but those of the other species dramatically decreased. To confirm this result, stem-loop RT-PCR was used to determine the expression of four mature miR169s in the root under N, P, and S starvation (Figure 2B). Consistent with the sequencing results, miR169d–g, but not the other miR169 species, increased specifically upon N starvation. The differential expression implies that different miRNA species may have roles in specialized functions under N starvation conditions. Differential expression was also observed among members of the miR167, miR171, miR172, and miR319 families (Table S2). To understand the functions of different miRNA species, further research should focus on the spatial and temporal expression patterns of these miRNA species under N deficiency.
Figure 2. Differential expression of different miR169 species.
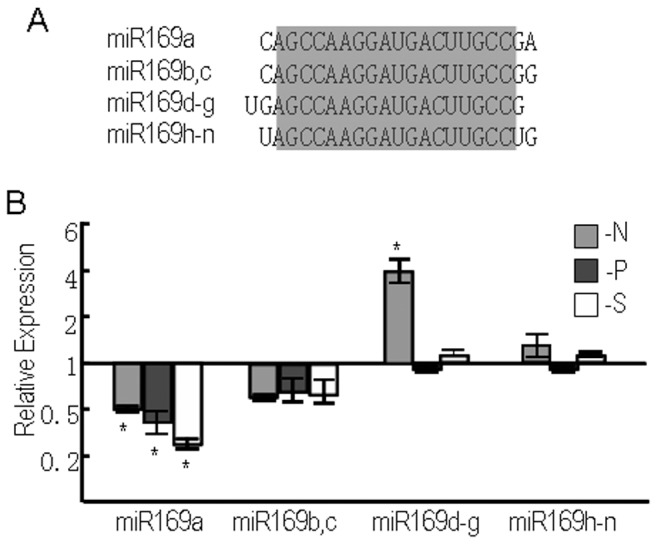
(A) Four different mature miR169 species. (B) Expression of different miR169 members in response to different nutrient deficiencies. Gene expression values shown are relative to the expression in plants grown under normal MS medium, for which the value is set to 1. Error bars indicate ± SE obtained from three biological repeats. Values marked by an asterisk are significantly different from the corresponding control value with Student's t-test (p<0.01; n = 3).
Targets of N-starvation-responsive miRNAs
Under N-starvation conditions, miR399, miR395, miR850, miR857, miR863, and miR827 were significantly down-regulated, whereas miR160, miR826, miR839, and miR846 were dramatically up-regulated. These N-starvation-responsive miRNAs were further classified into two groups, N-starvation-suppressed (NSS) miRNAs and N-starvation-induced (NSI) miRNAs. To analyze the functions of these miRNAs, it is important to identify their targets, since plant miRNAs mainly negatively regulate expression of their target genes. Plant miRNAs recognize their target genes through complementary base-pairing, so it is easy to identify miRNA target genes by computational predictions. For miR156, miR160, miR169, miR171, miR172, miR395, miR397, miR398, miR399, miR408, miR775, miR780.1, miR827, miR842, miR846, miR857, and miR2111, their targets have been predicted and most of them were validated previously (Table 2). To identify candidate miRNA target genes of the other N-starvation-responsive miRNAs, the target prediction software package, WMD3 [20], was used. To improve the reliability of predicted targets, three mismatches or less for miRNA-target duplex was set. The potential targets were listed in the Table 2. No candidate targets were predicted for five miRNAs (miR829.1, miR839, miR850, miR851, and miR861). Although miR158 has been predicted to target a transcript encoding a pentatricopeptide (PPR) repeat-containing protein [21], we predicted two new targets of miR158, At2g03210 and At2g03220, both of which encode fucosyltransferase proteins.
Table 2. N-starvation-responsive miRNAs and their targets.
| Class | Families | Members | Target gene families | Target genes | Potential roles | Other nutrient stimuli | References |
| NSS | miR158 | a | Pentatricopeptide repeat (PPR) | At1g64100 | [21], [22] | ||
| Fucosyltransferase | At2g03210 | Glycosylation | |||||
| At2g03220 | |||||||
| miR169 | a–c,h–n | CAAT binding factor | At1g17590 | Nitrogen homeostasis | -P | [12], [17], [25], [61] | |
| At1g54160 | |||||||
| At1g72830+ | |||||||
| At3g05690+ | |||||||
| At3g20910 | |||||||
| At5g06510+ | |||||||
| At5g12840+ | |||||||
| miR172 | a,b | AP2 transcription factor | At5g60120 | Flower development | [58]–[60] | ||
| At4g36920 | |||||||
| At2g28550 | |||||||
| At2g28550 | |||||||
| At5g67180 | |||||||
| miR395 | a–f | ATP sulfurylase | At3g22890+ | Sulfate homeostasis | -S -P | [10], [12], [57] | |
| Sulfate transporter | At4g14680 | ||||||
| At5g43780+ | |||||||
| At5g10180 | |||||||
| miR397 | a,b | Laccase | At2g29130+ | Copper homeostasis | -Cu | [15], [56] | |
| At2g38080+ | |||||||
| At5g60020+ | |||||||
| miR398 | b,c | Cu/Zn superoxide dismutase; | At1g08830+ | Copper homeostasis | -Cu -P -K | [12], [15], [56] | |
| Cytochrome oxidase c; | At2g28190+ | ||||||
| Copper chaperone | At3g15640 | ||||||
| At1g12520 | |||||||
| miR399 | a–f | Ubiquitin conjugase E2 | At2g33770+ | Phosphate homeostasis | -P | [11] | |
| miR408 | Laccase; Plantacyanin | At2g30210+ | Copper homeostasis | -Cu | [15] | ||
| At5g05390− | |||||||
| At5g07130+ | |||||||
| At2g02850+ | |||||||
| miR775 | Fucosyltransferase | At1g53290 | Glycosylation | [35] | |||
| miR783 | Protein of unknown | At4g01090 | Unknown | ||||
| miR825 | * | * | * | ||||
| miR827 | E3 ligase with RING and SPX | At1g02860+ | Nitrogen/Phosphorus metabolism | -P | [12], [27] | ||
| miR841 | Histone H2A | At2g38810 | Unknown | [55] | |||
| At4g13570 | |||||||
| miR849 | CXC domain-containing protein | At4g29000 | Unknown | ||||
| miR850 | * | * | * | ||||
| miR851 | 3p | * | * | * | |||
| miR854 | a–d | Oligouridylate binding protein1b | At1g17370 | Transcription regulation | [62] | ||
| miR857 | Laccase | At3g09220+ | Copper homeostasis | -Cu | [15] | ||
| miR861 | 5p | * | * | * | |||
| miR863 | 3p | Transducin/WD40 repeat-like other RNA | At2g40360+ | rRNA process | [23], [25] | ||
| At4g13495 | Unknown | ||||||
| miR1886.3 | Dentin sialophosphoprotein-related | At5g07970 | Unknown | ||||
| miR1888 | Haloacid dehalogenase-like hydrolase | At5g65140 | Trehalose biosynthesis | [22] | |||
| SAUR-like auxin-responsive protein | At2g16580 | Response to auxin stimulus | |||||
| miR2111 | a,b | Kelch repeat-containing F-box | At3g27150 | Phosphate metabolism | -P | [12], [13] | |
| NSI | miR156 | h | SPL transcription factors | At1g53160 | Vegetative phase change | -P -K | [12], [21], [54] |
| At2g33810 | |||||||
| At3g15270 | |||||||
| At5g43270 | |||||||
| At1g27360 | |||||||
| At1g27370 | |||||||
| At1g69170 | |||||||
| At2g42200 | |||||||
| At3g57920 | |||||||
| At5g50570 | |||||||
| miR160 | a–c | Auxin response factors | At2g28350− | Root/Flower development | [21], [43] | ||
| At4g30080− | |||||||
| At1g77850− | |||||||
| miR171 | b,c | SCL transcription factors | At2g45160− | Root development | [45] | ||
| At3g60630− | |||||||
| At4g00150− | |||||||
| miR780.1 | Na+/H+ antiporter family | At5g41610 | Sodium ion export | [35] | |||
| At4g33260 | |||||||
| miR826 | Alkenyl hydroxalkyl producing 2 | At4g03060− | Glucosinolate synthesis | [22], [55] | |||
| miR829.1 | * | * | * | ||||
| miR829.2 | AP2 domain ethylene response | At5g18560 | Root development | [52], [55] | |||
| miR839 | * | * | * | ||||
| miR842 | Jacalin lectin family protein | At1g60130 | Unknown | [22], [55] | |||
| At1g57570 | |||||||
| At5g38550 | |||||||
| miR846 | Jacalin lectin family protein | At5g49850 | Unknown | [55] | |||
| At2g25980 | |||||||
| At5g49870 |
Boldface letters indicate the previously validated targets. “*”indicates that no target was predicted. “+” and “−” indicate that up-regulation and down-regulation by N-starvation.
To establish the targets of one miRNA, it is necessary to find the corresponding cleaved products of its target transcripts. Recently, many miRNA targets were identified in rice and A. thaliana by degradome sequencing technology [22]–[24]. To confirm the predicted targets, the released A. thaliana degradome dataset was searched for potential cleavage sites of target transcripts. The new target of miR158, At2g03220, which encodes a fucosyltransferase, was cleaved in the miR158 recognition site. We found that miR775 was predicted to target genes coding fucosyltransferases responsible for transferring fucose groups. N starvation can leads to hexose accumulation in Arabidopsis [25]. Fucosyl residues have a role in the symbiotic interaction between legumes and Rhizobium species. Therefore, miR158 and miR775 may be involved in the symbiotic interaction in N limitation. In addition, the target cleavage products of miR826, miR863 and miR1888 were also identified (Table 2). However, the roles of these targets in N starvation cannot be predicted.
Many NSS miRNAs are involved in P, S, and Cu homeostasis
Several miRNAs have been identified to regulate nutrient metabolism [10], [11], [15], [17], [26]. We identified 23 NSS miRNAs, 9 of which directly take part in nutrient homeostasis (Table 2). Three miRNAs (miR399, miR827, and miR2111) were identified to function in P homeostasis. miR399 is specifically induced by P starvation and promotes phosphate uptake by cleavage of PHO2 transcripts. pho2 mutant plants showed phosphate-toxic phenotypes similar to miR399 over-expressing plants [11]. P starvation also simultaneously induced miR827 and miR2111 [12], [13], both of which were also clearly repressed by N limitation (Figure 3; Table 2). Very recently, miR827 was identified to function in phosphate absorption by targeting NITROGEN LIMITATION ADAPTATION (NLA) under N starvation [27]. When plants were subjected to N-deficient and P-sufficient conditions, nla mutant plants accumulated more phosphate than wild-type plants, displayed phosphate-toxic symptoms, and showed a decrease in anthocyanin synthesis. We revealed that both miR399 and miR827 were expressed at low levels under N-starvation conditions, with concomitant increase in expression of their targets, PHO2 and NLA (Figure 3). This may be necessary for plants to prevent over-uptake of phosphate. miR2111 was confirmed to be induced by P starvation and to target a gene encoding a Kelch domain-containing F-box protein [12]. However, it is still unclear why miR2111 is suppressed by N starvation, but induced by P-starvation.
Figure 3. Expression of targets of different nutrient responsive miRNAs under N starvation conditions.
Gene expression values shown are relative to the expression in plants grown under normal MS medium, for which the value is set to 1. Error bars indicate ± SE obtained from three biological repeats. Values marked by an asterisk are significantly different from the corresponding control value with Student's t-test (p<0.01; n = 3).
Both N and S are essential for plant growth and development, and the two assimilatory pathways are very similar and well coordinated. A deficiency in one element always results in suppression of the other pathway [28], [29]. miR395, a sulfate starvation-inducible miRNA, regulates sulfate accumulation and allocation in Arabidopsis leaves [10]. The over-expression of miR395 led to over-accumulation of sulfate in Arabidopsis leaves. In the present study, the abundance of miR395 decreased dramatically under N-starvation conditions, with moderate increases of its targets, APS1 and APS4 (Figure 3). Therefore, N starvation can reduce the accumulation of sulfate in leaves by repressing miR395. These results provide a new insight into the regulatory network between N and S assimilation pathways.
In addition to P or S starvation-induced miRNAs, Cu (copper) starvation-induced miRNAs were also down-regulated under N-starvation conditions. When plants were deprived of Cu, the abundance of miR397, miR398, miR408, and miR857 increased significantly. The target genes of these four miRNAs encode three kinds of copper-rich proteins (copper/zinc superoxide dismutase (CSD), plantacyanin, and laccase). Under Cu-starvation conditions, the up-regulation of these miRNAs can reduce the synthesis of copper-rich proteins, which facilitates the release of Cu to meet the Cu demand of plants. Under N starvation conditions, these four Cu-starvation-induced miRNAs were expressed at low levels and their target genes at high levels (Figure 3), suggesting that the Cu-starvation signal was inhibited by N deficiency. Similar results were also obtained in N starved maize roots [18], [30].
When subjected to low nutrient conditions, plants can sense and transmit nutrient deficiency signals. However, it is unclear how plants perceive and transmit low-N signals. miRNAs have been proved to be important signal molecules. Recent sequence analysis of miRNAs from Brassica napus [31] revealed that miR395 and miR399 are abundant in the phloem under low-sulfate and low-phosphate conditions, respectively. For example, miR399 was induced under low-phosphate conditions, and moves from shoots to roots in Arabidopsis [32]. In B. napus, miR395 is translocated through graft unions from scions to rootstocks under S-starvation conditions [33]. When exposed to Cu deficiency stress, rice and Arabidopsis over-accumulate miR397 and miR408 in the phloem. These miRNAs that accumulate in the phloem play roles in signal transduction. These nutrient deficiency-signaling molecules were significantly repressed by N starvation, which may decrease the amount of P, S, and Cu uptake. Deep sequencing analyses also suggested that miR169, miR395, and miR398 were expressed at low levels under P-deficient conditions [12]. Therefore, this miRNA-regulation mechanism is crucial for plants to maintain nutrient homeostasis and to adapt to nutrient deficiency stresses.
Putative functions of N-starvation-induced (NSI) miRNAs
Two non-homologous NSI-miRNAs, miR842 and miR846, are processed from the same polycistron [34] and potentially target several Jacalin Lectin family genes. 5′-RACE experiments revealed that two jacalin lectin genes are targets of miR842 and miR846, respectively [35]. However, the functions of these Jacalin Lectin genes during N starvation remain unclear. The miR829 precursor can produce two different miR829 species, both of which were induced by N starvation. The putative targets of miR829.1 cannot be predicted; miR829.2 is predicted to target PUCHI, encoding an AP2 domain ethylene response factor, which is required for morphogenesis in the early lateral root primordium of Arabidopsis [36]. Thus, miR829 might regulate Arabidopsis root development under N starvation. The miR156 family contains three different mature miRNAs, all of which were up-regulated under N-deficient conditions. The fold-change of miR156h was the greatest, suggesting that there may be spatial and/or temporal differences in the functions of different miR156 members. In contrast, miR172 was repressed by N starvation (Table 2), which is consistency with the fact that miR156 negatively regulated the expression of miR172. miR156 and miR172 can prolong and promote the expression of juvenile vegetative traits in Arabidopsis, respectively [37], implying that N-starvation delays the transition of Arabidopsis from the vegetative to the reproductive phase by modulating the their abundance. miR780 targets a gene encoding a protein with sodium/hydrogen antiporter activity. The strong induction of miR780 could be responsible for the regulation of pH.
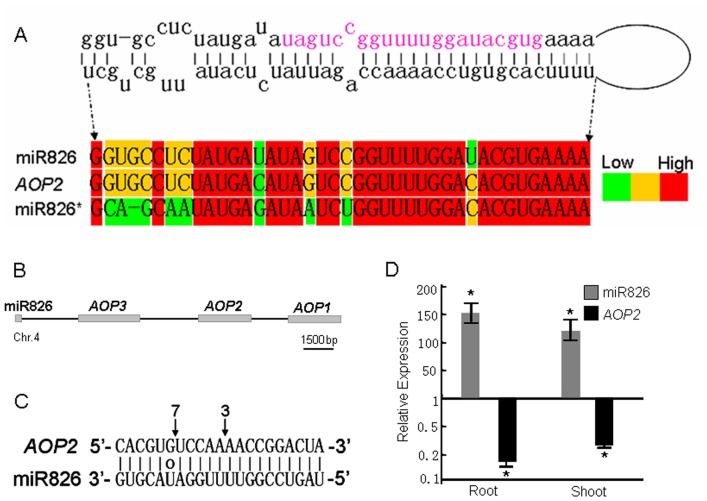
The abundance of miR826 significantly increased under N deficiency (Figure 1). miR826 was predicted to target the AOP2 gene, which encodes a 2-oxoglutarate-dependent dioxygenase involved in glucosinolate biosynthesis. In Arabidopsis, miR826 is closely linked on the chromosome with its predicted target AOP2, as well as AOP1 and AOP3 (homologous genes of AOP2) (Figure 4B). Comparative genome analysis revealed that the miR826 precursor sequence is highly similar to that of the AOP2 gene (Figure 4A). It is likely that the miR826 gene originated from duplication of its target gene [38]. Corresponding AOP2 cleavage products matched with miR826 were also identified (Figure 4C). Interestingly, AOP2 encodes a truncated and null-function protein due to a 5-bp deletion in the AOP2 transcript of the Col ecotype [39]. The Cvi ecotype, which has normal AOP2 transcripts, was used for further expression analysis. Consistent with the sequencing data, miR826 was dramatically induced in roots and shoots by N starvation. Correspondingly, N starvation significantly repressed the expression of its target, AOP2 (Figure 4D). Glucosinolates are a group of plant secondary metabolites produced mainly in Brassicas and these compounds are rich in nitrogen and sulfur. Therefore, the suppression of AOP2 by miR826 could decrease the production of glucosinolates, which would decrease the demand for N. However, further investigation is required to clarify the functions of miR826 during N starvation.
Figure 4. miR826 is a N-starvation-induced miRNA.
(A)Comparative analysis of miR826 precursor and its target AOP2 sequences. The sequence in pink indicates mature miR826 sequence. (B) The position of miR826, AOP1, AOP2, and AOP3 genes in chromosome. (C) The cleavage sites of AOP2 transcripts. (D) Expression of miR826 and AOP2 in response to N starvation. Gene expression values shown are relative to the expression in plants grown under normal MS medium, for which the value is set to 1. Error bars indicate ± SE obtained from three biological repeats. Values marked by an asterisk are significantly different from the corresponding control value with Student's t-test (p<0.01; n = 3).
Are miRNAs involved in root plasticity under N starvation?
N deficiency affects the morphogenesis of plant roots and often results in a stronger root system, such as more lateral roots. However, the mechanism underlying root development in response to N starvation is unclear. Recent studies suggested that miRNAs are regulators of root development and architecture [40]. For example, when Arabidopsis was subjected to nitrate treatment, miR167 and miR393 were induced to modulate root development [41], [42].
In the sequencing results, the abundance of miR160 increased 6-fold under N-deficient conditions, compared with that under N-sufficient conditions. The expression of miR160a and its targets was determined under N-deficient conditions. With the induction of miR160a, its targets ARF16 and ARF17 were down-regulated under N deficiency (Figure 5B). Studies suggested that miR160 controls lateral root formation by mediating regulation of ARF16 [43]. To investigate whether the increased abundance of miR160 facilitates lateral root formation during N starvation, miR160a-overexpressing transgenic plants (35S::miR160a) were constructed. As expected, all miR160 over-expressing plants produced more lateral roots than wild type plants (Figure 5A). These results implied that N deficiency induced expression of miR160, which then mediated the cleavage of ARF16 and promoted lateral root production in Arabidopsis.
Figure 5. miR160, miR167, and miR171 are involved in development of root system under N starvation coditions.

(A) More lateral roots in 35S::miR160a plants than WT (wild-type) plants. (B) Expression of miR160, miR167, miR171, and their targets under N starvation conditions. Gene expression values shown are relative to the expression in plants grown under normal MS medium, for which the value is set to 1. Error bars indicate ± SE obtained from three biological repeats. Values marked by an asterisk are significantly different from the corresponding control value with Student's t-test (p<0.01; n = 3). (C) A putative work model for miRNA-mediated root growth under N starvation conditions.
miR167 takes part in the plasticity of roots by mediating the regulation of ARF8 in response to N treatment [41]. The sequencing data showed that miR167 was repressed under N-deficient conditions. As expected, the inhibition of ARF8 by miR167 was relieved (Figure 5B), which could promote lateral root outgrowth. In addition to the plasticity of lateral roots, miR160 and miR167 also play crucial roles in adventitious root development [44]. miR160 facilitates adventitious root outgrowth via repressing ARF17, whereas miR167 negatively regulates adventitious root initiation via cleavage of ARF6 and ARF8. Therefore, both the increased miR160 and decreased miR167 favor root growth under N-starvation conditions.
Recent research showed that miR171 decreased primary root elongation by cleaving three SCL6 genes [45], [46]. Under N-deficient conditions, the abundance of miR171 was three-fold higher than that under N-sufficient conditions. Quantitative RT-PCR analyses showed that the expression of miR171c was clearly up-regulated in the root under N starvation (Figure 5B). Correspondingly, the three targets of miR171 were down-regulated significantly by N starvation. It is very likely that induction of miR171 results in decreased expression of its targets (SCL6-II, SCL6-III, and SCL6-IV) (Figure 5B), which then suppresses the elongation of primary roots during N starvation in Arabidopsis.
miR160, miR167, and miR171 are involved in the signaling pathways triggering root system development. From these results, a conclusion was drawn that in response to N deficiency, plants may enhance their root systems by inducing expression of miR160 and decreasing those of miR167 and miR171 (Figure 5C).
Novel miRNAs and their targtets
In addition to the known miRNAs, 20 novel miRNAs were also identified based on standard annotation criteria [47], nine of which were responsive to N starvation (Table 3). These new miRNAs were found in both N-deficient and N-sufficient samples. The read number of most novel miRNAs was much lower than that of the conserved miRNAs. This is consistent with the conclusion that non-conserved miRNAs are usually expressed at lower levels and with a spatial and temporal pattern. Precursors of these novel miRNAs were identified and their putative secondary hairpin structures were predicted (Table S4). The corresponding miRNA* sequences of four novel miRNAs (miRN01, miRN03, miRN06, miRN14) were found in the sequencing data (Table S4).
Table 3. Novel miRNAs identified from deep sequencing data.
| miRNA | Sequence | Length | +N | −N | Fold(log2)(−N/+N) | Target | Target Annotation |
| miRN01 | TGAGAGAAGGAATTAGATTCA | 21 | 22 | 18 | −0.3 | At3g60830 | Unknown |
| At4g28760 | |||||||
| miRN02 | CTTGAGGAGGTGTATAGAGGTTA | 23 | 6 | 8 | 0.4 | - | |
| miRN03 | TTTTACTGCTACTTGTGTTCC | 21 | 9 | 13 | 0.5 | At3g09010 | Protein kinase |
| miRN04 | ATACTGAAGATGAAACTAGCT | 21 | 6 | 6 | 0 | At1g75280 | Isoflavone reductase |
| miRN05 | TAGTGTTTTTTATGGATCGTCTA | 23 | 12 | 28 | 1.2 | - | |
| miRN06 | AAAGATGCAGATCATATGTCC | 21 | 17 | 38 | 1.2 | - | |
| miRN07 | TGGCGAGGATGAATAATGCTAA | 22 | 28 | 153 | 2.4 | - | |
| miRN08 | TTCGGTTCGGTTCGGTTCGGTTA | 23 | 14 | 27 | 0.9 | At1g74930 | Chloroplast function |
| At1g67623 | |||||||
| At1g26520 | |||||||
| miRN09 | TTGGTAGTGGATAAGGGGGCA | 21 | 71 | 226 | 1.7 | At1g80740 | Chromomethylase 1 |
| miRN10 | TACAGAGTAGTCAAGCATGACC | 22 | 21 | 26 | 0.3 | - | |
| miRN11 | TTGTCGATGTTTTTTTTACGGTA | 23 | 13 | 13 | 0 | - | |
| miRN12 | TGGGGTATTGTTGGAGTTTATTA | 23 | 9 | 7 | −0.4 | - | |
| miRN13 | TTGACTGCATTAACTTGATCG | 21 | 24 | 9 | −1.4 | At1g26450 | Carbohydrate-binding protein |
| miRN14 | TGACATCCAGATAGAAGCTTT | 21 | 1145 | 82 | −3.8 | At3g59210 | F-box family protein |
| At1g58310 | |||||||
| At5g41840 | |||||||
| At3g59230 | |||||||
| At4g00320 | |||||||
| At3g59200 | |||||||
| miRN15 | AGACACGGAGAAATCGGGAGATC | 23 | 9 | 13 | 0.5 | - | |
| miRN16 | ACAGTGGTCATCTGGTGGGCT | 21 | 8 | 22 | 1.5 | - | |
| miRN17 | AAAGAATCGTTGTTCAAGCTA | 21 | 8 | 36 | 2.2 | - | |
| miRN18 | TTTGAATTTTTAGAGCATGTCCA | 23 | 12 | 11 | −0.1 | - | |
| miRN19 | TTAGTCGAATATGTTTTGGTTTA | 23 | 5 | 6 | 0.3 | - | |
| miRN20 | TACAAGGAGTCAAGCATGACC | 21 | 14 | 81 | 2.5 | At3g24610 | F-box family protein |
Boldface letters indicate that the predicted target cleavages were identified from Arabidopsis degradome data.
“-” Indicates that no target was predicted. The miRNAs with a relative change ratio greater than 2 were underlined.
To explore the functions of these novel miRNAs, we predicted their potential target genes. However, the targets of only eight novel miRNAs were predicted with no predicted targets for the remainder (Table 3). To confirm the prediction results, we identified target cleavage sites from degradome data [22], [23]. Target gene cleavage products of five miRNAs (miRN04, miRN08, miRN13, miRN14, and miRN20) were identified (Figure 6). miRN04 targets an isoflavone reductase encoding gene, but its abundance was not affected by N starvation. Out of six predicted target genes for miRN08, corresponding cleavage of three target genes were found. At1g26520 encodes a protein with unknown functions. The products of both At5g24020 and At5g24120 function in chloroplast. At1g26450 is the potential targets of miRN13, which encode proteins located in membrane to bind carbohydrate. It is possible that the decrease of miRN13 facilitate energy supply of cells during N starvation. Both miRN14 and miRN20 target F-box protein encoding genes. P-starvation-induced miRNAs (miR399, miR827, and miR2111) are involved in ubiquitin-mediated protein degradation [11]–[13], [27] and all of them were repressed greatly by N starvation. Notably, the abundance of miRN14 decreased in response to N starvation, and its target genes also were involved in ubiquitin-mediated protein degradation. Therefore, posttranslational regulation of protein abundance may be crucial for the adaptation of plants to N starvation environment. However, further investigation is required to understand the functions of these novel miRNAs under N starvation conditions.
Figure 6. Identified targets of novel miRNAs from Arabidopsis degradome data.
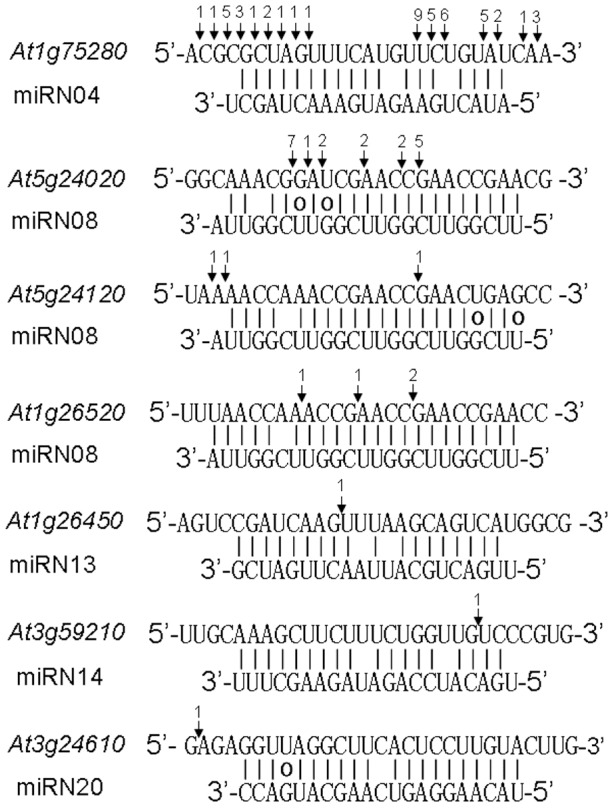
Vertical arrows indicate the target cleavage positions. The number indicates the number of corresponding cleavage products.
Conclusion
Recently, several miRNAs were identified to be responsive to N limitation in Arabidopsis, which includes miR156, miR167, miR169, and miR398 [13], [17]. However, it is unclear how other plant miRNAs respond to N starvation. This study presented an extensive survey of the miRNAs showing differential expression in response to N starvation in Arabidopsis. These N-responsive miRNAs and their target genes are likely involved in the development or regulation of the adaptive response to N starvation. The identification of these N-starvation-responsive miRNAs provides new insights into the molecular mechanism of adaptation of plants to N deficiency.
Nutrient balance and homeostasis are crucial for plant growth and development [48]. The deficiency of one mineral element often affects absorption of other mineral elements. P, S, Cu-starvation-induced miRNAs were suppressed by N starvation, suggesting that these miRNAs may mediate the crosstalk among N, P, S, and Cu under N-starvation conditions. The mobility of these miRNAs means that they are candidate molecules for nutrient-deprivation signal transduction [13], [32]. Their up-regulation or down-regulation coordinates and balances the demands of different nutrients. Homeostasis of N in growing plants requires a sustained uptake of N into root cells. In most situations, adjustment of N acquisition by the roots according to the nutrient demands of the plant is hampered by the limiting and fluctuating availability of N in soil. Higher plants modulate their root uptake capacity to compensate for changes in external N concentrations. Our results suggested that changes in the expression levels of miR160, miR167, and miR171 may be important for the enhancement of plant root system under nitrogen deficiency.
Previously, most researches on nitrogen sensing focused on metabolism and morphological responses to the addition of nitrate [49]–[52]. Recently, experiments that focus on the switch from N-sufficient to -deficient conditions have received more attention [18], [25], [30]. Our work will help us to further understand the responses and signal transduction pathways for N starvation.
Materials and Methods
Plant materials and growth conditions
Arabidopsis (A. thaliana ecotype Columbia) wild-type plants were used to construct small RNAs libraries. Seeds were surface-sterilized with 20% (v/v) bleach and washed three times with distilled water before plating on MS medium (containing 0.8% agar). Three days after vernalization, plates with seeds were moved to a growth cabinet. For N-sufficient (+N) conditions, plants were grown on MS medium; for N-deficient (−N) conditions, plants were grown on modified MS medium (without NH4NO3 and with KCl instead of KNO3). For P-deficient (−P) conditions, plants were grown on modified MS medium in which PO3 3− was replaced by Cl−; for S-deficient (−S) conditions, plants were grown on modified MS medium in which SO4 2− was replaced by Cl−. Ten-day-old seedlings were used for RNA extraction.
RNA preparation and small RNA sequencing
The whole seedlings grown in +N or −N medium were sampled to generate small RNA libraries. Total RNA was extracted using Trizol reagent (Invitrogen). Low molecular weight RNA was isolated from 200 µg total mRNAs by PEG8000/NaCl precipitation. Small RNAs in the size range of 20 to 30 nucleotides were purified from 15% denaturing polyacrylamide gels and ligated first with the 5′ RNA adaptor and then with the 3′ RNA adaptor. At each step, the ligated products were purified by electrophoretic separation on polyacrylamide gels. After first-strand synthesis and 18 cycles of PCR amplification, the final bands were purified on PAGE gels and submitted for sequencing. Sequencing was performed at the Beijing Genomics Institute (BGI).
Computational analysis of sequencing data
The raw sequencing data were trimmed by removing adaptor sequences and mapped to the Arabidopsis genome (The Arabidopsis Information Resource (TAIR) release version 10; http://www.arabidopsis.org/). Reads perfectly matching those in the Arabidopsis genome, excluding those matching tRNAs, rRNAs, snRNA, and snoRNAs, were used for further analysis. Arabidopsis mature miRNAs and their precursors were retrieved from miRBase (version 14; http://www.mirbase.org).
miRNA identification and target prediction
New miRNAs were identified as described by Fahlgren et al. [35]. WMD3 software package was used for the target prediction of miRNAs. The degradome data were from experiments described in Addo-Quaye et al. [23] and German et al. [22] (http://www.ncbi.nlm.nih.gov/geo/; GEO accession number GSE11007 and GSE11093).
Generation of transgenic plants
The miR160a precursor sequence was inserted into the pOCA30 binary vector. The binary vector was then transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis was transformed by the floral-dip method. Transgenic plants were selected on MS medium supplemented with 50 µg/mL kanamycin.
Gene expression analysis
Root samples were harvested separately from Arabidopsis and frozen in liquid nitrogen for storage at −80°C. Total RNA was isolated by Trizol reagent (Invitrogen) and digested by DNaseI (Fermentas).
Expression of miRNAs was detected by stem-loop RT-PCR. To produce miRNA-fused stem-loop cDNA, 0.5 µg total RNA was used for the reverse transcription with miRNA mature-sequence-specific stem-loop RT primers according to the stem-loop RT-PCR protocol [53]. For mRNA cDNA, 0.5 µg total RNA was reverse-transcribed using oligo(dT)18 primer according to the reverse transcription protocol (Fermentas). A 20-µl reaction mixture was used for the production of cDNA. After heat inactivation, a 1-µl aliquot was used as a template for real-time quantitative RT-PCR. An miRNA-specific primer and a universal primer were used to amplify miRNA-fused cDNA. Two specific primers were used to amplify each miRNA target gene. All primers used in this study are listed in Table S3. Arabidopsis ACT2 (At3g18780) was used as an internal control for real-time RT-PCR. All quantitative RT-PCR analyses were performed using a SYBR Premix Ex Taq™ kit (TaKaRa) on a Roche LightCycler 480 real-time PCR machine, according to the manufacturer's instructions.
Supporting Information
Summary of deep sequencing data.
(DOC)
Summary of miRNAs.
(DOC)
Normalized abundances of miRNAs.
(DOC)
Primers used in this study.
(DOC)
Putative secondary structure of novel miRNA precursors.
(TXT)
Funding Statement
This work was supported by the National Natural Science Foundation of China (90817003) and the Science Foundation of the Ministry of Agriculture of the People's Republic of China (2009ZX08009-066B). http://www.nsfc.gov.cn. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marschner H (1995) Mineral Nutrition of Higher Plants. London: Academic press.
- 2. Voinnet O (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687. [DOI] [PubMed] [Google Scholar]
- 3. Dong Z, Han MH, Fedoroff N (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu B, Yang Z, Li J, Minakhina M, Yang M, et al. (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Gene Dev 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336–338. [DOI] [PubMed] [Google Scholar]
- 7. Bartel DP (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 218–297. [DOI] [PubMed] [Google Scholar]
- 8. Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, et al. (2005) Cloning and characterizing of micro-RNAs from moss. Plant J 43: 837–848. [DOI] [PubMed] [Google Scholar]
- 9. Molnar A, Schwach F, Studholme DJ, Thuenemann E, Baulcombe D (2007) miRNAs control gene expression in single cell alga Chlamydomonas reinhardtii . Nature 447: 1126–1129. [DOI] [PubMed] [Google Scholar]
- 10. Liang G, Yang FX, Yu DQ (2010) MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana . Plant J 62: 1046–1057. [DOI] [PubMed] [Google Scholar]
- 11. Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, et al. (2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis . Plant Cell 18: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsieh LC, Lin SI, Shih ACC, Chen JW, Lin WY, et al. (2009) Uncovering small RNA-Mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol 151: 2120–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pant BD, Musialak-lange M, Nuc Pr, May P, Buhtz A, et al. (2009) Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol 150: 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamasaki H, Abdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, et al. (2007) Regulation of copper homeostasis by micro-RNA in Arabidopsis . J Biol Chem 282: 16369–16378. [DOI] [PubMed] [Google Scholar]
- 15. Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis . J Biol Chem 283: 15932–15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Combier JP, Frugier F, de Billy F, Booalem A, El-Yahyoui F, et al. (2006) MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Gene Dev 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao M, Ding H, Zhu JK, Zhang F, Li WX (2011) Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis . New Phytol 190: 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao M, Tai H, Sun S, Zhang F, Xu Y, et al. (2012) Cloning and characterization of maize miRNA involved in response to nitrogen deficiency. Plos ONE 7 1 e29669 doi:10.1371/journal.pone.0029669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li X, Wang X, Zhang S, Liu D, Duan Y, et al. (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 7 6 e39650 doi:10.1371/journal.pone.0039650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossowski S, Fitz J, Schwab R, Riester M, Weigel D (2010) personal communication.
- 21. Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, et al. (2002) Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- 22. German MA, Pillay M, Jeong DH, Hetawal A, Luo S, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotech 8: 941–946. [DOI] [PubMed] [Google Scholar]
- 23. Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y, Zheng Y, Addo-Quaye C, Zhang L, Saini A, et al. (2010) Transcriptome-wide identification of microRNA targets in rice. Plant J 62: 742–759. [DOI] [PubMed] [Google Scholar]
- 25. Krapp A, Berthome R, Orsel M, Mercey-Boutet S, Yu A, et al. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sunkar R, Kapoor A, Zhu JK (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18: 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kant S, Peng M, Rothstein SJ (2011) Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis . Plos Genet 7 3 e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reuveny Z, Dougall DK, Trinity PM (1980) Regulatory coupling of nitrate and sulfate assimilation pathways in cultured tobacco cells. Proc Natl Acad Sci USA 77: 6670–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neuenschwander U, Suter M, Brunold C (1991) Regulation of sulfate assimilation by light and O-acetyl-L-serine in Lemna minor L. Plant Physiol 97: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trevisan S, Nonis A, Begheldo M, Manoli A, Palme K, et al. (2012) Expression and tissue-specific localization of nitrate-responsive miRNAs in roots of maize seedlings. Plant Cell Environ 35: 1137–1155. [DOI] [PubMed] [Google Scholar]
- 31. Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus . Plant J 53: 739–749. [DOI] [PubMed] [Google Scholar]
- 32. Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J 53: 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buhtz A, Pieritz J, Springer F, Kehr J (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol10: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Merchan F, Boualem A, Crespi M, Frugier F (2009) Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol 10: R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirota A, Kato T, Fukaki H, Aida M, Tasaka M (2007) The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis . Plant Cell 19: 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu G, Park MY, Conway SR, Wang J, Weigel D, et al. (2009) The sequential action of miR156 and miR172 regulates development timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, et al. (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290. [DOI] [PubMed] [Google Scholar]
- 39. Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchel-Olds T (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis . Plant Cell 13: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khan GA, Declerck M, Sorin C, Hartmann C, Crespi M, et al. (2011) MicroRNAs as regulators of root development and architecture. Plant Mol Biol 77: 47–58. [DOI] [PubMed] [Google Scholar]
- 41. Gifford ML, Dean A, Gutieerez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vidal EA, Araus V, Lu C, Parry G, Coruzzi GM, et al. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana . Proc Natl Acad Sci USA 107: 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, et al. (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis . Plant cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gutierrez L, Bussell JD, Pacurar DI, Schwambach J, Pacurar M, et al. (2009) Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of auxin response factor transcripts and microRNA abundance. Plant Cell 21: 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- 46. Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ (2010) MicroRNA171c-Targeted SCL6-II, SCL6-III, and SCL6-IV Genes Regulate Shoot Branching in Arabidopsis . Mol Plant 3: 794–806. [DOI] [PubMed] [Google Scholar]
- 47. Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, et al. (2008) Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schachtman DP, Shin R (2007) Nutrient sensing and signaling: NPKS. Annu Rev Plant Biol 58: 47–69. [DOI] [PubMed] [Google Scholar]
- 49. Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224. [DOI] [PubMed] [Google Scholar]
- 52. Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, et al. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Varkonyi-Gasi E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, et al. (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49: 683–693. [DOI] [PubMed] [Google Scholar]
- 55. Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2007) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Gen Dev 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol Cell14: 787–799. [DOI] [PubMed] [Google Scholar]
- 57. Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- 58. Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, et al. (2003) P1/HC-Pro, a viral suppressor of RNA silencing interferes with Arabidopsis development and miRNA function. Dev Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- 60. Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li WX, Oono Y, Zhu J, He XJ, Wu JM, et al. (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20: 2238–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arteaga-Vazquez M, Caballero-Perez J, Vielle-Calzada JP (2006) A family of microRNAs present in plants and animals. Plant Cell 18: 3355–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of deep sequencing data.
(DOC)
Summary of miRNAs.
(DOC)
Normalized abundances of miRNAs.
(DOC)
Primers used in this study.
(DOC)
Putative secondary structure of novel miRNA precursors.
(TXT)