Abstract
Objective
To perform long QT syndrome and catecholaminergic polymorphic ventricular tachycardia cardiac channel postmortem genetic testing (molecular autopsy) for a large cohort of cases of autopsy-negative sudden unexplained death (SUD).
Methods
From September 1, 1998, through October 31, 2010, 173 cases of SUD (106 males; mean ± SD age, 18.4±12.9 years; age range, 1-69 years; 89% white) were referred by medical examiners or coroners for a cardiac channel molecular autopsy. Using polymerase chain reaction, denaturing high-performance liquid chromatography, and DNA sequencing, a comprehensive mutational analysis of the long QT syndrome susceptibility genes (KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2) and a targeted analysis of the catecholaminergic polymorphic ventricular tachycardia type 1–associated gene (RYR2) were conducted.
Results
Overall, 45 putative pathogenic mutations absent in 400 to 700 controls were identified in 45 autopsy-negative SUD cases (26.0%). Females had a higher yield (26/67 [38.8%]) than males (19/106 [17.9%]; P<.005). Among SUD cases with exercise-induced death, the yield trended higher among the 1- to 10-year-olds (8/12 [66.7%]) compared with the 11- to 20-year-olds (4/27 [14.8%]; P=.002). In contrast, for those who died during a period of sleep, the 11- to 20-year-olds had a higher yield (9/25 [36.0%]) than the 1- to 10-year-olds (1/24 [4.2%]; P=.01).
Conclusion
Cardiac channel molecular autopsy should be considered in the evaluation of autopsy-negative SUD. Several interesting genotype-phenotype observations may provide insight into the expected yields of postmortem genetic testing for SUD and assist in selecting cases with the greatest potential for mutation discovery and directing genetic testing efforts.
Abbreviations and Acronyms: CPVT, catecholaminergic polymorphic ventricular tachycardia; LQTS, long QT syndrome; SCD, sudden cardiac death; SUD, sudden unexplained death; SUDEP, sudden unexplained death in epilepsy
Sudden cardiac death (SCD) is a major cause of death in developed countries. An estimated 300,000 to 400,000 individuals die suddenly each year in the United States, with most deaths involving elderly people.1 In comparison, sudden death in infants, children, adolescents, and young adults is relatively uncommon, with an incidence between 1.3 and 8.5 per 100,000 patient-years.2 Nevertheless, an estimated 1000 to 5000 individuals between 1 and 35 years of age die suddenly each year. Fortunately, the cause and manner of death can be explained in many cases by a comprehensive medicolegal investigation that includes an autopsy.3,4
A conventional autopsy investigation may detect a noncardiac basis for the sudden death, such as pulmonary embolism, asthma, or epilepsy. However, SCD is the most common cause of sudden death in young people, with structural cardiovascular abnormalities often identifiable at autopsy.4,5 Nevertheless, standard forensic autopsy investigations often fail to reveal the underlying cause of the SCD. In fact, even after gross and histologic examination, at least 3% and perhaps as much as 53% of sudden deaths involving previously healthy children, adolescents, and young adults have no identifiable morphological abnormalities found at autopsy, remain unexplained, and are classified as autopsy-negative sudden unexplained death (SUD).3,4,6-8
Long QT syndrome (LQTS) and catecholaminergic polymorphic ventricular tachycardia (CPVT) are potentially lethal, heritable channelopathies associated with structurally normal hearts that leave no evidentiary clue to be gleaned during a comprehensive medicolegal autopsy. This absence of evidence leaves medical examiners, coroners, and forensic pathologists to only surmise that a lethal arrhythmia may have precipitated the SUD.3,9-13 However, postmortem genetic testing, specifically a cardiac channel molecular autopsy, may potentially elucidate such a pathogenic mechanism and establish probable cause and manner for an individual's death.14-17 In fact, recent guidelines for autopsy investigations of SUD in young people have suggested that postmortem cardiac channel genetic testing should become the new standard of care in the evaluation of SUD cases.18-20
Since the availability of preliminary case reports of molecular autopsies,15 investigators have sought to determine the spectrum and prevalence of pathogenic cardiac ion channel mutations in the now 9 unique molecular autopsy series of SUD cases, providing a range of 15% to 35% mutation detection yield when testing for the 3 major LQTS susceptibility genes (KCNQ1, KCNH2, SCN5A) and/or the major CPVT-associated gene (RYR2).17,21-28 However, these 9 molecular autopsy series, now totaling only 207 cases (49 of which we have reported on previously), mostly represent very small case series (<20 cases), used a largely unreliable source (formalin-fixed paraffin-embedded tissue) of high-quality DNA for comprehensive mutational analysis, or performed a limited mutational analysis (ie, only assessed a partial list of the major LQTS and CPVT genes).
Given the recent guidelines for autopsy-negative SUD investigations and the relatively small cohort size of these previously published molecular autopsy series, an extensive analysis to better define the expected yield of mutation detection and perhaps offer possible genotype-phenotype correlations that may assist in guiding phenotype-directed cost-effective mutation detection efforts in future cases of SUD is much needed. We report the spectrum and prevalence of possible LQTS- and CPVT-associated channel mutations among 173 consecutive cases of SUD that were referred by medical examiners, coroners, and forensic pathologists from throughout North America during the past decade for a cardiac channel molecular autopsy. In an effort to guide and inform the proper use of such postmortem genetic testing, we examined the effect of different phenotypic parameters on the yield of LQTS and CPVT genetic testing from the largest molecular autopsy series on SUD published to date.
Methods
Medical Examiner–Referred Cases of Autopsy-Negative SUD
From September 1, 1998, through October, 31, 2010, 173 medical examiners' cases of autopsy-negative SUD (106 males; mean ± SD age, 18.4±12.9 years; range, 1-69 years) were referred to Mayo Clinic's Windland Smith Rice Sudden Death Genomics Laboratory for a cardiac channel molecular autopsy. After receipt of written consent from the decedent's appropriate next of kin for this Mayo Foundation Institutional Review Board–approved protocol, a postmortem genetic mutational analysis (ie, cardiac channel molecular autopsy) was performed. The first 49 of these 173 consecutively referred cases have been reported previously22,29 and are included here in this extended molecular autopsy series. None of the decedents or their family members had an established diagnosis of LQTS, CPVT, or any other sudden death–predisposing cardiac condition at the time of either their death or at enrollment for postmortem genetic testing.
Cardiac Channel Molecular Autopsy
Genomic DNA was extracted from frozen necropsy tissue or autopsy blood using the Puregene DNA Isolation Kit (Qiagen Inc, Valencia, CA). Comprehensive coding, open reading frame, splice-site mutational analysis of the entire coding region (60 exons) of the 3 major LQTS susceptibility genes (KCNQ1, KCNH2, and SCN5A), 2 minor LQTS susceptibility genes (KCNE1 and KCNE2), and a targeted molecular screen of the 64 exons (3-28, 36-50, and 83-105) previously reported to encompass the 3 major mutation-clustering domains of the 105 exon CPVT type 1 (CPTV1) susceptibility gene RYR230 was performed using polymerase chain reaction, denaturing high-performance liquid chromatography, and direct DNA sequencing as previously described.31
To be considered a putative, pathogenic, SUD-associated mutation, the genetic variant had to (1) be a nonsynonymous variant (synonymous single-nucleotide polymorphisms were excluded from consideration); (2) involve a highly conserved residue; and (3) be absent from either more than 2600 reference alleles previously examined for LQTS mutations or more than 800 reference alleles previously examined for RYR2 mutations32-34 and an additional 400 reference alleles derived from 100 healthy white and 100 healthy black controls. This latter control genomic DNA was obtained from the Human Genetic Cell Repository sponsored by the National Institute of General Medical Sciences and the Coriell Institute for Medical Research (Camden, NJ). Mutations were annotated using the single-letter nomenclature whereby W379R, for example, denotes a nonsynonymous single-nucleotide polymorphism producing a missense mutation involving a substitution of tryptophan (W) by arginine (R) at amino acid W379R. To assess whether mutations were sporadic or familial, direct DNA sequencing was used to confirm the presence or absence of the putative mutations in DNA extracted from the parents of the decedents whenever possible and after written informed consent. Primer sequences and polymerase chain reaction and denaturing high-performance liquid chromatography conditions are available on request.
Results
Cohort Description
Demographic characteristics for this consecutively referred SUD cohort are given in Table 1. Briefly, there were 173 cases (106 males and 67 females; mean ± SD age, 18.4±12.9 years; range, 1-69 years; 89% white). The mean ± SD ages of the males (18.2±13 years; range, 1-69 years) and females (18.8±13 years; range, 1-52 years) were similar. Most (161/173 [93.1%]) were 40 years or younger at their SUD. The circumstance (events or triggers) surrounding the SUD was grouped into 1 of 3 categories, with the most prevalent being death during sleep (n=70, 30 males [40.5%]), followed by a nonspecific event (n=52, 27 males [30.1%]) and exertion (n=46, 30 males [26.6%]). In addition, 1 individual (0.6%) experienced an auditory-triggered sudden death, and 4 (2.3%) died suddenly during extreme emotion. The distribution of triggers or events was not statistically different between males and females, respectively (sleep, 45.3% [48/106] vs 32.8% [22/67], P=.11; nonspecific, 25.5% [27/106] vs 37.3% [25/67], P=.12; exertion, 28.3% [30/106] vs 23.9% [16/67], P=.60).
TABLE 1.
Clinical Characteristics of the Autopsy-Negative SUD Cohorta
| Characteristic | All cases | LQTS-positive cases | CPVT1-positive cases | Negative cases |
|---|---|---|---|---|
| No. of individuals | 173 | 25 | 20 | 128 |
| Sex, No. | ||||
| Male | 106 | 8 | 11 | 87 |
| Female | 67 | 17 | 9 | 41 |
| Age at SUD (y), mean ± SD (range) | ||||
| Overall | 18.4±12.9 (1-69) | 17.6±9.4 (2-43) | 16.5±10.2 (2-36) | 18.9±13.9 (1-69) |
| Females | 18.8±13 (1-52) | 18.11±10.4 (4-43) | 18.9±11 (5-35) | 19.3±14 (1-52) |
| Males | 18.2±13 (1-69) | 16.6±7.2 (3-24) | 14.5±9.6 (2-36) | 18.8±14 (1-69) |
| Reported ethnicity, No. | ||||
| White | 154 | 24 | 16 | 113 |
| Black | 9 | 1 | 3 | 6 |
| Hispanic | 6 | 0 | 1 | 5 |
| Asian | 3 | 0 | 0 | 3 |
| Unknown | 1 | 0 | 0 | 1 |
| Events at SUD | ||||
| Sleep | 70 (40.5) | 12 (48.0) | 1 (5.0) | 57 (44.5) |
| Nonspecific | 52 (30.1) | 6 (24.0) | 8 (40.0) | 38 (29.7) |
| Exertion | 46 (26.6) | 5 (20.0) | 11 (55.0) | 30 (23.4) |
| Auditory | 1 (0.6) | 1 (4.0) | 0 | 0 |
| Emotion | 4 (2.3) | 1 (4.0) | 0 | 3 (2.3) |
| Personal history before SUD | ||||
| Positive | 42 (24.3) | 8 (32.0) | 7 (35.0) | 27 (21.1) |
| Negative | 124 (71.7) | 15 (60.0) | 13 (65.0) | 96 (75.0) |
| Unknown | 7 (4.0) | 2 (8.0) | 0 | 5 (3.9) |
| Family history of cardiac events | ||||
| Positive | 34 (19.7) | 5 (20.0) | 7 (35.0) | 22 (17.2) |
| Negative | 127 (73.4) | 19 (76.0) | 12 (60.0) | 96 (75.0) |
| Unknown | 12 (6.9) | 1 (4.0) | 1 (5.0) | 10 (7.8) |
| Family history of SCD | 20 (11.5) | 2 (8.0) | 7 (35.0) | 11 (8.6) |
| Personal or family history | ||||
| Seizures | 12 (6.9) | 1 (4.0) | 2 (10.0) | 9 (7.0) |
| Syncope | 25 (14.5) | 9 (36.0) | 4 (20.0) | 12 (9.4) |
CPVT1 = catecholaminergic polymorphic ventricular tachycardia type 1; LQTS = long QT syndrome; SUD = sudden unexplained death.
Data are presented as No. (percentage) unless indicated otherwise.
Importantly, no decedent or relative had received a clinical diagnosis of a suspected cardiac channelopathy or heritable arrhythmia syndrome (ie, LQTS or CPVT) before the SUD or at the time necropsy tissue was received to commence the molecular autopsy. However, either a positive personal or family history of syncope, seizures, cardiac arrest, near drowning or unexplained drowning (in a family member), or prolonged QT interval was documented explicitly by the medical examiner in 70 of the 173 cases (40.5%). A personal or family history of syncope (n=25 [14.4%], personal history in 20) or seizures (n=13 [7.5%], personal history in 11) was identified. A family history of premature SCD before 50 years of age, occurring before the current patient's SUD, was reported in 20 cases (11.6%). A personal or family history of a cardiac event occurring during the postpartum period was reported in 6 cases. Status concerning personal history was not available in 7 cases (4.0%), and family history information was unavailable in 12 cases (6.9%).
Spectrum and Prevalence of Rare, Putatively Channelopathic Mutations in SUD
Overall, 45 rare, putative pathogenic mutations (20 RYR2 [44.5%, 14 novel], 11 KCNQ1 [24.5%, 4 novel], 6 KCNH2 [13.3%, 5 novel], 6 SCN5A [13.3%, 2 novel], and 2 KCNE2 [4.4%]) (Figures 1 and 2, and Table 2) were identified in 45 cases (26.0%) of autopsy-negative SUD. Most of the mutations (n=40) were missense, with only 5 (12.5%) radical mutations (4 frameshift [2 KCNQ1, 2 KCNH2] and 1 splicing error [KCNH2]). Two decedents hosted multiple mutations (case 16, L799sp-KCNH2, T1107M-RyR2; case 27, H240R-RyR2, T4158P-RyR2). Mutations involving LQTS-associated genes (KCNQ1, KCNH2, SCN5A, and KCNE2) were identified in 25 cases (14.5%) overall compared with 21 cases (12.1%) hosting mutations involving the CPVT1-associated RYR2 gene.
FIGURE 1.
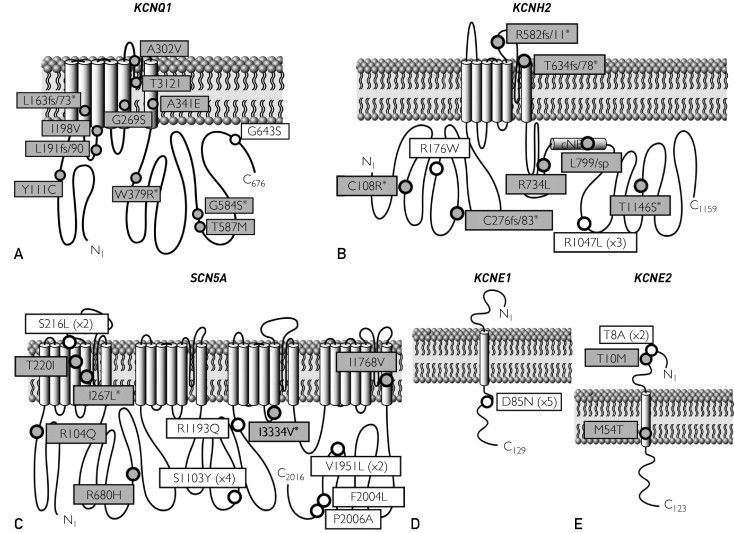
Summary of cardiac channel mutations in long QT syndrome (LQTS)–associated genes identified in a series of autopsy-negative sudden unexplained death (SUD). The putative, pathogenic SUD-associated mutations (gray circles) and additional nonsynonymous, functional polymorphisms (white circles) identified in this study are depicted with their proximate location on the linear topologies (not drawn to scale) of the LQT1-associated KCNQ1-encoded cardiac Kv7.1/IKs potassium channel α-subunit (A), the LQT2-associated KCNH2 encoded cardiac Kv11.1/IKr potassium channel α-subunit (B), the LQT3-associated SCN5A encoded cardiac Nav1.5/INa sodium channel α-subunit (C), the LQT5-associated KCNE1-encoded cardiac Kv7.1 potassium channel β-subunit (D), and the LQT6-assoicated KCNE2-encoded cardiac Kv11.1 potassium channel β-subunit (E). Asterisk indicates novel mutation absent in the published literature. The numbers within parentheses represent the number of times the variant was seen in cases. For example, R1047L (×3) is a functional polymorphism seen in 3 cases.
FIGURE 2.
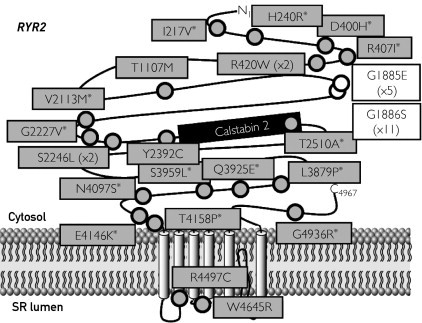
Summary of cardiac channel mutations in the catecholaminergic polymorphic ventricular tachycardia type 1 (CPVT1)–associated RYR2-encoded calcium release channel (cardiac ryanodine receptor) identified in a series of autopsy-negative sudden unexplained death (SUD). The putative, pathogenic SUD-associated mutations (gray circles) and nonsynonymous polymorphisms (white circles) identified in this study are depicted with their proximate location on the linear topologies (not drawn to scale) of the CPVT1-associated RYR2-encoded calcium release channel (cardiac ryanodine receptor) α-subunit. Asterisk indicates novel mutation. The numbers within parentheses represent the number of times the variant was seen in cases.
TABLE 2.
Summary of Putative Channelopathic Mutations and Channelopathy-Positive SUD Casesa
| Case No. | Sex | Age (y) | Ethnicity | Gene | Exon | Nucleotide change | Mutation (amino acid change) | Nonsynonymous polymorphisms | SUD event | Sentinel event | Family history |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 3 | W | KCNQ1 | 1 | 332 A>G | Y111C | Sleep | SUD | Positive for syncope | |
| 2 | F | 21 | W | KCNQ1 | 3 | del488T | L163FS/X236b | T8A-KCNE2 | Nonspecific | Syncopec | Negative |
| 3 | M | 17 | W | KCNQ1 | 3 | del572_576 | L191FS/90 | Sleep | SUD | Negative | |
| 4 | M | 20 | W | KCNQ1 | 3 | 592 A>G | I198V | Sleep | SUD | Negative | |
| 5 | F | 43 | W | KCNQ1 | 6 | 805 G>A | G269S | Exertion | SUD | Negative | |
| 6 | M | 22 | W | KCNQ1 | 6 | 905 C>T | A302V | G1886S-RYR2 | Sleep | Seizures | Negative |
| 7 | F | 8 | W | KCNQ1 | 7 | 935 C>T | T312I | Exertion | SUD | Positive for SCDc | |
| 8 | M | 24 | W | KCNQ1 | 7 | 1022 C>A | A341E | Sleep | SUD | Negative | |
| 9 | F | 17 | W | KCNQ1 | 9 | 1135 T>C | W379Rb | Emotion | SUD | Negative | |
| 10 | F | 5 | W | KCNQ1 | 15 | 1750 G>A | G584Sb | Exertion | Syncope | Negative | |
| 11 | F | 4 | W | KCNQ1 | 15 | 1760 C>T | T587M | Nonspecific | SUD | Negative | |
| 12 | F | 14 | W | KCNH2 | 3 | 322 T>C | C108Rb | V1951L-SCN5A | Nonspecific | SUD | Positive for SCD |
| 13 | F | 29 | W | KCNH2 | 4 | del823T | C276fsX359b | Nonspecific | SUDc | Positive for syncope | |
| 14 | F | 15 | W | KCNH2 | 7 | insGC1746_1747 | R582FS/11b | Sleep | Syncope | Negative | |
| 15 | F | 16 | W | KCNH2 | 7 | del1902C | T634FS/78b | Sleep | SUD | Negative | |
| 16 | F | 13 | W | KCNH2, RYR2 | 9, 28 | 2398+5 G>T, 3321 C>T | L799/SP, T1107M-RYR2 | Auditory | Syncope | Negative | |
| 17 | F | 15 | W | KCNH2 | 15 | 3436 A>T | T1146Sb | Sleep | Syncope | Negative | |
| 18 | F | 16 | W | SCN5A | 3 | 311 G>A | R104Q | Sleep | SUD | Negative | |
| 19 | F | 19 | W | SCN5A | 6 | 659 C>T | T220I | Nonspecific | SUD | Negative | |
| 20 | M | 12 | W | SCN5A | 7 | 799 A>C | I267Lb | Nonspecific | SUD | Negative | |
| 21 | M | 23 | B | SCN5A | 14 | 2039 G>A | R680H | S1103Y-SCN5A | Exertion | SUD | Negative |
| 22 | F | 39 | W | SCN5A | 23 | 4000 A>G | I3334Vb | Sleep | Syncope | Positive for syncope and CA | |
| 23 | F | 16 | W | SCN5A | 28 | 5302 A>G | I1768V | Sleep | SUD | Negative | |
| 24 | M | 12 | W | KCNE2 | 2 | 29 C>T | T10M | D85N-KCNE1, | Exertion | Syncope | Negative |
| 25 | F | 18 | W | KCNE2 | 2 | 161 T>C | M54T | Sleep | SUD | Negative | |
| 26 | M | 21 | W | RYR2 | 9 | 649 A>G | I217Va | G1885E-RYR2, G1886S-RYR2, | Nonspecific | Syncope | Positive for SCD |
| 27 | M | 2 | W | RYR2 | 10, 90 | 719 A>G, 12472 A>C | H240R,b T4158Pb | Exertion | SUD | Negative | |
| 28 | F | 35 | W | RYR2 | 14 | 1198 G>C | D400Hb | Nonspecific | SUD | Positive for SCD | |
| 29 | M | 9 | H | RYR2 | 14 | 1220 G>T | R407Ib | Nonspecific | Syncope | Negative | |
| 30 | M | 17 | W | RYR2 | 14 | 1258 C>T | R420W | Exertion | Syncope | Negative | |
| 31 | F | 22 | W | RYR2 | 14 | 1258 C>T | R420W | Nonspecific | SUD | Positive for SCD | |
| 32 | M | 36 | W | RYR2 | 41 | 6337 G>A | V2113Mb | Exertion | SUD | Negative | |
| 33 | M | 16 | W | RYR2 | 43 | 6680 G>T | G2227Vb | Nonspecific | SUD | Negative | |
| 34 | M | 5 | W | RYR2 | 44 | 6737 C>T | S2246L | G643S-KCNQ1, D85N-KCNE1 | Exertion | SUD | Negative |
| 35 | M | 4 | B | RYR2 | 44 | 6739 C>T | S2246L | Exertion | SUD | Negative | |
| 36 | F | 25 | W | RYR2 | 47 | 7175 A>G | Y2392C | R1047L-KCNH2 | Exertion | SUD | Positive for SCD |
| 37 | F | 17 | W | RYR2 | 50 | 7528 A>G | T2510Ab | Nonspecific | SUD | Negative | |
| 38 | M | 17 | W | RYR2 | 86 | 11636 T>C | L3879Pb | Exertion | Syncope | Negative | |
| 39 | F | 9 | W | RYR2 | 87 | 11773 C>G | Q3925Eb | Exertion | Seizures | Negative | |
| 40 | F | 15 | W | RYR2 | 88 | 11876 C>T | S3959Lb | Exertion | SUD | Negative | |
| 41 | M | 18 | W | RYR2 | 90 | 12290 A>G | N4097Sb | Nonspecific | SUD | Positive for SCD | |
| 42 | M | 14 | W | RYR2 | 90 | 12436 G>A | E4146Kb | Sleep | SUD | Positive for SCD | |
| 43 | F | 34 | B | RYR2 | 93 | 13610 C>T | R4497C | Nonspecific | SUD | Positive for SCD | |
| 44 | F | 5 | W | RYR2 | 96 | 13933 T>A | W4645R | Exertion | SVT | Negative | |
| 45 | F | 8 | W | RYR2 | 104 | 14803 G>A | G4936Rb | Exertion | Seizures | Negative |
B = black; CA = cardiac arrest; F = female; H = Hispanic; M = male; SCD = sudden cardiac death; SUD = sudden unexplained death; SVT = sustained ventricular tachycardia; W = white.
Novel to this cohort.
Postpartum period.
Among the 11 mutation-positive SUDS cases (of 45) for which the families chose to participate in genetic pedigree analysis, the SUD-associated mutation was established as a familial mutation in every family (4 RYR2, 4 KCNQ1, 1 KCNH2, 1 SCN5A, and 1 KCNE2) despite having no family history of cardiac events in 10 of 11 cases (Table 2). One of the decedents had a family history of SCD in a paternal uncle. The SUD was the sentinel event in 8 of the 11 families. Among this small subset of participating families, 3 of the decedents had a personal history of syncope.
Besides these aforementioned rare putative channelopathic mutations, several common nonsynonymous, functional polymorphisms, previously associated with LQTS, Brugada syndrome, drug-induced torsades de pointes, or ventricular arrhythmias or SCD, were also identified (Figure 1 and Tables 2 and 3), including G1886S-RyR235,36 (11 cases), G1885E-RyR236 (5 cases), D85N-KCNE137 (5 cases, all males), S1103Y-SCN5A38,39 (4 cases), R1047L-KCNH240 (3 cases), V1951L-SCN5A41 (2 cases), S216L-SCN5A41,42 (2 cases), T8A-KCNE243 (2 cases), R176W-KCNH244(1 case), G643S-KCNQ145 (1 case), R1193Q-SCN5A46,47 (1 case), F2004L-SCN5A41 (1 case), and P2006A-SCN5A41,48 (1 case). Common polymorphisms with a heterozygous frequency greater than 20%, including K897T-KCNH2, H558R-SCN5A, G38S-KCNE1, and Q2958R-RyR2, and less common polymorphisms lacking functional data to suggest proarrhythmic susceptibility (DUP 52-54 API-KCNQ1 [1 case], K393N-KCNQ1 [1 case], P448R-KCNQ1 [1 case], V648I-KCNQ1 [1 case], R148W-KCNH2 [1 case], R18W-SCN5A [1 case], S1787N-SCN5A [1 case], A1136V-RyR2 [3 cases], and R4037C-RyR2 [1 case]) have been excluded from Figure 1 and Tables 2 and 3.
TABLE 3.
Summary of Additional SUD Patients Hosting Only a Common Functional, Nonsynonymous Channel Polymorphisma
| Case No. | Sex | Age (y) | Ethnicity | Gene | Nonsynonymous polymorphism | SUD event | Sentinel event | Family history |
|---|---|---|---|---|---|---|---|---|
| 46 | M | 27 | W | KCNE1 | D85N-KCNE1 | Nonspecific | SUD | Positive for CA |
| 47 | M | 17 | W | KCNE1 | D85N-KCNE1 | Sleep | Syncope | Negative |
| 48 | M | 46 | W | KCNE1 | D85N-KCNE1 | Nonspecific | SUD | Negative |
| 49 | F | 43 | W | KCNE2 | T8A-KCNE2 | Sleep | SUD | Negative |
| 50 | F | 30 | W | KCNH2 | R176W-KCNH2 | Nonspecific | Syncopeb | Negative |
| 51 | M | 15 | W | KCNH2 | R1047L-KCNH2 | Sleep | SUD | Positive for SCD |
| 52 | M | 20 | W | KCNH2 | R1047L-KCNH2, S216L-SCN5A | Sleep | SUD | Negative |
| 53 | M | 21 | W | SCN5A | S216L-SCN5A | Sleep | SUD | Negative |
| 54 | M | 1 | B | SCN5A | S1103Y-SCN5A | Sleep | SUD | Negative |
| 55 | F | 2 | B | SCN5A | S1103Y-SCN5A | Sleep | SUD | Negative |
| 56 | M | 15 | H | SCN5A | S1103Y-SCN5A | Emotion | SUD | Negative |
| 57 | M | 19 | W | SCN5A | R1193Q-SCN5A | Sleep | Syncope | Negative |
| 58 | M | 18 | H | SCN5A | V1951L-SCN5A | Sleep | SUD | Negative |
| 59 | M | 21 | W | SCN5A | F2004L-SCN5A | Nonspecific | SUD | Negative |
| 60 | M | 1 | W | SCN5A | P2006A-SCN5A | Sleep | SUD | Negative |
| 61 | F | 16 | W | RYR2 | G1885E-RYR2 | Emotion | Seizures | Negative |
| 62 | M | 38 | W | RYR2 | G1885E-RYR2 | Sleep | SUD | Negative |
| 63 | M | 19 | B | RYR2 | G1885E-RYR2 | Nonspecific | SUD | Negative |
| 64 | M | 12 | W | RYR2 | G1885E-RYR2 | Exertion | Syncope | Positive for syncope |
| 65 | M | 2 | W | RYR2 | G1886S-RYR2 | Sleep | SUD | Negative |
| 66 | M | 12 | W | RYR2 | G1886S-RYR2 | Exertion | SUD | Positive for arrhythmias |
| 67 | F | 12 | W | RYR2 | G1886S-RYR2 | Exertion | Syncope | Negative |
| 68 | M | 18 | W | RYR2 | G1886S-RYR2 | Exertion | SUD | Positive for arrhythmias |
| 69 | M | 18 | W | RYR2 | G1886S-RYR2 | Nonspecific | SUD | Negative |
| 70 | F | 4 | W | RYR2 | G1886S-RYR2 | Exertion | Syncope | Negative |
| 71 | F | 8 | W | RYR2 | G1886S-RYR2 | Nonspecific | SUD | Negative |
| 72 | M | 30 | W | RYR2 | G1886S-RYR2 | Sleep | SUD | Negative |
| 73 | M | 12 | W | RYR2 | G1886S-RYR2 | Sleep | SUD | Negative |
B = black; CA = cardiac arrest; F = female; H = Hispanic; M = male; SCD = sudden cardiac death; SUD = sudden unexplained death; W = white.
Postpartum period.
Most of these polymorphisms do not appear to be overrepresented within this SUD cohort. However, the common proarrhythmic and sudden death–associated SCN5A polymorphism S1103Y38 with an expected heterozygote frequency of 13% among healthy black controls was present in 3 of 9 black (33.3%) and 1 of 6 Hispanic (16.7%) SUD patients. The drug-induced LQT- and arrhythmia-associated D85N-KCNE1 polymorphism was identified in 5 of 154 (3.2%, all male) white SUD patients compared with the expected prevalence of 1.0% in the general white population.33 Interestingly, although only 40.5% (70/173) of the overall cohort died during sleep, 77.8% (7/9) putative, pathogenic mutation–negative SUD patients, who were identified as having an SCN5A functional polymorphism (S216L, S1103Y, R1193Q, V1951L, F2004L, and P2006A), died during sleep (P=.04).
Importantly, although these proarrhythmic channel polymorphisms may have contributed to the patient's death, we excluded them from the calculations of SUD molecular autopsy yield and from the genotype-phenotype correlative studies detailed below. Accordingly, the 26.0% prevalence of channelopathy-mediated sudden death in this autopsy-negative SUD cohort could be underestimated.
Genotype-Phenotype Correlations
Effect of Sex and Age on Yield
Female SUD patients (26/67, 38.8%) were more likely to host a channel mutation than male patients (19/106, 17.9%; P=.004). Moreover, mutation-positive females were more likely to host an LQTS-associated mutation rather than a CPVT1-associated RYR2 mutation (65.4% [17/26] LQTS vs 34.6% [9/26] CPVT), whereas mutation-positive males were more likely CPVT1 positive (57.9% [11/19]) than LQTS mutation positive (42.1% [8/19], Figure 3). In fact, the distribution of genes hosting mutations was different between females and males (Figure 4). Specifically, although 57.9% (11/19) of the mutations seen in males were in RYR2, only 34.6% (9/26) of the mutations in females involved RYR2. In females, 23.1% (6/26) of the mutations identified involved KCNH2, whereas 0 of 19 male SUD patients had a KCNH2 mutation (P=.03).
FIGURE 3.
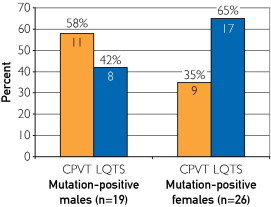
The percent distribution of mutations identified in the catecholaminergic polymorphic ventricular tachycardia (CPVT)–associated RYR2 gene compared with mutations identified in the long QT syndrome (LQTS)–associated genes for the 19 mutation-positive males and the 26 mutation-positive females. The numbers in the bars represent the numbers of cases with a mutation. For example, 11 of 19 (57.9%) mutation-positive males had a CPVT-associated RYR2 mutation compared with 8 males (42.1%) who had a mutation in an LQTS-associated gene (KCNQ1, KCNH2, SCN5A, KCNE1, or KCNE2).
FIGURE 4.
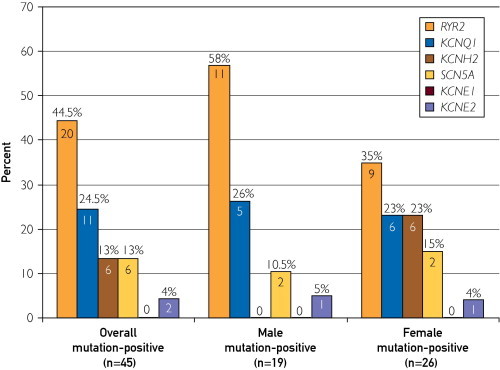
Genotype distribution in sudden unexplained death cases among the mutation-positive overall cohort (n=45), mutation-positive males (n=19), and mutation-positive females (n=26). Again, individuals with a functional polymorphism that might have contributed to the sudden unexplained death were excluded from this calculation. Instead, only the 45 decedents with a rare, potentially channelopathic mutation were counted.
The yield of the molecular autopsy was not different between SUD patients who were 1 to 20 years of age (32/118, 27.1%) compared with decedents older than 20 years (13/55, 23.6%; P=.71; Figure 5). Albeit small in number, none of the 3 SUD cases (1 female and 2 male) who were older than 50 years at the time of death were mutation positive. At all age groups, a trend was seen toward a higher mutation detection yield in females, with the highest differential occurring in the 11- to 20-year range (12/25, 48.0% in females vs 9/50, 18.0% in males; P=.01).
FIGURE 5.
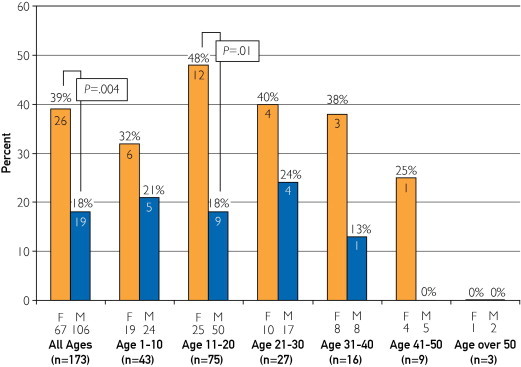
The yield of mutation detection for different age groups in 10-year intervals (1-10, 11-20, 21-30, 31-40, 41-50, and >50 years) for both males (M) and females (F). The numbers in parentheses represent the total number of cases, and the numbers below the F or M represent the numbers of females or males in the respective category. The numbers in the bars represent the absolute number of cases with a mutation, with the percent yield of mutation detection highlighted above the bars.
Effect of Event or Trigger on Yield
The mutation detection yield was associated with the circumstance (events or triggers) surrounding the death, with the highest yield occurring in those with a documented exertional trigger (34.8% [16/46]) compared with a nonspecific trigger (26.9% [14/52]) or death during a period of sleep (18.6% [13/70], Figure 6). This trend held true for females and males, with females consistently having a higher yield than males within the 3 event or trigger categories (exertion, 50.0% [8/16] vs 26.7% [8/30], P=.19; nonspecific trigger, 36.0% [9/25] vs 18.5% [5/27], P=.21; sleep, 31.8% [7/22] vs 12.5% [6/48], P=.09). Again, however, because of the relatively small sizes in each of these event or trigger subsets, these sex and event or trigger trends did not achieve statistical significance.
FIGURE 6.
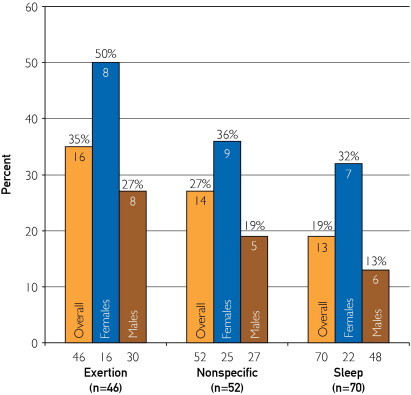
The different yield of mutation detection among the 3 main categories of sudden unexplained death–associated triggers or events (exertion, nonspecific, and sleep) for the overall cohort, females, and males at all ages. The numbers below the bars represent the number of cases in each category. The numbers within the bars represent the actual number of cases with a mutation, with the percent yield of mutation detection highlighted above the bars.
Although cases of exercise-related SUD had the highest mutation detection rate overall, an interesting age effect emerged when examining the yield in specific triggered event categories in different age groups. Overall, SUD patients aged 1 to 10 years with an exertion-induced death had a mutation detection yield (8/12, 66.7%) that was significantly higher than that of 11- to 20-year-olds with exertion-induced death (4/27, 14.8%; P=.002). In contrast, for those with death during a period of sleep, the 11- to 20-year-olds had a higher yield (9/25, 36.0%) than the 1- to 10-year-olds (1/24, 4.2%; P=.01; Figure 7).
FIGURE 7.
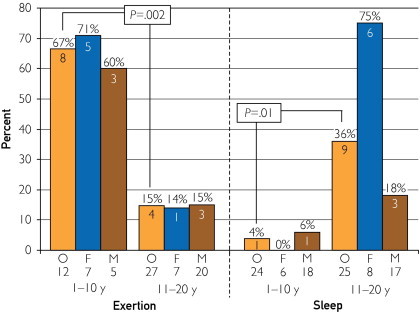
The difference in mutation detection between children aged 1 to 10 years and adolescents aged 11 to 20 years with either exertion- or sleep-associated sudden death. The numbers below O, F, and M represent the number of cases in each category. The numbers within the bars represent the absolute number of cases with a mutation, with the percent yield of mutation detection highlighted above the bars. F = females; M = males; O = overall.
The mutation-positive gene distribution among females (62.5% [5/8] RYR2 and 37.5% [3/8] KCNQ1) and males (75.0% [6/8] RYR2, 12.5% [1/8] SCN5A, and 12.5% [1/8] KCNE2) with exertion was similar and somewhat anticipated based on previous studies showing KCNQ1 LQTS susceptibility mutations and RYR2 CPVT susceptibility mutations most often associated with exertion. Interestingly, although the gene distribution among females (42.9% [3/7] SCN5A, 42.9% [3/7] KCNH2, and 14.3% [1/7] KCNE2) with SUD during sleep correlated with previous observations that suggested that KCNQ1 and RYR2 mutations are infrequently associated with periods of rest and that SCN5A mutations are associated more frequently with cardiac events during rest, the male gene distribution was strikingly different from the female distribution and unanticipated (83.3% [5/6] KCNQ1 and 16.7% [1/6] RYR2, Figure 8).
FIGURE 8.
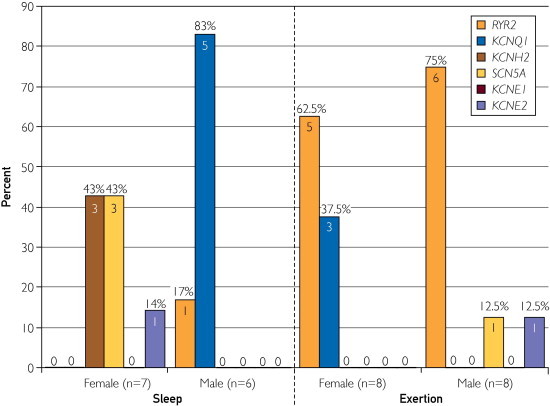
The differences observed in genotype distribution among mutation-positive female (n=7) and males (n=6) with sleep-associated sudden unexplained death (SUD) and female (n=8) and male (n=8) with exertional SUD.
Effect of Personal History and Positive Family History on Yield
As anticipated, the mutation detection yield was significantly higher in patients with SUD who had either a positive personal or family history of cardiac events when compared with those in whom no personal or family history was elicited or documented (37.1% [26/70] vs 19.1% [17/89]; P=.01; Figure 9). Moreover, nearly half (45.0% [9/20]) of patients with SUD who had a positive family history of premature SCD and who were younger than 50 years had a positive cardiac channel molecular autopsy with identification of a rare, potentially channelopathic mutation.
FIGURE 9.
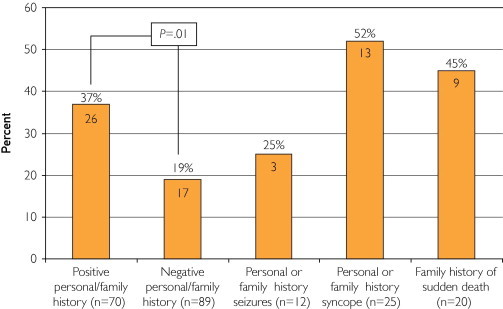
The difference in mutation detection yield between patients with sudden unexplained death (SUD) with either a negative (n=89) or positive (n=70) personal or family history of cardiac events or other “warning” signs, including seizures (n=12), syncope (n=25), or sudden cardiac death (n=20). The numbers in the bars represent the numbers of cases with a mutation, with the percent yield of mutation detection highlighted above the bars.
Discussion
Postmortem genetic testing (the cardiac channel molecular autopsy) has not yet been transformed fully from a research-based effort into a routine, standard part of the conventional autopsy when the coroner, medical examiner, or forensic pathologist is faced with a case of SUD in the young. However, recently, Basso and colleagues,18 on behalf of the Association for European Cardiovascular Pathology, have strongly recommended postmortem genetic analysis in both structural and nonstructural genetically determined heart disease as part of the requirements for the adequate postmortem assessment of SCD. In 2008, members of the Trans-Tasman Response AGAinst sudden Death in the Young (TRAGADY), endorsed by the Royal College of Pathologists of Australasia, put forward guidelines to ensure standardization of autopsy practice in young sudden unexpected deaths, ancillary testing, and retention of appropriate material for all cases of SCD to be used for genetic testing. In 2011, the Heart Rhythm Society and the European Heart Rhythm Association provided an expert consensus statement on the state of genetic testing, providing consensus-based guidelines and recommendations for postmortem genetic testing in cases of SUD.20 Accordingly, the key recommendations of the Heart Rhythm Society/European Heart Rhythm Association guidelines state that “in the setting of autopsy negative SUD, comprehensive or targeted (KCNQ1, KCNH2, SCN5A, RYR2) ion channel genetic testing may be considered in an attempt to establish probable cause and manner of death and to facilitate the identification of potentially at-risk relatives and is recommended if circumstantial evidence points toward a clinical diagnosis of LQTS or CPVT specifically.”
Unfortunately, for several reasons, it has been extremely difficult for the medical examiner, coroner, or forensic pathologist to provide this level of care. Perhaps chief among the reasons is that insurance companies or other third-party payers largely do not accept any responsibility for providing coverage for the molecular autopsy of a deceased person, regardless of the implications to surviving relatives. This position, not surprisingly, limits postmortem genetic testing to only families who can afford the out-of-pocket expense for commercially available genetic testing because few medical examiner offices have the internal budget to cover such a molecular autopsy. Alternatively, the medical examiner, coroner, or forensic pathologist and the grieving family are left with the option of enrolling the deceased's sample into research-based genetic testing, where although the price is right (ie, free), the process can be painfully slow.49 Given the expensive and time-consuming nature of postmortem genetic testing, it is currently necessary for the medical examiner, coroner, or forensic pathologist to be case selective in pursuing a molecular autopsy.50
In 2007, we completed our original molecular autopsy series of 49 medical examiner–referred cases of autopsy-negative SUD.23 Comprehensive mutational analysis of all 60 translated exons in the LQTS-associated genes, KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2, along with targeted analysis of the CPVT1-associated RYR2-encoded cardiac ryanodine receptor revealed that more than one-third of SUD cases hosted a seemingly pathogenic cardiac channel mutation.22 We have extended this cohort to now include more than 170 cases of SUD to provide a more extensive analysis to better define the expected yield of mutation detection and offer possible genotype-phenotype correlations that may assist in guiding phenotype-directed mutation detection efforts in future cases of SUD.
In this expanded molecular analysis of 173 SUD cases, we provide molecular evidence that suggests that just more than one-fourth (26.0%) of these SUD cases host putative pathogenic mutations in critical ion channel genes associated with the potentially lethal arrhythmia syndromes LQTS and CPVT. Besides these 26.0% of cases that host putative pathogenic mutations, another 16.2% of this cohort hosted nonsynonymous, amino acid–altering, functional polymorphisms previously associated with LQTS, Brugada syndrome, drug-induced torsades de pointes, or ventricular arrhythmias or SCD polymorphisms. Thus, the overall contribution of channelopathies or sudden death–predisposing or proarrhythmic, functional channel polymorphisms is likely underestimated in this study. For example, we excluded 10 SUD patients (5.8%) who were positive for D85N-KCNE1, R1047L-KCNH2, or S1103Y-SCN5A. These proarrhythmic polymorphisms have been associated convincingly with increased risk for sudden death, particularly in the setting of drugs with an unwanted QT-prolonging effect. Despite our efforts, we were unable to elicit a reliable history of pre-event medication exposures to determine whether a case for drug exposure plus functional polymorphism could be made. Nevertheless and perhaps more importantly than this overall point estimate for the yield of such postmortem genetic testing in the setting of an SUD, this cohort has provided us with several interesting genotype-phenotype observations that may provide insight into the expected yields of postmortem genetic testing for SUD and assist in selecting cases with the greatest potential for mutation discovery and directing genetic testing efforts.
Interestingly, although males represented two-thirds of the cohort, females were more likely to host a channel mutation than males, especially if the SUD was during adolescence (48.0% females vs 18.0% males). Moreover, there was a cardiac channelopathy–sex effect, with two-thirds of the mutation-positive females having an LQTS-associated mutation compared with only one-third having a CPVT-associated RYR2 mutation, whereas mutation-positive males most often had a CPVT (approximately 60%) mutation rather than an LQTS mutation (approximately 40%). On average, males who had mutations in LQTS- or CPVT-associated genes were younger than females who were genotype positive. This sex predilection is consistent with previous work by Priori et al,51 in which among their cohort of 30 CPVT index cases, most of the RYR2 mutations were identified in young males (11/13 [85%]) compared with females (3/17 [18%]), and male sex in RYR2-mediated CPVT was associated with a relative risk of 4.2 (95% confidence interval, 1.2-15) of developing syncope compared with females. Accordingly, one might a priori expect a higher yield of LQTS-associated mutation detection in a female adolescent or young adult compared with a higher expected yield of CPVT-associated mutations among prepubertal boys.
The circumstances surrounding the death also had an effect on the mutation detection yield, in which decedents had the highest mutation detection rate when the death was associated with exertion compared with a nonspecific triggered death or death during sleep. This trend held true for females and males, with females consistently having higher yields than males within the 3 event or trigger categories. Although half of all females and 26.7% of males with exercise-associated sudden death were identified as mutation positive, only 31.8% and 12.5%, respectively, had mutations when the death occurred during sleep. However, an interesting age effect was revealed when examining the effect on yield when comparing exercise-induced death with death during sleep. For example, 71.4% of the 1- to 10-year-old girls with exercise-induced death were found to be mutation positive compared with 0% of the 1- to 10-year-old girls found dead in bed. Conversely, although only 14.3% (1 of 7) of 11- to 15-year-old girls with exercise-induced death were mutation positive, three-fourths of the adolescent girls with death during sleep were identified as mutation positive. Although failing to reach statistical significance, this trend was also observed among males.
Previously, specific genotype-phenotype associations in LQTS have been established, suggesting relatively gene-specific triggers.52 Exertion-induced cardiac events are associated strongly with mutations in KCNQ1 (LQT1), whereas auditory triggers and events occurring during the postpartum period most often occur in patients with LQT2 (KCNH2). Although exertion- or emotional stress–induced events are most common in LQT1 (KCNQ1), events occurring during periods of sleep or rest are most common in LQT3 (SCN5A). CPVT1 (RYR2) classically manifests with exercise-induced syncope or sudden death51 and rarely manifests during periods of rest.
These particularly well-vetted genotype-phenotype associations have led some investigators to selectively direct genetic testing efforts based on trigger-associated events, particularly in LQTS cases.53 For example, if an LQTS individual presents with exertion-induced syncope or cardiac arrest, then a genetic test for KCNQ1 (LQT1) mutations should be performed. If the syncope or cardiac arrest occurred during periods of rest or sleep, then a test for SCN5A (LQT3) should be performed. Although the observed mutation-positive gene distribution in this SUD cohort matched the anticipated results for those males and females with exercise-induced death (ie, overwhelmingly RYR2 or KCNQ1 mutations) and females with death during sleep (ie, mostly SCN5A and no RYR2 or KCNQ1 mutations), strikingly and unexpectedly, all of the mutation-positive males with death during sleep had mutations in either KCNQ1 (83.3%) or RYR2 (16.7%).
Both LQTS and CPVT are hereditary disorders that manifest within families with potential warning signs, such as syncope, seizures, survived cardiac arrest, near drowning, or prolonged QT interval, that may lead to a clinical diagnosis of the disorder and circumvent the sudden death of a prophylactically treated family member. However, tragically these particular warning signs often go unheeded or unrecognized as evident by the remarkable presence of a positive family history of cardiac events in 40.5% of our SUD cohort, including nearly 11.6% of the patients with a positive family history of a prior sudden death. Yet, no decedent or relative had received a clinical diagnosis of a suspected cardiac channelopathy or heritable arrhythmia syndrome (ie, LQTS or CPVT) before the SUD of the current patient in our study. As expected, SUD cases with such a positive personal or family history had a significantly higher mutation detection yield (37.1%) than those with no personal or family history (19.1%) of cardiac events. In fact, among the subset of patients with a positive family history of otherwise unexplained sudden death before the age of 50 years, nearly half were positive for a putative channelopathic mutation.
Interestingly, there was a trend toward a higher mutation detection yield in SUD patients with a personal or family history of syncope compared with a history of seizures. Although not as common as syncope, seizure-like symptoms are experienced by some patients with LQTS or CPVT. Our data suggest that SUD in an individual with a personal or family history of seizure activity may in fact be a result of a neurologic disorder, akin to SUD in epilepsy (SUDEP), rather than as a result of a primarily cardiogenic disorder, such as LQTS or CPVT.
Although mutations in LQTS and CPVT genes have been illustrated as the pathogenic basis from some SUDEP cases, most SUDEP appears to not be related to mutations in major genes associated with theses 2 disorders. In 2010, Tu et al54 analyzed a cohort of 68 SUDEP cases for mutations in 3 major LQTS genes (KCNQ1, KCNH2, and SCN5A) and only identified 2 patients (3%) with putative pathogenic mutations (R176W-KCNH2 and P1090L-SCN5A), which were absent in their control population. However, both of these genetic variants have been observed in other control populations and represent functionally significant polymorphisms that confer an increased risk for arrhythmia.32,33 Together, these data suggest that sudden death cases with a personal or family history of seizures may warrant mutational analysis of ion channel encoding genes, such as SCN1A and KCNA1, which are abundantly expressed in the brain and have been implicated as candidates for SUDEP,55,56 rather than starting with an initial interrogation of LQTS- and CPVT-associated genes. Whether our genotype-negative SUD cases with a personal or family history of seizure activity have mutations in these SUDEP candidate genes is currently unknown. In contrast, a personal or family history of unexplained syncope, especially exercise-induced syncope, may reflect a strong indicator for LQTS and CPVT genetic testing for the SUD case.
Although a positive personal or family history is associated with a 2-fold greater likelihood of a positive postmortem genetic test result than when there is no history, a negative personal or family history alone should not preclude genetic testing with the intent to identify mutations that may be familial and therefore provide an increased risk for sudden death of an unsuspecting surviving family member of the decedent. In fact, among the 11 mutation-positive SUD families that chose to participate in genetic pedigree analysis in our study, all 11 SUD cases were identified as having a familial mutation, rather than a sporadic one, despite having a negative personal or family history of cardiac events in 72.7% (8/11) of these families, enabling personal risk stratification and prophylactic therapies for the mutation-positive family members who are still living.
The major limitation of this study pertains to the issue of mutation calling.34 Although most mutations identified in this study are either radical (ie, frameshift or splicing errors) or localize to critical ion channel domains (ie, transmembrane region or channel pore regions), with a predicted probability of pathogenicity exceeding 90%, most mutations identified in this study, in particular those that are novel, have not been characterized functionally. Heterologous expression studies for every ion channel variant identified to assess in vitro functionality are not feasible practically. Instead, strict absence from our internal set of ethnic-matched reference alleles was required. Nevertheless, it is possible that some of these variants listed as putative pathogenic mutations may be innocuous functionally. Therein lies the difficulty in determining the “actionableness” of these rare, nonfunctionally characterized genetic variants for surviving family members when the probability of pathogenicity for that given variant is not 100%. However, this issue of mutation calling is present in premortem genetic testing as well and underscores the reality that, whether the patient is dead or alive, a genetic test result must be interpreted with great caution.
Conclusion
More than one-fourth of autopsy-negative SUD cases may stem from either LQTS- or CPVT-associated channelopathic mutations. A cardiac channel molecular autopsy should be considered as a standard part of the evaluation of autopsy-negative SUD, especially among children with exercise-induced SUD, adolescent girls, and those with a positive personal or family history of cardiac events. A 2-tiered approach starting with the LQTS-susceptibility genes for female patients with SUD and the CPVT susceptibility genes for males with SUD may be prudent.
Footnotes
Grant Support: This work was supported by the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program, the Sheikh Zayed Saif Mohammed Al Nahyan Fund in Pediatric Cardiology Research, the Dr. Scholl Fund, the Hannah M. Wernke Memorial Fund, and the National Institutes of Health, grant number HD42569.
Potential Competing Interests: M.J.A. is a consultant for Biotronik, Boston Scientific, Medtronic, St. Jude Medical Inc, and Transgenomic. Intellectual property derived from M.J.A.'s research program resulted in license agreements in 2004 between Mayo Clinic Health Solutions (formerly Mayo Medical Ventures) and PGxHealth (formerly Genaissance Pharmaceuticals, recently acquired by Transgenomic). D.J.T., A.M.-D., M.L.W., and C.M.H. have no conflicts to disclose. M.J.A. had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Virmani R., Burke A.P., Farb A. Sudden cardiac death. Cardiovasc Pathol. 2001;10(6):275–282. doi: 10.1016/s1054-8807(01)00108-9. [DOI] [PubMed] [Google Scholar]
- 2.Liberthson R.R. Sudden death from cardiac causes in children and young adults. N Engl J Med. 1996;334(16):1039–1044. doi: 10.1056/NEJM199604183341607. [DOI] [PubMed] [Google Scholar]
- 3.Chugh S.S., Kelly K.L., Titus J.L. Sudden cardiac death with apparently normal heart. Circulation. 2000;102(6):649–654. doi: 10.1161/01.cir.102.6.649. [DOI] [PubMed] [Google Scholar]
- 4.Maron B.J., Shirani J., Poliac L.C., Mathenge R., Roberts W.C., Mueller F.O. Sudden death in young competitive athletes: clinical, demographic, and pathological profiles. JAMA. 1996;276(3):199–204. [see comment] [PubMed] [Google Scholar]
- 5.Corrado D., Basso C., Thiene G. Sudden cardiac death in young people with apparently normal heart. Cardiovasc Res. 2001;50(2):399–408. doi: 10.1016/s0008-6363(01)00254-1. [DOI] [PubMed] [Google Scholar]
- 6.Behr E., Wood D.A., Wright M. Cardiological assessment of first-degree relatives in sudden arrhythmic death syndrome. Lancet. 2003;362(9394):1457–1459. doi: 10.1016/s0140-6736(03)14692-2. [DOI] [PubMed] [Google Scholar]
- 7.Morentin B., Suarez-Mier M.P., Aguilera B. Sudden unexplained death among persons 1-35 years old. Forensic Sci Int. 2003;135(3):213–217. doi: 10.1016/s0379-0738(03)00212-3. [DOI] [PubMed] [Google Scholar]
- 8.Puranik R., Chow C.K., Duflou J.A., Kilborn M.J., McGuire M.A. Sudden death in the young. Heart Rhythm. 2005;2(12):1277–1282. doi: 10.1016/j.hrthm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Tester D.J., Ackerman M.J. The role of molecular autopsy in unexplained sudden cardiac death. Curr Opin Cardiol. 2006;21(3):166–172. doi: 10.1097/01.hco.0000221576.33501.83. [DOI] [PubMed] [Google Scholar]
- 10.Wever E.F., Robles de Medina E.O. Sudden death in patients without structural heart disease. J Am Coll Cardiol. 2004;43(7):1137–1144. doi: 10.1016/j.jacc.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 11.Ackerman M.J. Cardiac channelopathies: it's in the genes. Nat Med. 2004;10(5):463–464. doi: 10.1038/nm0504-463. [DOI] [PubMed] [Google Scholar]
- 12.Ackerman M.J. Cardiac causes of sudden unexpected death in children and their relationship to seizures and syncope: genetic testing for cardiac electropathies. Semin Pediat Neurol. 2005;12(1):52–58. doi: 10.1016/j.spen.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Goldenberg I., Moss A.J., Zareba W. Sudden cardiac death without structural heart disease: update on the long QT and Brugada syndromes. Curr Cardiol Rep. 2005;7(5):349–356. doi: 10.1007/s11886-005-0088-1. [DOI] [PubMed] [Google Scholar]
- 14.Priori S.G., Napolitano C., Giordano U., Collisani G., Memmi M. Brugada syndrome and sudden cardiac death in children. Lancet. 2000;355(9206):808–809. doi: 10.1016/S0140-6736(99)05277-0. [DOI] [PubMed] [Google Scholar]
- 15.Ackerman M.J., Tester D.J., Driscoll D.J. Molecular autopsy of sudden unexplained death in the young. Am J Forensic Med Pathol. 2001;22(2):105–111. doi: 10.1097/00000433-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Ackerman M.J., Siu B.L., Sturner W.Q. Postmortem molecular analysis of SCN5A defects in sudden infant death syndrome. JAMA. 2001;286(18):2264–2269. doi: 10.1001/jama.286.18.2264. [DOI] [PubMed] [Google Scholar]
- 17.Chugh S.S., Senashova O., Watts A. Postmortem molecular screening in unexplained sudden death. J Am Coll Cardiol. 2004;43(9):1625–1629. doi: 10.1016/j.jacc.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 18.Basso C., Burke M., Fornes P. Guidelines for autopsy investigation of sudden cardiac death. Virchows Arch. 2008;452(1):11–18. doi: 10.1007/s00428-007-0505-5. [DOI] [PubMed] [Google Scholar]
- 19.Semsarian C., Hamilton R.M. Key role of the molecular autopsy in sudden unexpected death. Heart Rhythm. 2012;9(1):145–150. doi: 10.1016/j.hrthm.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Ackerman M.J., Priori S.G., Willems S. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8(8):1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Di Paolo M., Luchini D., Bloise R., Priori S.G. Postmortem molecular analysis in victims of sudden unexplained death. Am J Forensic Med Pathol. 2004;25(2):182–184. doi: 10.1097/01.paf.0000127406.20447.8a. [DOI] [PubMed] [Google Scholar]
- 22.Tester D.J., Spoon D.B., Valdivia H.H., Makielski J.C., Ackerman M.J. Targeted mutational analysis of the RyR2-encoded cardiac ryanodine receptor in sudden unexplained death: a molecular autopsy of 49 medical examiner/coroner's cases. Mayo Clin Proc. 2004;79(11):1380–1384. doi: 10.4065/79.11.1380. [DOI] [PubMed] [Google Scholar]
- 23.Tester D.J., Ackerman M.J. Postmortem long QT syndrome genetic testing for sudden unexplained death in the young. J Am Coll Cardiol. 2007;49(2):240–246. doi: 10.1016/j.jacc.2006.10.010. [see comment] [DOI] [PubMed] [Google Scholar]
- 24.Doolan A., Langlois N., Semsarian C. Causes of sudden cardiac death in young Australians. Med J Australia. 2004;180(3):110–112. doi: 10.5694/j.1326-5377.2004.tb05830.x. [DOI] [PubMed] [Google Scholar]
- 25.Creighton W., Virmani R., Kutys R., Burke A. Identification of novel missense mutations of cardiac ryanodine receptor gene in exercise-induced sudden death at autopsy. J Mol Diagn. 2006;8(1):62–67. doi: 10.2353/jmoldx.2006.050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishio H., Iwata M., Suzuki K. Postmortem molecular screening for cardiac ryanodine receptor type 2 mutations in sudden unexplained death: R420W mutated case with characteristics of status thymico-lymphatics. Circ J. 2006;70(11):1402–1406. doi: 10.1253/circj.70.1402. [DOI] [PubMed] [Google Scholar]
- 27.Gladding P.A., Evans C.A., Crawford J. Posthumous diagnosis of long QT syndrome from neonatal screening cards. Heart Rhythm. 2010;7(4):481–486. doi: 10.1016/j.hrthm.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Skinner J.R., Crawford J., Smith W. Prospective, population-based long QT molecular autopsy study of postmortem negative sudden death in 1 to 40 year olds. Heart Rhythm. 2011;8(3):412–419. doi: 10.1016/j.hrthm.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Tester D.J., Ackerman M.J. Postmortem long QT syndrome genetic testing for sudden unexplained death in the young. J Am Coll Cardiol. 2007;49(2):240–246. doi: 10.1016/j.jacc.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Medeiros-Domingo A., Bhuiyan Z., Tester D. Comprehensive open reading frame mutational analysis of the RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome. J Am Coll Cardiol. 2009;54(22):2065–2074. doi: 10.1016/j.jacc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tester D.J., Will M.L., Haglund C.M., Ackerman M.J. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2(5):507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 32.Ackerman M.J., Splawski I., Makielski J.C. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1(5):600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman M.J., Tester D.J., Jones G., Will M.K., Burrow C.R., Curran M. Ethnic differences in cardiac potassium channel variants: implications for genetic susceptibility to sudden cardiac death and genetic testing for congenital long QT syndrome. Mayo Clin Proc. 2003;78(12):1479–1487. doi: 10.4065/78.12.1479. [DOI] [PubMed] [Google Scholar]
- 34.Kapa S., Tester D.J., Salisbury B.A. Genetic testing for long-QT syndrome: distinguishing pathogenic mutations from benign variants. Circulation. 2009;120(18):1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran Y., Chen J., Li N. Common RyR2 variants associate with ventricular arrhythmias and sudden cardiac death in chronic heart failure. Clin Sci. 2010;119(5):215–223. doi: 10.1042/CS20090656. [DOI] [PubMed] [Google Scholar]
- 36.Koop A., Goldmann P., Chen S.R.W., Thieleczek R., Varsanyi M. ARVC-related mutations in divergent region 3 alter functional properties of the cardiac ryanodine receptor. Biophys J. 2008;94(12):4668–4677. doi: 10.1529/biophysj.107.122382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishio Y., Makiyama T., Itoh H. D85N, a KCNE1 polymorphism, is a disease-causing gene variant in long QT syndrome. J Am Coll Cardiol. 2009;54(9):812–819. doi: 10.1016/j.jacc.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Splawski I., Timothy K.W., Tateyama M. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297(5585):1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 39.Burke A., Creighton W., Mont E. Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation. 2005;112(6):798–802. doi: 10.1161/CIRCULATIONAHA.104.482760. [DOI] [PubMed] [Google Scholar]
- 40.Sun Z., Milos P.M., Thompson J.F. Role of KCNH2 polymorphism (R1047L) in dofetilide-induced Torsades de Pointes. J Mol Cell Cardiol. 2004;37(5):1031–1039. doi: 10.1016/j.yjmcc.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Wang D.W., Desai R.R., Crotti L. Cardiac sodium channel dysfunction in sudden infant death syndrome. Circulation. 2007;115(3):368–376. doi: 10.1161/CIRCULATIONAHA.106.646513. [DOI] [PubMed] [Google Scholar]
- 42.Marangoni S., Di Resta C., Rocchetti M. A Brugada syndrome mutation (p.S216L) and its modulation by p.H558R polymorphism: standard and dynamic characterization. Cardiovasc Res. 2011;91(4):606–616. doi: 10.1093/cvr/cvr142. [DOI] [PubMed] [Google Scholar]
- 43.Abbott G.W., Sesti F., Splawski I. MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell. 1999;97(2):175–187. doi: 10.1016/s0092-8674(00)80728-x. [DOI] [PubMed] [Google Scholar]
- 44.Marjamaa A., Salomaa V., Newton-Cheh C. High prevalence of four long QT syndrome founder mutations in the Finnish population. Ann Med. 2009;41(3):234–240. doi: 10.1080/07853890802668530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubota T., Horie M., Takano M. Evidence for a single nucleotide polymorphism in the KCNQ1 potassium channel that underlies susceptibility to life-threatening arrhythmias. J Cardiovasc Electr. 2001;12(11):1223–1229. doi: 10.1046/j.1540-8167.2001.01223.x. [DOI] [PubMed] [Google Scholar]
- 46.Vatta M., Dumaine R., Varghese G. Genetic and biophysical basis of sudden unexplained nocturnal death syndrome (SUNDS), a disease allelic to Brugada syndrome. Hum Mol Genet. 2002;11(3):337–345. doi: 10.1093/hmg/11.3.337. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q., Chen S., Chen Q. The common SCN5A mutation R1193Q causes LQTS-type electrophysiological alterations of the cardiac sodium channel. J Med Genet. 2004;41(5):e66. doi: 10.1136/jmg.2003.013300. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinlapawittayatorn K., Du X.X., Liu H., Ficker E., Kaufman E.S., Deschenes I. A common SCN5A polymorphism modulates the biophysical defects of SCN5A mutations. Heart Rhythm. 2011;8(3):455–462. doi: 10.1016/j.hrthm.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ackerman M.J. State of postmortem genetic testing known as the cardiac channel molecular autopsy in the forensic evaluation of unexplained sudden cardiac death in the young. PACE. 2009;32(suppl 2):S86–S89. doi: 10.1111/j.1540-8159.2009.02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliva A., Brugada R., D'Aloja E. State of the art in forensic investigation of sudden cardiac death. Am J Forensic Med Pathol. 2011;32(1):1–16. doi: 10.1097/PAF.0b013e3181c2dc96. [DOI] [PubMed] [Google Scholar]
- 51.Priori S.G., Napolitano C., Memmi M. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106(1):69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 52.Ackerman M.J. Genotype-phenotype relationships in congenital long QT syndrome. J Electrocardiol. 2005;38(4, suppl):64–68. doi: 10.1016/j.jelectrocard.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 53.Van Langen I.M., Birnie E., Alders M., Jongbloed R.J., Le Marec H., Wilde A.A.M. The use of genotype-phenotype correlations in mutation analysis for the long QT syndrome. J Med Genet. 2003;40(2):141–145. doi: 10.1136/jmg.40.2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tu E., Bagnall R.D., Duflou J., Semsarian C. Post-mortem review and genetic analysis of sudden unexpected death in epilepsy (SUDEP) cases. Brain Pathol. 2010;21(2):201–208. doi: 10.1111/j.1750-3639.2010.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glasscock E., Yoo J.W., Chen T.T., Klassen T.L., Noebels J.L. Kv1.1 potassium channel deficiency reveals brain-driven cardiac dysfunction as a candidate mechanism for sudden unexplained death in epilepsy. J Neurosci. 2010;30(15):5167–5175. doi: 10.1523/JNEUROSCI.5591-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Gal F., Korff C.M., Monso-Hinard C. A case of SUDEP in a patient with Dravet syndrome with SCN1A mutation. Epilepsia. 2010;51(9):1915–1918. doi: 10.1111/j.1528-1167.2010.02691.x. [DOI] [PubMed] [Google Scholar]


