Abstract
Early detection and isolation of circulating tumor cells (CTC) can enable better prognosis for cancer patients. A Hele-Shaw device with aptamer functionalized glass beads is designed, modeled, and fabricated to efficiently isolate cancer cells from a cellular mixture. The glass beads are functionalized with anti-epidermal growth factor receptor (EGFR) aptamer and sit in ordered array of pits in PDMS channel. A PDMS encapsulation is then used to cover the channel and flow through cell solution. The beads capture cancer cells from flowing solution depicting high selectivity. The cell-bound glass beads are then re-suspended from the device surface followed by the release of 92% cells from glass beads using combination of soft shaking and anti-sense RNA. This approach ensures that the cells remain in native state and undisturbed during capture, isolation and elution for post-analysis. The use of highly selective anti-EGFR aptamer with the glass beads in an array and subsequent release of cells with antisense molecules provide multiple levels of binding and release opportunities that can help in defining new classes of CTC enumeration devices.
INTRODUCTION
Isolation of rare, pure and viable circulating tumor cells from blood has high diagnostic potential.1 A number of strategies for isolation of tumor cells have been reported.2–8 Although the sorting based on affinity interactions may yield higher efficiency and greater specificity,9 owing to limitations of biomarkers that can be used for selective capture10 and high levels of off-target cross-reactivity of antibodies used for such capture,11 most approaches fail to satisfy clinical validity or utility. Aptamers have been shown to have affinities and specificities that are comparable to those of antibodies.12 Moreover, aptamers can be chemically synthesized, site-specifically labeled, and homogeneously immobilized on the substrate surfaces. Only recently, aptamers have been used in lab-on-chip devices to sort, isolate and detect tumor cells.12–15 Aptamers have been proven to specifically recognize, capture and isolate human glioblastoma (hGBM) cells, known to over-express epidermal growth factor receptor (EGFR), from a mixture of fibroblasts.12
Releasing captured tumor cells from sensor surface for subsequent molecular analysis or cell culture is also an important and challenging step. The strong adhesion forces between cells and antibody functionalized surfaces need to be overcome to detach cells. Methods like thermodynamic release, electrochemical desorption and proteolytic enzyme degradation have been used to achieve this.2,16,17 However, these require either elaborate design of sensors or specific enzymes that target cell receptors and/or antibodies. In addition, the spreading and flatness of captured cells on the surface require extremely high threshold of detachment force which has to be applied for completely eluting captured cells from the surfaces.16,18,19 All of these are invasive, have the potential to harm the completeness of cell structure and greatly disturb the cell microenvironment.
This article addresses a number of issues like affinity between a novel aptamer against EGFR, the effect of fluid flow velocity through a microfluidic channel and the associated shear stress faced by the cell, relevance of the flow behavior with the binding forces between surface-bound aptamers and EGFR expression on the cell walls, and non-invasive recovery of the captured cells.
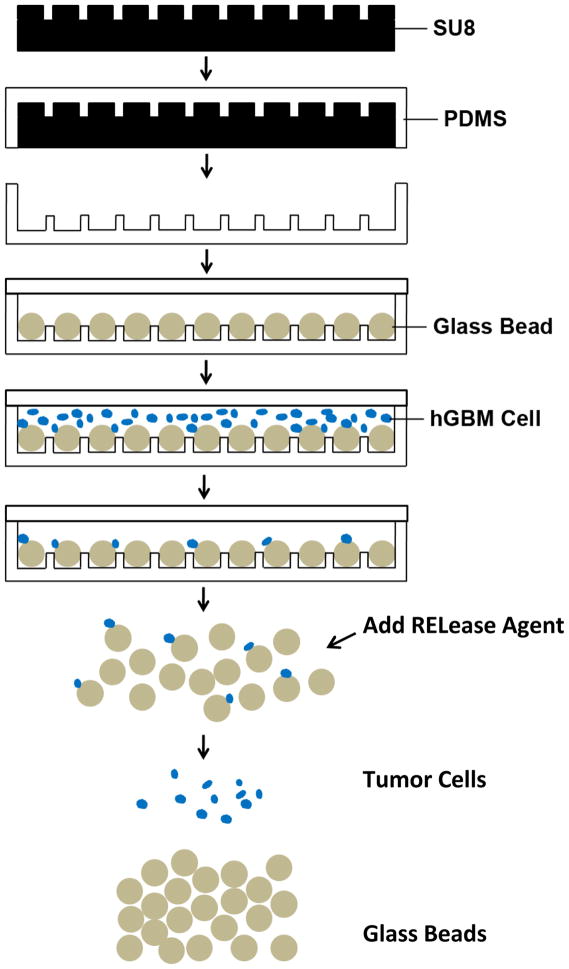
Hele-Shawmicrofluidic devices with plain surface of the channel and with array of pits on the channel floor are then presented (Fig. 1). The pits were filled with anti-EGFR aptamer functionalized glass beads (GB). The curved hemispherical surfaces of GBs presented altered flow in the channel with varying shear stresses around the GBs. The benefit of Hele-Shaw design was that the special geometry gave linearly reducing fluidic shear stress along the longitudinal axis of the devicelength.20 This helped in varying overall shear stress at various regions and resulted into sweet spots to balance affinity forces of anti-EGFR aptamer and EGFR on cells. Such design can possibly provide cell sub-populations with varying amount of EGFR over-expression as that would be a factor in how far the cells travel under linearly decreasing shear stress.
Figure 1.
Schematics showing steps of fabrication and experiments. SU-8 photoresist is spin-cast on silicon wafer, exposed and wells are developed to form the desired pattern. PDMS is poured on SU-8 master, baked, and peeled off. 50 μm diameter GBs are loaded into 25 μm deep pits and the substrate is covered with a flat PDMS slab. Cancer cell suspension is flowed through the device, and cells are captured by aptamer functionalized GBs. Captured cells are finally released from the GB surface after GBs are collected from the device.
A mixture of hGBM and normal cells was passed through the GB device and capture of tumor cells on GBs was achieved. The release of GBs from the arrayed pits was simple and straight-forward; placing the device upside down released the GB in solution. The GBs were collected and cancer cells were then detached from GBs. The detachment of the most of the cancer cells occurred with gentle shaking but upto 92% cells were recovered when an anti-sense RNA molecule was used. The anti-sense, or what is called here RELease molecule, competitively hybridized with the aptamer thus reducing its affinity to EGFR.
This device provides an important platform that can possibly be used to isolate circulating tumor cells (CTC) from peripheral blood samples. In a practical scenario, the human blood can be drawn in annual physical check-up, red blood cells lysed, cell sub-population fractionated with simple centrifugation, supernatant re-suspended in a buffer and ran through GB device.
EXPERIMENTAL SECTION
Oligonucleotide Preparation
The aptamer preparation has been described in detail previously.14,15 In brief, the anti-EGFR aptamer was isolated by iteratively selecting binding species against purified human EGFR from a pool that spanned a 62-nucleotide random region. After PCR amplification from a double stranded DNA (dsDNA) template, the products were transcribed into RNA. The high-affinity anti-EGFR aptamer (Kd = 2.4 nmol/L) was extended with a linker sequence used to hybridize with surface bound capture probe. The sequences for the extended anti-EGFR aptamer, extended mutant aptamer, capture probe DNA and release RNA agent were as follows: anti-EGFR aptamer (5′-

 AUU UAA UCG CCG UAG AAA AGC AUG UCA AAG CCG GAA CCG UGU AGC ACA GCA GA GAA UUA AAU GCC CGC CAU GAC CAG-3′); mutant aptamer (5′-GGC GCU CCG ACC UUA GUC UCU GUU CCC ACA UCA UGC ACA AGG ACA AUU CUG UGC AUC CAA GGA GGA GUU CUC GGA ACC GUG UAG CAC AGC AGA GAA UUA AAU GCC CGC CAU GAC CAG-3′); Capture probe DNA (5′-amine/biotin-CTG GTC ATG GCG GGC ATT TAA TTC-3′); and release RNA agent (5′-
AUU UAA UCG CCG UAG AAA AGC AUG UCA AAG CCG GAA CCG UGU AGC ACA GCA GA GAA UUA AAU GCC CGC CAU GAC CAG-3′); mutant aptamer (5′-GGC GCU CCG ACC UUA GUC UCU GUU CCC ACA UCA UGC ACA AGG ACA AUU CUG UGC AUC CAA GGA GGA GUU CUC GGA ACC GUG UAG CAC AGC AGA GAA UUA AAU GCC CGC CAU GAC CAG-3′); Capture probe DNA (5′-amine/biotin-CTG GTC ATG GCG GGC ATT TAA TTC-3′); and release RNA agent (5′-

 GA GGG AAG GAA GUA AG-3′). The extended linker sequence is underlined, and complementary parts of aptamer and release RNA agent are framed. The aptamer was 2′-F-Py modified which made it resistant to degradation. Such 2′-fluoro pyrimidine modified RNA aptamers are known to be stable in serum for hours.21
GA GGG AAG GAA GUA AG-3′). The extended linker sequence is underlined, and complementary parts of aptamer and release RNA agent are framed. The aptamer was 2′-F-Py modified which made it resistant to degradation. Such 2′-fluoro pyrimidine modified RNA aptamers are known to be stable in serum for hours.21
Aptamer Functionalized Glass Beads Preparation
The covalent immobilization of capture probe DNA onto 50 μm diameter GBs (Cospheric) was achieved using previously described method.22 Briefly, GBs were cleaned with nitric acid before derivatization with 2% 3-aminopropyltriethoxysilane (APTES) in ethanol. GBs were baked at 110 °C for 1 h, and then immersed in a dimethylformamide (DMF) solution containing 10% pyridine and 1 mmol/l phenyldiisothiocyanate (PDITC) for 2 h. After rinsing with DMF and 1,2-dichloroethane, 10 μM annealed complex of capture probe DNA/aptamer was added to 30 mg activated GBs. After immobilization, GBs were washed with 1× PBS to remove non-bound oligonucleotides. The GBs were then dried in vacuum.
Collection and Culture of Cancer Cells
Human glioblastoma samples were obtained from consenting patients at the University of Texas Southwestern Medical Center (Dallas, TX, USA) as per the process described before.12 While acknowledging the controversy of whether only CD133+ve are truly tumorigenic, for the purposes of this study we used CD133+ve cells isolated from surgically resected samples that were also found to overexpress wild-type EGFR. Specimens averaging in size to be >50 mm3 were removed from brain and immediately placed into ice cold hank’s buffered salt solution (HBSS). The hGBM tumor tissue was gently dissociated with papain and dispase, triturated, labeled with a CD133/2 (293C3)-PE antibody and sorted with FACSCalibur machine. Cells were suspended in a chemically defined serum-free Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium, consisting of 20 ng/ml mouse EGF (Peprotech), 20 ng/ml of bFGF (Peprotech), 1× B27 supplement (Invitrogen), 1× Insulin-Transferrin-Selenium-X (Invitrogen), 100 units/ml:100 μg/ml of Penicillin:Streptomycin (HyClone) and plated at a density of 3×106 live cells/60 mm plate.
EGFR overexpressing A431 cells were also grown in DMEM with 10% FBS (Invitrogen). A431 is a human carcinoma cell line overexpressing EGFR wild type receptor. These cells were grown to 70% confluence, trypsinized, washed and counted. These cells were used to measure effect of RELease molecules. The RELease molecule was ultimately applied to release hGBM cells from GBs.
Flow Cytometry of Aptamer and Release RNA Agent
These experiments were done to characterize the aptamer release sequence, named RELease (Fig. 2). Anti-EGFR aptamer was annealed with biotin modified capture probe DNA in BND buffer (10 mM MgCl2 in PBS) for 4 min at 80 °C then cooled to 4 °C at a slow ramp of 0.1 °C per second. This aptamer-DNA-biotin construct was then incubated for 10 min with streptavidin-phycoerythrin (SAPE, Invitrogen). Phycoerythrin provided a strong fluorescence emission signal at 575 nm that was used to measure binding of the aptamer with cell surface EGFR.
Figure 2.
Schematics showing cells exposed to anti-EGFR aptamer complex, binding between cells and aptamer-DNA-biotin-SAPE complex and introduction of RELease molecule. Fluorescence came from SAPE bound to anti-EGFR aptamer. After washing, cells show fluorescence due to selective binding with aptamer. The addition of RELease RNA agent competitively hybridizes with anti-EGFR aptamer, opening up its hairpin structure. It can release 76% of anti-EGFR aptamers from cellular surfaces.
The A431 cells were washed, resuspended in PBS and incubated at 4 °C for 30 min with the aptamer DNA biotin SAPE construct (Fig. 2). Controls included A431 cells only; the DNA biotin SAPE construct only (no aptamer); and A431 cells incubated with mutant aptamer-DNA-biotin-SAPE. The two cell samples incubated with aptamers were washed to remove free aptamer and treated with 2 equivalents of the RELease RNA molecules and allowed to incubate for 30 min at 4 °C or 37 °C respectively. All samples were then washed of any free aptamer, suspended in PBS and analyzed on a FACSCalibur flow cytometer (BD Biosciences).
Once the RELease effect was quantified, it was applied to GB bound hGBM cells. Anti-EGFR aptamer functionalized GBs were divided into two groups (15 mg each). hGBM cells (24×104 cells per ml of PBS) were seeded on each group of GBs. After 30 min incubation at 37 °C, RELease RNA agent was added into the first group, and the mixture was incubated for another 30 min. At the end, GBs from two groups were softly resuspended by shaking solution with 20 μl micropipette; supernatant fluid was removed after GBs’ precipitation; and GBs were resuspended in PBS. The cells which remained adherent to the GBs in the two groups were manually counted.
Computational Analysis and Fabrication of Microfluidic Device
A Hele-Shaw microfluidic device was fabricated for cancer cell isolation.3 The computer simulation was used to optimize the distance between individual GBs and fluid flow behavior. A view of the simulated microfluidic device and Hele-Shaw device design is shown in Figure 1 of Electronic Supplementary Materials (ESI). For simulations, GBs of 50 μm diameter were distributed in an array at the bottom plane of the device in an equilateral triangle fashion. To find the optimum distance between individual GBs for cell capture, simulations were performed by varying the GBs center-to-center separation from 70 to 100 μm (with distance between boundary of one GB to the next one ranging between 20–50 μm). The left and right boundaries of the domain were assumed to have an inlet velocity to satisfy a whole device flow rate of 1 ml/h and an open boundary, respectively. The boundaries at two sides were assumed to be symmetrical, thus the modeled domain represented a portion of the actual device. The top and bottom surface walls were modeled as non-slip boundaries. A total of around 20,000 polygons were used in the finite element mesh.
The microfluidic devices were fabricated using soft lithography.23 In short, a desired SU-8 photoresist layer was spin-coated on a silicon wafer, and then microfluidic device design from mask was transferred to photoresist (Fig. 1). After post-exposure bake, development, rinse in isopropyl alcohol and hard bake, the dried photoresist pattern yielded the master mold. Polydimethylsiloxane (PDMS) was poured on it to produce microfluidic device. The PDMS mold was then peeled off from the master. Two types of devices were fabricated. One was just a flow-through channel with 60 μm height, used to estimate and measure competition between forces of shear-stress and binding between EGFR and anti-EGFR aptamer. The other device was Hele-Shaw structure with pit array (Fig. 1). The pits were filled with aptamer-functionalized glass beads.
Shear-stress Studies
The relationship between shear stress and axial position was calculated using the equation derived by Usami et al.24 PDMS chamber of 60 μm height was used for flow experiments. Anti-EGFR aptamer and mutant aptamer were attached on separate glass slides using the protocol described before.12 The PDMS chamber was treated with plasma and then immediately bonded to glass surface. The hGBM cells were injected at 30×104 cells per ml density into the device with a syringe pump (Harvard Apparatus) at 1 ml/h for 30 min, and then PBS was used to elute nonspecifically bound cells at the same flow rate for 10 min. Pictures were taken with Leica LED microscope (Leica Microsystems). Captured cells were manually counted at selected points along the flow axis.
Glass Bead Array Hele-Shaw Device and Cell Capture
The dimensions of the fabricated Hele-Shaw devices are depicted in Fig. 1 of ESI. The solidified PDMS chamber with arrayed pits was treated with oxygen plasma to make it hydrophilic. Aptamer functionalized GBs were suspended in pure ethanol, and then were added into the chamber. GBs automatically fell into pits with proper shaking. Flat PDMS cover was bonded to Hele-Shaw device at last. PBS was used to perfuse the whole chamber to completely remove ethanol. A 100 cells per ml spiked hGBM cell suspension was flown through the GB array at flow rate of 1 ml/h for 1 h, and then PBS was used to elute nonspecifically bound cells at same flow rate for 10 min. Captured cells were manually counted under microscope.
RESULTS AND ANALYSIS
Computational Analysis
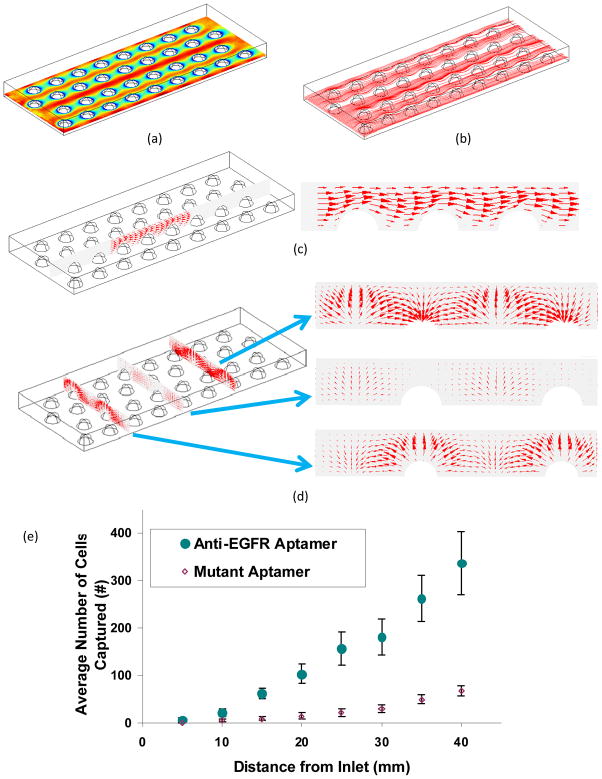
Fig. 3 shows the velocity fields obtained from numerical simulations. Due to the existence of GBs, the fluid in the device was squeezed into the gaps between adjacent GBs, which caused a non-uniformly distributed fluid flow and velocity between GBs (Fig. 3(a)). The stream lines are shown in Fig. 3(b). The flow field near the GBs is examined in detail in Figs. 3(c) and 3(d). The vectors depict the direction and amplitude of the velocity flow. In the flow direction, the streamlines were distorted due to the GBs (Fig. 3(c)). Such distorted flow pattern was favorable for cell capture because the flow would move the cells around in the suspension, thus increasing the probability of contact between cells and anti-EGFR aptamers coated GBs. Considering the velocity pattern in the direction perpendicular to flow, as shown in Fig. 3(d), cells would move up to the top and then move down to the bottom when encountering GBs. Such velocity pattern is expected to increase the hydrodynamic efficiency of the device in capturing cells.
Figure 3.
(a) Velocity patterns in the device at 10 μm height; (b) Stream lines for the velocity field at 10 μm height. The flow rate is set as 1 ml/h. (c) Velocity pattern along the inlet flow. The direction and weight of each arrow depict fluid movement and amplitude of the force, respectively; (d) Velocity pattern perpendicular to the flow with arrow size depicting the amplitude of the velocity vector. The positions of the three slices are at the central plane, and 15 μm before and after the central plane of beads. (e) Average number ± standard deviation of captured hGBM cells at specific distances from inlet on anti-EGFR and mutant aptamer functionalized plain Hele-Shaw devices.
Shear stress would also play an important role in cell capture. Fig. 2 in ESI shows the shear stress in the flow direction. At the top of the GB, the shear stress reaches its maximum. The large shear stress would stretch and rotate the cells so that they would roll downwards and adhere to the lower part of the GB. By reducing the distance between adjacent GBs from 50 μm to 20 μm, it was seen that the shear stress increased from 0.237 Pa to 0.303 Pa. In order to quantify capture efficiency, 2000 cells with randomly distributed initial positions were simulated for injection at the inlet of microfluidic device with various GB distance configurations. The cells were subjected to both bulk fluid flow and Brownian motion. Periodic boundary conditions were applied at the inlet and outlet. Trajectories of a few cells in the device are shown in Fig. 3(a) of ESI. For the computation, the binding efficiency was defined as the ratio of the number of cells bound with GBs and the total number of cells ultimately released. This changed as a function of time for devices with GB separation distances from 20 to 50 μm (ESI Fig. 3(b)). The binding efficiency increased very fast in the first 0.3 sec and quickly reached equilibrium. This indicated that most captured cells were located in flow streamlines adjacent to the GBs, and Brownian motion did not significantly influence cell capture. The binding efficiency also increased from 1% to 4% as the separation distance decreased from 50 to 20 μm. This was due to the packing of many more GBs in the device at 20 μm separation distance and the more available surface for contact and binding. The 20 μm spacing was thus employed in the fabricated Hele-Shaw devices (Fig. 4).
Figure 4.
SEM micrographs of Hele-Shaw microfluidic device with pits and loaded glass beads. (a) Shows the equilateral triangle array of pits (50 μm diameter, 25 μm depth, 20 μm spacing); (b) Shows the glass beads loaded in the arrayed pits made in PDMS. The scale bars are 100 μm in both micrographs. The inset to (b) shows a close-up view of a re-suspended glass bead with hGBM cell. Flow rate of cell suspension was set as 1 ml/h.
Shear-stress Studies using Linear-shear Hele-Shaw Chamber
Optimizing shear stress is important for flow-through devices meant to capture targets based on probe-target affinity. Too high of shear-stress results into lost sensitivity (no cells are captured) and too small of shear stress looses selectivity (all cells care captured). The stress has to equilibrate the force of binding between affinity aptamer and the EGFR. Cells have different EGFR concentrations so optimized shear stress should give the best sensitivity and selectivity. The results in the Hele-Shaw device showed the density of captured cells increased as the shear stress decreased (ESI Fig. 4). The optimum shear stress not only significantly improved the isolation specificity but also ensured maximum cell capture (sensitivity), thus meeting the goal for a high-throughput device.
The average size of hGBM cells was 12.2 μm (S.D: 2.31). The density of EGFR on hGBM cell membrane has been calculated before and as a lower limit there is roughly 1 EGFR per 100 nm2.25 This number comes by calculating the surface area of the cell and the total amount of EGFR on tumor cells.26
Tumor cells are known to have up to millions of proteins per cell in some tissue culture cell lines.27 The anti-EGFR aptamer/EGFR binding process can be considered to be instantaneous. Once EGFR firmly binds to anti-EGFR aptamer, the total binding force can be predicted from the Bell’s model.28 According to this model, the binding force between one anti-EGFR aptamer and EGFR is around 8×10−6 dynes. Considering the EGFR density on hGBM cells to be 1×1015/m2, the total binding force for 12 μm diameter hGBM cell firmly bound to EGFR aptamer functionalized surface is around 0.63 dynes. Because the expression levels of EGFR in cancer cells can be over 100 times higher than those in normal cell (such as white blood cells which average in size to be around 10 μm), the total binding force for normal cell would be only 0.16 dynes, or even less. It is important to note here that cells, normal or diseased, show a statistical distribution in EGFR expression. The difference of expression between normal and diseased cells is however high.15 Such difference was used for cancer cell isolation from the cell suspension under suitable shear stresses. Shear stress differences coupled with the remarkable difference in EGFR expression between normal and cancerous cells would result in the sweet spots where shear stress was just enough to sweep away normal cells (loosely bound) but not the tumor cells. The binding force between aptamer and EGFR is thus an important factor to estimate shear stress qualitatively.
In this experiment, a plain surface Hele-Shaw microfluidic device was used to test the influence of changing flow rate (and shear stress) on cell capture. As mentioned before, the adhesion force of hGBM cells was almost four times than that of normal cells. We deduce that if the shear stress could be kept higher than 0.16 dynes, the nonspecific adhesion of normal cells would be minimized. When the flow rate was set as 1 ml/h, at 40 mm position along the flow axis the shear stress was close to 0.2 dynes which was still higher than adhesion force of normal cell (ESI: Fig. 1(b)). With higher flow rate, the isolation specificity could be further improved; meanwhile, lower flow rate would yield higher capture efficiency at the expense of selectivity. In a practical application, the selection of flow rate through practical CTC isolation chip will depend on the competing goals of isolation efficiency (sensitivity) and specificity (selectivity).
The captured cells were counted every 5 mm along the flow axis from the inlet. The corresponding shear stress value and average cell densities measured on two different aptamer functionalized substrates are shown in Fig. 4 of ESI. It should be noted here that flow had little diffusion component under these conditions. The calculated Péclet numbers given in Table 1 of ESI show that the flow was mainly convective and not diffusive. Péclet number (Pe) is defined as: Pe=L U/D where L is the characteristic length of the channel, U is the corresponding fluid velocity, and D is the cell diffusion coefficient. Here, L was 30 μm, U was the velocity at each point in the channel and D was 4.4×10−14 m2/s for cells. The calculated Péclet number for cell transport in the channel was on the order of 105, which is ≫1. This meant that the flow was dominantly convective.
On anti-EGFR and mutant aptamer functionalized substrates, cell densities (cells per mm2) decreased linearly with distance. However, number of cells captured was very different and the trend was distinctly different (going from 5 to 40 mm). The results show that the density of captured cells significantly increased as the shear stress decreased along the axis, and the maximum density was achieved at 40 mm position (where shear stress was 0.2 dynes) (Fig. 3(e)). Philips et al.13 demonstrated that more cells can be isolated if higher concentration of aptamers are used during immobilization. However, on smooth glass slide, the best DNA probe density is around 1012 molecules/cm2.29 Higher density only causes increased steric effect which can inhibit hybridization between capture probe DNA and aptamer. Nevertheless, on 100 nm2 area the number of aptamers can reach 20 to 25 molecules which are sufficient for only one EGFR in the same area. In other words, with an optimal density of aptamers, it is only the EGFR density of cell membrane that becomes the most important factor for cell isolation. Use of nano-textured substrates can increase the number of aptamers available but then again, the limiting factor remains the EGFR concentration on the cell wall. Nano-textured surfaces has been seen to increase aptamer efficiency in capturing tumor cells but for entirely other reasons.15 Most important feature of nano-texturing is presentation of biomimetic surface emulating nanostructured characteristics of the basement membrane that is known to improve cell adhesion and growth.30 The use of GBs has its own advantages in lab-on-a-chip settings.
Cell Capture with Glass Bead Array
Hele-Shaw microfluidic device with pits (25 μm depth) were fabricated using PDMS molding (Fig. 4(a)). The diameters of pits were set to be 50 μm for loading GBs, and the equilateral triangle pattern had 20 μm spacing following simulation results (ESI Fig. 3(b)). From the cross-section micrographs, the wall of each pit was seen to be vertical, and the depth and diameter conformed to the original design. The aptamer functionalized GBs carried overall negative charges. The repulsion between GBs ensured that these neither formed clumps nor aggregated in buffer solution. The loading efficiency was seen to be very low if hydrophilic aptamer functionalized GBs were prepared in PBS. Most of GBs remained in PBS instead of falling into the pits, or these were easily removed from the pits just with gentle shaking. This was possibly due to low surface energy of PDMS.31 The surface energy of the PDMS dictated the wetting degree when the GB solution came in contact with the surface.32 A solution that provided low surface tension as well as kept DNA/aptamer hybridized was preferred. Pure ethanol, which met both requirements, was thus adopted.33 PDMS device was also treated with oxygen plasma to make its surface hydrophilic. The device contained an array of 250,000 pits within 516 mm2 surface, and 40 mg GBs were prepared to fill up the pits. The GBs were added to the device, and a flat PDMS cover was bonded to the device after steady shaking. The loading efficiency (the ratio of the number of loaded pits to the whole number of pits on the surface) was 98% (S.D.: 0.2). Excessive ethanol was completely removed by PBS rinsing; which also removed unloaded GBs.
To determine the cell isolation efficiency, 100 hGBM cells, suspended in PBS, were injected, and the capture efficiency with aptamers was seen to be 44.3% (S.D.: 2.5). Although antibodies have also been used before to capture tumor cells based on EGFR overexpression, it is known that this aptamer provides better affinity and selectivity under same conditions as aptamer.12 Fig. 4(b) shows two hGBM cells captured at the 40 mm position along the axis. The higher surface energy and hydrophilic surface on PDMS was achieved by oxygen plasma.34 No cells were captured at the top of GBs due to higher shear stress (3.03 dynes per cm2). The shear stress was much lower at the bottom of GBs (ESI Fig. 2), thus it led to higher chance of cell adhesion there. Cell morphology on GBs was also significantly different from what it looked like on flat glass or nano-textured PDMS surfaces.12,15 Cells did not change from spherical shape (in suspension) to flat on the aptamer grafted GB surface, but kept semi-elliptical shapes. Cells are known to resist higher shear stress when flattened,18,19 however, the height of cell did not decrease too much on GBs due to curved surface, and this might have been one cause for relatively lower capture efficiency.
The GBs with captured cells were easily removed from their pits by simply placing the device upside down. Fig. 4(b) inset shows the collected GBs which carried captured hGBM cells. At this step eluting or proteolytic enzyme treatment to remove cells had the potential to harm the completeness of cell structure. The proteolytic enzyme method could totally digest the receptors on cell membrane. Instead, to remove the cancer cells from GBs we used gentle pipetting and an anti-sense RELease molecule.
Cell Detachment by RELease Oligonucleotide
The cell attachment on GB surfaces was seen to be not as stable as it was reported previously on flat glass or nano-textured PDMS surfaces.12,15 These cells could be eluted from the GB surfaces by gently pipetting the solution in and out. During the soft resuspending process, the agitation resulted into higher shear stresses on the GBs. An estimate of the shear stress stemming from pipetting can be made from lower limit of 0.63 dynes. From pipetting a flow rate of 5 ml/sec could be achieved which when compared to 1 ml/hr flow rate results into as high a shear stress as 18000 more than the capture flow rate. The 1 ml/hr flow that was used during capture resulted into a shear stress of 0.2 dynes at 40 mm (ESI Fig. 1(b)). This gentle pipetting removed cells from GBs and these could be harvested from the supernatant. However, the releasing efficiency was not satisfactory. Vigorous shaking could have hurt the cell completeness. To retrieve all the cells that were bound to GBs and retrieve these without damaging the cell walls, the RELease RNA agent was used.
The RELease moleculee hybridized with cell-bound anti-EGFR aptamer to release cells from GB surface. The interactions of RELease molecule and the anti-EGFR aptamer were analyzed using native polyacrylamide gel electrophoresis (PAGE). The results showed that the RELease molecule could bind with the aptamer (Fig. 5(a)). Note mobility shift of the anti-EGFR aptamer+FACS+RELease to a higher weight relative to anti-EGFR aptamer without the REL. The schematic shown in Fig. 2 shows the FACS experiment strategy.
Figure 5.
(a) Native polyacrylamide gel electrophoresis (PAGE) shows the release effect of the RELease sequence. The FACS extension binds to the capture probe and when RELease sequence is used the band for anti-EGFR aptamer shifts showing structure changes in the aptamer. The structure change looses binding capability with the cellular EGFR. (b) Fluorescence intensity of FACS flow cytometer data shows the shift in signal from just cells to the cell bound with anti-EGFR aptamer construct and then upon introduction of RELease molecule. RELease agent significantly affects the fluorescence intensity before and after removing the anti-EGFR aptamer.
Cells were also treated with the aptamer-DNA-biotin-SAPE complex and then washed to remove free EGFR aptamer. The control experiments here showed that DNA complementary to RNA aptamer was not able to release the aptamer from the cells. It could be that DNA-RNA interactions were insufficiently strong to denature the RNA-EGFR interactions. The Fig. 5(b), however, shows flow cytometry results indicating that anti-EGFR aptamer binds to cells but can be unbound with the RELease RNA that is not only complementary to aptamer but also carries a toehold part (labeled as anti-EGFR aptamer + REL). In that case a 76% reduction in the median fluorescence was seen. On the other hand, Mutant aptamer (Mutant Apt) or the FACS extension (DNA-alone) produced fluorescence levels just slightly above background. The fluorescence for complex of anti-EGFR aptamer and RELease was slightly above this nonspecific binding level. Without RELease, the aptamer with toehold bound the cells comparably to the original aptamer (without toehold). Furthermore, random RNA did not cause release of the EGFR aptamer (Anti EGFR Apt + RNA). This also showed that aptamer did not get internalized to the cell otherwise it would have not shown selectivity to RELease RNA with respect to random RNA. The cell incubation experiment was also performed at 4 °C (which would inhibit internalization) and the results were identically the same (thus data not shown).
There were on average 73 cells (S.D.:11) adhered to 100 GBs after flow-through capture of hGBM cells. In control group which was not incubated with RELease, there were on average 23 cells (S.D.: 6) still attached on 100 GBs after soft resuspending. In other words, soft suspending could remove around 69% captured cells from GBs surface. However, the GBs which were further treated with RELease, only 6 cells (S.D.: 3) could remain on 100 GBs surface. The release efficiency was thus improved to 92%. This should be noted here that apparently the RELease only resulted in 23% more cells to be released but it would have probably released many more cells that were bound if no soft suspending was employed. Two tiered release thus increased the recovery of isolated cells by 23% which would be much more important and evident if we would want to isolate and recover 5–10 CTCs from a poll of billion normal cells.
Once RELease hybridized with anti-EGFR aptamer, it completely opened up aptamer’s hairpin structure. The aptamer could thus no longer specifically bind to EGFR. The results from flow cytometry revealed that 76% of anti-EGFR aptamers could be released from the surface. The rest might not have interacted with RELease, or these aptamers might have entered into the cytoplasm via EGFR mediated endocytosis. Overall, because of RELease effect on the hairpin structure of anti-EGFR aptamer, the cell adhesion force significantly decreased; therefore cells could be easily detached from GBs during the harvesting process.
CONCLUSION
It has been shown that anti-EGFR aptamer grafted glass bead array can recognize and capture hGBM cells that overexpress EGFR, which is an important biomarker. The device flow through velocity of 1 ml/h gives a single-pass capture of tumor cells. The cells are detached from the glass beads for further analysis using an antisense release oligonucleotide which increases the recovery of tumor cells to 92%. The ultimate goal of such a microfluidic device is to isolate circulating tumor cells from peripheral blood, given translation of the approach from controlled samples in lab to real human samples is smooth. There may be challenges like varying and diverse overexpression of biomarkers, varying recovery of sub-populations of diseased cells, molecular degradation activities, and such. In any case, the presented framework can dramatically facilitate cell harvest, intervention and prognosis monitoring of known metastasis.
Supplementary Material
Acknowledgments
We are thankful to Robert Bachoo from Department of Neurology at University of Texas Southwestern Medical Center, Dallas, Texas for patient samples. This work was supported with NSF CAREER grant (ECCS-0845669) to SMI. YL acknowledges support from NIH grant EB009786 and NSF grant CBET-1067502. YTK acknowledges support from UTA Nano-bio cluster program. ADE acknowledges support from Welch Foundation grant F-1654, NIH 1-R21-HG005763-01 and National Cancer Institute Award Number 5R01CA119388-05. PBA acknowledges support from NIH fellowship GM095280.
References
- 1.Paterlini-Brechot P, Benali NL. Cancer letters. 2007;253:180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Journal of the American Chemical Society. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He W, Wang H, Hartmann LC, Cheng JX, Low PS. Proceedings of the National Academy of Sciences. 2007;104:11760. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald MP, Neale S, Paterson L, Richies A, Dholakia K, Spalding GC. Journal of biological regulators and homeostatic agents. 2004;18:200–205. [PubMed] [Google Scholar]
- 6.Tibbe AGJ, de Grooth BG, Greve J, Dolan GJ, Rao C, Terstappen LWMM. Cytometry Part A. 2002;47:163–172. doi: 10.1002/cyto.10060. [DOI] [PubMed] [Google Scholar]
- 7.Lee H, Sun E, Ham D, Weissleder R. Nature medicine. 2008;14:869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng J, Sheldon EL, Wu L, Heller MJ, James POC. Analytical chemistry. 1998;70:2321–2326. doi: 10.1021/ac971274g. [DOI] [PubMed] [Google Scholar]
- 9.Toner M, Irimia D. Biomedical Engineering. 2005:7. doi: 10.1146/annurev.bioeng.7.011205.135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. The Journal of Cell Biology. 192:373. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman K, Pullen N, Graham M, Ragan I. Nature Reviews Drug Discovery. 2007;6:120–126. doi: 10.1038/nrd2242. [DOI] [PubMed] [Google Scholar]
- 12.Wan Y, Kim Y-t, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Cancer Research. 2010;70:11. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 13.Phillips JA, Xu Y, Xia Z, Fan ZH, Tan W. Analytical chemistry. 2008;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Fabry B, Schiffrin EL, Wang N. American Journal of Physiology- Cell Physiology. 2001;280:1475–1484. doi: 10.1152/ajpcell.2001.280.6.C1475. [DOI] [PubMed] [Google Scholar]
- 15.Wan Y, Mahmood MAI, Li N, Allen PB, Kim Y-t, Bachoo R, Ellington AD, Iqbal SM. CANCER. 2012;118:1145–1154. doi: 10.1002/cncr.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ernst O, Lieske A, Jäger M, Lankenau A, Duschl C. Lab Chip. 2007;7:1322–1329. doi: 10.1039/b708619a. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Yan J, Revzin A. Colloids and Surfaces B: Biointerfaces. 2008;64:260–268. doi: 10.1016/j.colsurfb.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 18.Lu H, Koo LY, Wang WM, Lauffenburger DA, Griffith LG, Jensen KF. Analytical chemistry. 2004;76:5257–5264. doi: 10.1021/ac049837t. [DOI] [PubMed] [Google Scholar]
- 19.Cheung LSL, Zheng X, Stopa A, Baygents JC, Guzman R, Schroeder JA, Heimark RL, Zohar Y. Lab Chip. 2009;9:1721–1731. doi: 10.1039/b822172c. [DOI] [PubMed] [Google Scholar]
- 20.Vickers DAL, Kulik M, Hincapie M, Hancock WS, Dalton S, Murthy SK. Biomicrofluidics. 2012;6:024122. doi: 10.1063/1.4719979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White RR, Sullenger BA, Rusconi CP. Journal of Clinical Investigation. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng H, Ye BC. Applied biochemistry and biotechnology. 2009;152:54–65. doi: 10.1007/s12010-008-8245-9. [DOI] [PubMed] [Google Scholar]
- 23.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 24.Usami S, Chen HH, Zhao Y, Chien S, Skalak R. Annals of biomedical engineering. 1993;21:77–83. doi: 10.1007/BF02368167. [DOI] [PubMed] [Google Scholar]
- 25.Wan Y, Tan J, Asghar W, Kim Y-t, Liu Y, Iqbal SM. The Journal of Physical Chemistry B. 2011;115:13891–13896. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 26.Carpenter G, Stoscheck CM, Soderquist AM. Annals of the New York Academy of Sciences. 1982;397:11–17. doi: 10.1111/j.1749-6632.1982.tb43412.x. [DOI] [PubMed] [Google Scholar]
- 27.Carpenter G, Cohen S. Journal of Biological Chemistry. 1990;265:7709–7712. [PubMed] [Google Scholar]
- 28.Bell GI. Science. 1978;200:618. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 29.Fuentes M, Mateo C, Garcia L, Tercero JC, Guisan JM, Fernandez-Lafuente R. Biomacromolecules. 2004;5:883–888. doi: 10.1021/bm0343949. [DOI] [PubMed] [Google Scholar]
- 30.Ruoslahti E. Scientific American. 1996;275:72–78. doi: 10.1038/scientificamerican0996-72. [DOI] [PubMed] [Google Scholar]
- 31.Mata A, Fleischman AJ, Roy S. Biomedical microdevices. 2005;7:281–293. doi: 10.1007/s10544-005-6070-2. [DOI] [PubMed] [Google Scholar]
- 32.Nun E, Oles M, Schleich B. Wiley Online Library. 2002. pp. 677–682. [Google Scholar]
- 33.Dean JA. Lange’s handbook of chemistry. 1985. [Google Scholar]
- 34.Fuard D, Tzvetkova-Chevolleau T, Decossas S, Tracqui P, Schiavone P. Microelectronic Engineering. 2008;85:1289–1293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.