Abstract
The combination of pegylated interferon (PEG-IFN) and ribavirin is the standard treatment for chronic hepatitis C. Our recent clinical study suggests that ribavirin augments the induction of interferon-stimulated genes (ISGs) in patients treated for hepatitis C virus (HCV) infection. In order to further characterize the mechanisms of action of ribavirin, we examined the effect of ribavirin treatment on ISG induction in cell culture. In addition, the effect of ribavirin on infectious HCV cell culture systems was studied. Similar to interferon (IFN)-α, ribavirin potently inhibits JFH-1 infection of Huh7.5.1 cells in a dose-dependent manner, which spans the physiological concentration of ribavirin in vivo. Microarray analysis and subsequent quantitative polymerase chain reaction assays demonstrated that ribavirin treatment resulted in the induction of a distinct set of ISGs. These ISGs, including IFN regulatory factors 7 and 9, are known to play an important role in anti-HCV responses. When ribavirin is used in conjunction with IFN-α, induction of specific ISGs is synergistic when compared with either drug applied separately. Direct up-regulation of these antiviral genes by ribavirin is mediated by a novel mechanism different from those associated with IFN signaling and intracellular double-stranded RNA sensing pathways such as RIG-I and MDA5. RNA interference studies excluded the activation of the Toll-like receptor and nuclear factor κB pathways in the action of ribavirin.
Conclusion
Our study suggests that ribavirin, acting by way of a novel innate mechanism, potentiates the anti-HCV effect of IFN. Understanding the mechanism of action of ribavirin would be valuable in identifying novel antivirals
Ribavirin, a guanosine analog, demonstrates a broad spectrum of antiviral activity in tissue culture against numerous DNA and RNA viruses. Currently, it is used in conjunction with pegylated interferon (IFN)-α as standard treatment for chronic hepatitis C virus (HCV) infection.2 Ribavirin has transient antiviral activity in patients with HCV when used as monotherapy,3 but markedly improves treatment response when combined with IFN.4 Its role in improving treatment outcomes is well established, but the precise mechanism by which it acts synergistically with IFN has remained elusive. In patients, ribavirin appears to enhance the second and third phases of viral decay, thereby limiting relapse, particularly in patients with suboptimal responses to IFN.5 Although several mechanisms of action have been proposed for ribavirin’s antiviral activity, each with some experimental evidence, none has been convincingly shown to explain the combined effect of ribavirin with IFN in HCV antiviral therapy.6
There are five proposed mechanisms by which ribavirin exerts its antiviral activity.7 These can be subdivided into direct and indirect mechanisms based on whether they directly impact the virus life cycle. The direct mechanisms of action include ribavirin acting as a mutagen through its incorporation into the HCV genome, subsequently resulting in error catastrophe,8,9 and ribavirin as an inhibitor of the HCV RNA–dependent RNA polymerase.10 There are three indirect mechanisms of action that have been proposed. First, ribavirin is an inhibitor of the ionosine monophosphate dehydrogenase (IMPDH) enzyme, resulting in depletion of intracellular guanosine pools, which are needed for efficient viral replication.11,12 Ribavirin has also been shown to stimulate a T helper 1 antiviral response that favors viral clearance.13 Lastly, ribavirin has recently been shown to modulate the expression of interferon-stimulated genes (ISGs) in patients receiving a modified antiviral treatment regimen with a loading dose of ribavirin monotherapy given before the initiation of standard combination therapy and in a tissue culture model of respiratory syncytial virus infection.14–16 When we examined gene expression patterns of patients treated with this modified treatment regimen, we found that patients treated with ribavirin and pegylated interferon (PEG-IFN) had a higher level of ISG induction than patients who received PEG-IFN alone.14 Because ribavirin confers a significant therapeutic efficacy when given in combination with IFN,4 we hypothesized that ribavirin may augment the activity of IFN by interacting with its antiviral signaling cascade.
Type I IFNs are produced by most cells in the body in response to viral infection and other cellular stressors. After its production, IFN-α and IFN-β act in an autocrine and paracrine manner. Binding to the type I IFN receptor leads to activation of the Janus kinase–signal transducer and activator of transcription (STAT) pathway, resulting in phosphorylated STAT molecules translocating to the nucleus, combining with IFN regulatory factor (IRF) 9 to form a complex that binds to the IFN-sensitive response element, a promoter upstream of a large number of genes collectively known as ISGs. It is through the action of these genes that IFN-α is thought to exert its antiviral activity.17
Ultimately, the antiviral effect of ribavirin observed in HCV-infected patients may involve multiple mechanisms. However, little attention has been focused on the fact that ribavirin greatly enhances the clinical effect of IFN, and yet has minimal or no long-term antiviral effect of its own.18 This suggests that ribavirin functions by modulating the IFN response. Elucidating the interplay between these two agents will likely be important both for understanding and potentially augmenting their clinical effects.
Patients and Methods
The complete Patients and Methods are provided in the Supporting Information.
Results
Up-Regulation of IRF7 and IRF9 by Ribavirin in Patients
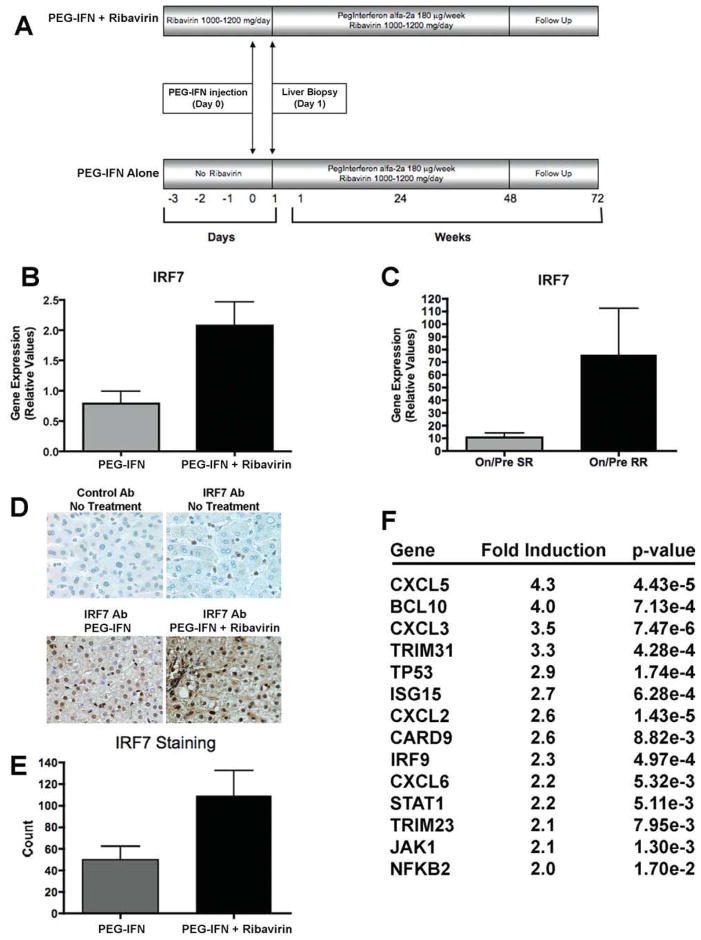
We previously conducted a clinical study in patients with HCV to examine the mechanisms of action of ribavirin in combination with PEG-IFN.14 Patients were separated into two cohorts. The first cohort received weight-based ribavirin monotherapy for 3 days before starting standard antiviral treatment. Twenty-four hours after their first injection of PEG-IFN, a liver biopsy was performed. The second cohort received PEG-IFN only and underwent a liver biopsy 24 hours later (Fig. 1A). To further examine these results, gene expression analysis revealed that IRF7 levels were higher in patients receiving ribavirin and PEG-IFN than in those on PEG-IFN alone (Fig. 1B); furthermore, higher IRF7 messenger RNA (mRNA) levels were present in patients who displayed a superior response to treatment (Fig. 1C). In addition to IRF7, other ISGs such as IRF9 were found to be higher in patients receiving combination therapy.14 To confirm the relevance of the microarray results, we performed immunohistochemical analysis on the liver biopsy specimens and found higher levels of IRF7 staining in patients receiving ribavirin and PEG-IFN than in patients receiving PEG-IFN alone (Fig. 1D, E).
Fig. 1.
IRF7 expression in liver biopsy samples from HCV-infected patients and induction of ISGs by ribavirin. (A) Treatment regimen of clinical protocol (IRB-UNC GCRC 1728). (B) IRF7 mRNA levels, determined by way of qPCR, in liver biopsies from patients receiving PEG-IFN (n = 5) versus PEG-IFN plus ribavirin (n = 6) (P = 0.035). (C) qPCR analysis of IRF7 mRNA induction by dividing on-treatment over pretreatment mRNA levels in liver biopsies from patients demonstrating a slow virological response (SR, n = 5) and a rapid virological response (RR, n = 6) (P = 0.004). Virological response was defined by posttreatment viral kinetics. (D) IRF7 staining in liver biopsies. Immunoglobulin G and IRF7 antibodies (Santa Cruz Biotechnology) were used to stain untreated liver biopsies. (E) Quantification of IRF7 staining using ImageJ software (P = 0.08). (F) Microarray analysis demonstrating gene induction of several ISGs after treatment with ribavirin for 24 hours at a dose of 100 μg/mL in uninfected Huh7.5.1 cells. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Up-Regulation of Select ISGs in Cultured Cells Treated with Ribavirin
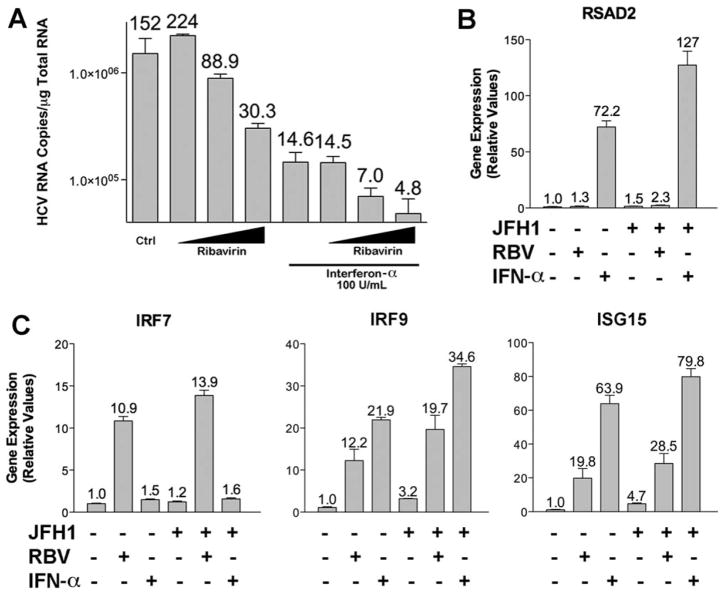
In order to study ribavirin’s action further, we used tissue culture models to assess the effect of ribavirin on mammalian cells with and without HCV infection. Microarray analysis of uninfected human hepatoma (Huh7.5.1) cells19 treated with ribavirin for 24 hours demonstrated that several ISGs, including IRF9 and ISG15, were induced by ribavirin treatment (Fig. 1F). In this experiment, ribavirin also up-regulated IRF7 mRNA levels as determined by quantitative polymerase chain reaction (qPCR) (data not shown). Because ISGs are known to contribute to the antiviral activity of IFN,17 we next determined whether ribavirin possessed antiviral activity against HCV using the JFH1 infectious clone in Huh7.5.1 cells.20 Ribavirin exhibited dose-dependent antiviral activity against HCV (Fig. 2A). Similarly, IFN-α used at a dose of 100 U/mL displayed significant antiviral activity that could be further increased by the addition of increasing doses of ribavirin (Fig. 2A). Notably, the two antiviral compounds had different effects on gene expression. As expected, IFN greatly up-regulated a host of classical ISGs (data not shown), whereas the effect of ribavirin was more selective, affecting the expression of a subset of these genes (Fig. 1F). IFN strongly induced RSAD2, but ribavirin had a minimal effect (Fig. 2B). In contrast, ribavirin increased the levels of IRF7, IRF9, and ISG15 (Fig. 2C). These data show that ribavirin induces a subset of ISGs and that the combined action of each agent has a potent inhibitory effect on HCV.
Fig. 2.
Antiviral activity of ribavirin and IFN-α on HCV infection and subsequent gene induction. (A) qPCR analysis of viral RNA after 28 hours of infection with JFH1 (multiplicity of infection = 0.5) in Huh7.5.1. with ribavirin (1, 10, 100 μg/mL) and IFN-α (100 U/ mL) separately or in combination for 24 hours. Numbers above the error bars represent HCV RNA copies × 104. (B,C) qPCR of mRNA levels after 24 hours of treatment with ribavirin (100 μg/mL) and IFN-α (100 U/mL) for RSAD2 (B) and IRF7, IRF9, and ISG15 (C) genes in cells with and without JFH1 infection. Data represent experiments performed in triplicate.
Synergistic Induction of ISGs by Ribavirin and IFN-α Treatment
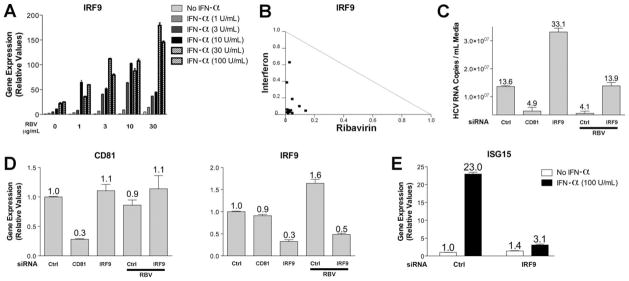
To further investigate ribavirin’s effect on gene expression, we performed dose-response and time course experiments with ribavirin. Supporting Fig. 1A demonstrates the reproducible up-regulation of IRF7 mRNA levels with increasing doses and length of treatment with ribavirin. We observed similar results in ribavirin-treated HepG2 cells (Supporting Fig. 1B). Because of the significant clinical benefit observed with the combination of IFN-α and ribavirin, we specifically evaluated whether ribavirin and IFN-α have a synergistic effect on gene expression in cultured cells. Treatment with IFN-α resulted in maximal gene induction at approximately 8 hours with lower levels of mRNA observed at 24 hours after treatment (Supporting Fig. 2). Because ribavirin and IFN have distinct kinetics with respect to gene induction, we chose to study gene expression after treatment with each drug, alone or in combination at various concentrations, at their maximal point of induction. IRF9 mRNA levels were extensively examined, because IRF9 was up-regulated by both IFN-α and ribavirin (Fig. 2C). Fig. 3A demonstrates greater gene induction of IRF9 by the combination of IFN and ribavirin than either compound alone, in a concentration-dependent manner, even at the lowest doses used (1 and 3 μg/mL). For example, IFN-α treatment alone resulted in a 25-fold induction of IRF9 mRNA levels at a dose of 100 U/mL, but when combined with 3 μg/mL of ribavirin, resulted in approximately 80-fold induction. A normalized isobologram was generated from the qPCR data, and it demonstrated synergy as combinations plotted to the left of the corresponding line of additivity (Fig. 3B). We also observed similar results for ISG15 mRNA levels after treatment with ribavirin and IFN and confirmed the ability of ribavirin to increase ISG induction by IFN in Huh7.5.1 and Huh7 cells (Supporting Fig. 3).
Fig. 3.
Synergistic up-regulation of IRF9 mRNA levels following ribavirin and IFN-α combination treatment and effect of IRF9 suppression on HCV infection and antiviral effects of ribavirin. (A) qPCR analysis of IRF9 induction after treatment with ribavirin (1, 3, 10, 30 μg/mL for 24 hours) and IFN-α (1, 3, 10, 30, 100 U/mL for 8 hours) in Huh7.5.1 cells. (B) Normalized isobologram demonstrating synergy between ribavirin and IFN-α on IRF9 mRNA levels for 14 combinations of the two drugs generated with Calcusyn software. (C) Effect of siRNA-mediated suppression, performed as described in the Materials and Methods, of IRF9 and CD81 on JFH1 replication (multiplicity of infection = 0.5 for 40 hours) with or without ribavirin (100 μg/mL for 36 hours). Nonspecific siRNA was used as a baseline control. Numbers above error bars represent HCV RNA copies × 106. (D) qPCR analysis of CD81 and IRF9 mRNA levels following siRNA mediated gene suppression. (E) qPCR analysis of ISG15 mRNA levels following treatment with IFN-α (100 U/mL for 24 hours) in cells treated with nonspecific and IRF9 siRNAs. Data represent experiments performed in triplicate.
Because ribavirin and IFN-α resulted in synergistic induction of IRF9 mRNA levels, we next determined the effect of IRF9 on HCV replication using RNA interference (RNAi). As a transfection and functional control, we used short interfering RNA (siRNA)-mediated suppression of CD81, which resulted in decreased levels of HCV RNA (Fig. 3C).21 In contrast, suppression of IRF9 levels by RNAi resulted in increased levels of HCV RNA, illustrating an antiviral role for IRF9 on HCV replication. In addition, suppression of IRF9 levels abrogated ribavirin’s antiviral effect compared with the nontargeting control siRNA (Fig. 3C). RNAi-mediated gene suppression was confirmed by qPCR for CD81 and IRF9 (Fig. 3D).
Because patients treated with combination therapy were already infected with HCV, we hypothesized that the presence of virus may also contribute to an increased effect of ribavirin and IFN-α on gene induction. Interestingly, high ISG levels have been observed in liver biopsies obtained from HCV-infected patients and chimpanzees.14,22,23 To mimic the effect seen in patients infected with HCV, we used polyinosinic:polycytidylic acid [poly(I:C)], a double-stranded RNA viral mimetic, in conjunction with ribavirin and IFN. We were able to detect low-level ISG induction by JFH1 in Huh7.5.1 cells in tissue culture (Fig. 2B, C) as described.24 Supporting Fig. 4A shows the effect of transfected poly(I:C) on gene induction with mRNA levels increasing over time for selected genes. Ribavirin, poly(I:C), and IFN-α treatment had very little effect on IFN-β mRNA levels (Supporting Fig. 4B). Poly(I:C) had a minimal effect on IFN-β expression because these cells harbor a defective RIG-I pathway.25 However, the addition of ribavirin augmented the induction of IFI44 and RSAD2 seen with poly(I:C) and/or IFN-α treatment separately or in combination (Supporting Fig. 4C).
Ribavirin Up-Regulates ISGs Through a Novel Mechanism
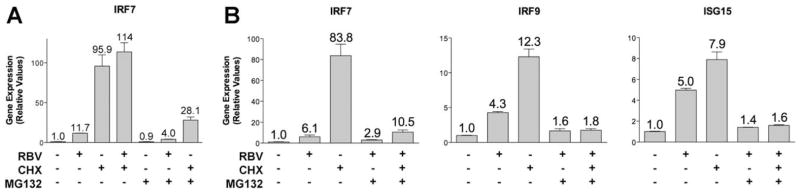
To investigate the mechanism by which ribavirin affects gene transcription, we first investigated whether de novo protein synthesis was required using cycloheximide, a molecule commonly used to block protein synthesis.26,27 Addition of cycloheximide 1 hour before ribavirin treatment did not block the induction of IRF7 mRNA levels, suggesting that new protein synthesis was not required. However, the addition of cycloheximide alone induced IRF7 expression, complicating the interpretation of this experiment (Fig. 4A). We next determined whether treatment with MG132, a proteosome inhibitor,27,28 could block the effect of ribavirin or cycloheximide on IRF7 induction. Indeed, treatment with MG132 greatly decreased the induction of IRF7 by either cycloheximide or ribavirin (Fig. 4A). Similarly, MG132 inhibited the effect of both ribavirin and cycloheximide on induction of IRF7, IRF9, and ISG15 (Fig. 4B). Together, these data suggest that a short-lived protein, possibly a transcriptional repressor, may be involved in the regulation of IRF7 and other ISGs and that ribavirin and cycloheximide may act on similar pathways.
Fig. 4.
MG132 blocks gene induction by cycloheximide and ribavirin. qPCR analysis of (A) IRF7 and (B) IRF7, IRF9, and ISG15 mRNA levels after treatment with ribavirin (100 μg/mL for 24 hours), cycloheximide (10 μg/mL for 25 hours), and MG132 (10 μM for 25 hours) in Huh7.5.1 cells.
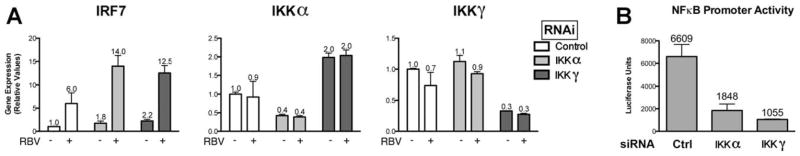
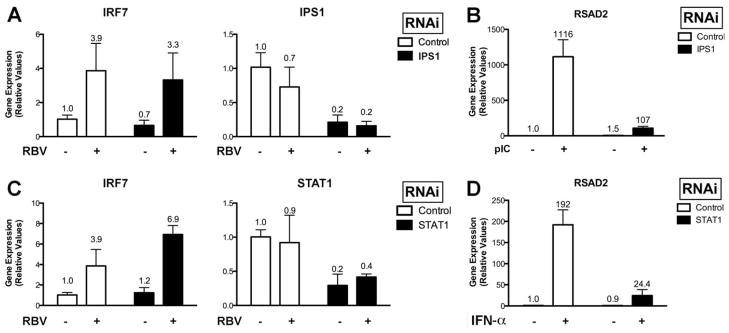
Because one consequence of MG132 treatment is the inhibition of the nuclear factor κB (NFκB)29 signaling pathway, we next investigated whether NFκB signaling was required for the induction of IRF7 mRNA levels by ribavirin. siRNA-mediated suppression of the NFκB signaling components I kappa B kinase (IKK) α and IKKγ resulted in a significant decrease in their respective mRNA levels and functional activity; however, ribavirin was equally effective in inducing IRF7 in these siRNA-treated cells (Fig. 5A, B). We have also used siRNAs targeting the transcription factors NFκB1, NFκB2, and RELA and saw no inhibition of ribavirin activity on gene induction (data not shown). Furthermore, Toll-like receptor 7 is involved in the recognition of guanosine analogues, an event that can lead to the activation of NFκB.30 However, we did not observe any detectable reduction in ribavirin activity in cells treated with siRNAs targeting TRIF and MyD88, critical adaptor molecules implicated in Toll-like receptor signaling and subsequent NFκB activation (Supporting Fig. 5).31 We also used RNA interference to determine if IFN signal transduction and intracellular double-stranded RNA sensing pathways such as RIG-I and MDA5 were involved in ribavirin-stimulated gene induction.32 siRNAs targeting IPS1 and STAT1, important signaling components of these pathways, had no effect (Fig. 6A–D). Therefore, ribavirin-stimulated induction of ISGs is not sensitive to inhibition of the aforementioned signaling pathways by siRNAs. In addition, we found that ribavirin was unable to stimulate both IFN-sensitive response element and NFκB promoter luciferase constructs (data not shown). Because both ribavirin and cycloheximide resulted in up-regulation of IRF7, IRF9, and ISG15, we determined the effect of these two molecules on the mRNA levels of the type I (α/β) and type III (IL28A, IL28B, and IL29) IFNs. Supporting Fig. 6A demonstrates that neither molecule up-regulated any of these cytokines, which are induced by similar signaling pathways.33 Furthermore, blocking type I IFN activity with a recombinant vaccinia virus-encoded neutralizing protein, B18R, had no effect on ribavirin-stimulated gene induction (Supporting Fig. 6B).
Fig. 5.
Gene induction by ribavirin does not involve NFκB signaling. (A) qPCR analysis of IRF7 mRNA levels in Huh7.5.1 cells first treated with siRNAs targeting IKKα and IKKγ and then treated with or without ribavirin (100 mg/mL) for 24 hours (with quantification of siRNA-mediated gene suppression). (B) NFκB promoter luciferase activity, 24 hours after plasmid transfection, in cells first treated with siRNAs for 3 days targeting IKKα and IKKγ
Fig. 6.
Gene induction by ribavirin does not involve RIG-I–like helicase or IFN signaling pathways. (A) qPCR analysis of IRF7 mRNA levels in Huh7.5.1 cells first treated with siRNAs targeting IPS1 and then treated with or without ribavirin. (B) qPCR analysis of RSAD2 mRNA levels following transfection of poly(I:C) (6 μg/mL for 8 hours) in cells treated with nonspecific and IPS1 siRNAs. (C) qPCR analysis of IRF7 mRNA levels in Huh7.5.1 cells first treated with siRNAs targeting STAT1 and then treated with or without ribavirin. (D) qPCR analysis of RSAD2 mRNA levels following treatment with IFN-α (100 U/mL) in cells treated with nonspecific and STAT1 siRNAs. Data represent experiments performed in triplicate.
Effects of Guanosine Analogues on ISG Expression
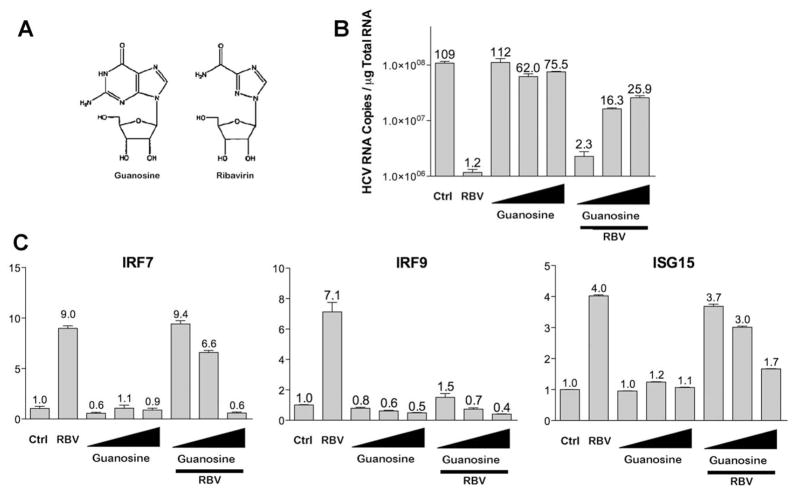
Because ribavirin is a guanosine analogue (Fig. 7A), we next determined whether guanosine itself could block ribavirin’s antiviral activity against the JFH1 infectious clone. We observed minimal antiviral activity after guanosine treatment; however, there was a dose-dependent reversal of ribavirin’s antiviral activity with concomitant addition of guanosine to the cell culture media (Fig. 7B). We next determined whether guanosine itself (Fig. 7A) could stimulate gene induction. Fig. 7C demonstrates up-regulation of IRF7, IRF9, and ISG15 by ribavirin; however, guanosine treatment had no effect on gene induction. We next determined whether addition of guanosine to the culture media could inhibit ribavirin-induced gene induction. qPCR analysis of Huh7.5.1 cells treated with ribavirin and guanosine clearly showed the ability of guanosine to reverse the effect of ribavirin on gene induction for IRF7, IRF9, and ISG15 (Fig. 7C). We have also performed experiments with the guanosine analogue loxoribine and saw no effect on ISG expression nor ribavirin antiviral activity (data not shown).
Fig. 7.
Guanosine does not stimulate ISG induction but can block ribavirin-stimulated gene induction and antiviral effects. (A) Chemical structures of guanosine and ribavirin. (B) qPCR analysis of viral RNA after 40 hours infection with JFH1 (multiplicity of infection = 0.5) in Huh7.5.1 cells. Four hours post infection, cells were treated with guanosine (1, 10, 100 μg/mL) and ribavirin (100 μg/mL) separately or in combination for 36 hours. Numbers above error bars represent HCV RNA copies × 106. (C–E) qPCR analysis of mRNA levels after treatment with guanosine (1, 10, 100 μg/mL for 24 hours) and ribavirin (100 μg/mL for 24 hours) separately or in combination for IRF7(C), IRF9 (D), and ISG15 (E). Data represent experiments performed in triplicate.
To additionally confirm that the effects of ribavirin are not cell type–specific, we also treated activated primary rat hepatic stellate cells with ribavirin. Activated hepatic stellate cells are thought to be the cell type responsible for the deposition of collagen in an injured liver,34 and ribavirin monotherapy has been shown to decrease liver inflammation as demonstrated by decreased alanine aminotransferase levels and improved liver histology in patients receiving ribavirin monotherapy.35 Furthermore, in our human study referenced above,14 we noted a substantial reduction of markers of hepatic stellate activation by ribavirin. Microarray analysis confirmed the induction of several ISGs, including OAS1, MX2, IRF7, and IRF9 (Supporting Fig. 7A), demonstrating that ribavirin’s effects on gene expression are not limited to hepatocyte-derived cells. Induction of IRF7 was also confirmed by real-time PCR (Supporting Fig. 7B) in these samples.
Discussion
Our data suggest that ribavirin may exert its antiviral activity in patients treated with combination therapy by potentiating the effect of IFN action on ISG induction, which results in enhanced antiviral responses. Although we observed the induction of several antiviral genes by ribavirin, IRF7 and IRF9 are of particular interest. Both IRF7 and IRF9 have been shown to be critical for antiviral responses in mice through their roles in endogenous IFN production and signaling.36,37 Furthermore, IRF9 (also known as ISGF3G) has been shown to have direct antiviral activity against HCV in tissue culture (Fig. 3C),38 and IRF7 mRNA levels from peripheral blood mononuclear cells have been shown to correlate with responses to antiviral therapy in HCV-infected patients.39 In addition to their direct antiviral effects, IRF7 and IRF9 may be of particular importance because of their role in amplifying the IFN signaling cascade. Improved IFN signaling through the up-regulation of these molecules would have the potential to significantly improve the effects of PEG-IFN treatment.
We have observed that treatment of cells in tissue culture with ribavirin leads to a dose-dependent induction of a subset of ISGs. This observation was unexpected given that the induction of ISGs is a tightly regulated process usually occurring only during viral infections and pathological states that result in the production of IFN.40 Furthermore, ribavirin was also shown to have antiviral effects on JFH1 infection (Fig. 2) and on various HCV replicon–containing cells12,41–43 when ribavirin was administered at similar doses used in this study.43 Several studies have demonstrated that ribavirin’s beneficial effect on HCV treatment outcomes is dose-dependent and that higher doses of ribavirin result in higher response rates.44–47 During antiviral therapy, the levels of ribavirin achieved in the serum are 1–4 μg/mL.48–50 Correspondingly, we have observed reproducible changes in gene expression and synergistic effects on gene induction by the combination of ribavirin and IFN with doses of ribavirin as low as 1 and 3 μg/mL in tissue culture, and these effects were dose-dependent (Fig. 3A, B; Supporting Figs. 1 and 3).
Previous pharmacodynamic studies have shown that an extended dosing period (≈4 weeks) is needed to reach steady-state blood concentration.51 Furthermore, the tissue concentration of ribavirin may be substantially higher, because ribavirin tends to concentrate in certain tissues. Indeed, several studies have shown that the intracellular concentration of ribavirin in erythrocytes can reach over 100 times greater than that that found in plasma52,53 and that hepatocytes have specific nucleoside transporters that facilitate uptake of ribavirin from plasma.54 In this study, we used ribavirin at doses up to 100 μg/mL in infected or uninfected cells for 24 hours in tissue culture experiments without any observable toxicity (Supporting Fig. 8 and data not shown).
The up-regulation of ISGs by ribavirin may be due to the inhibition of a transcriptional repressor. Other studies have shown that ISGs are under the control of transcriptional repressors, and this may be an increasingly important mechanism for ISG regulation.55 Our data also suggest that this repressor may be a short-lived protein degraded by the 26S proteosome. Consequently, treatment with proteosome inhibitors stabilize this protein, making cells resistant to the effect of ribavirin and cycloheximide on IRF7, IRF9, and ISG15 (Fig. 4A, B). The addition of guanosine to the medium can also block ribavirin-induced gene induction and antiviral activity in a dose-dependent fashion. The ability of guanosine to block ribavirin activity may point to involvement of the IMPDH pathway. IMPDH inhibition by ribavirin could induce guanosine depletion, and guanosine depletion may lead to IRF7/IRF9 induction, which may explain why gene induction can be reversed by supplementation of guanosine to the culture medium. Indeed, IMPDH inhibitors have shown antiviral activity in cell culture models of HCV infection.12,56 However, at this time, no IMPDH inhibitors have obtained U.S. Food and Drug Administration approval for the treatment of hepatitis C, indicating that this may not be the only mechanism by which ribavirin exerts its antiviral activity.57,58 Additional studies will be needed to determine if IMPDH inhibitors can effect ISG induction by IFN in cell culture models of virus infection as well as in patients.
Recognition of ribavirin’s ability to induce ISGs may explain ribavirin’s antiviral activity against numerous viruses and will facilitate the discovery of a class of therapeutic agents that possess more potent antiviral activity by specifically regulating antiviral genes. Importantly, ribavirin’s effect on ISG regulation appears to be IFN-independent (Fig. 6; Supporting Figs. 4B and 6). Several lines of evidence support the IFN-independent activity of ribavirin. First, we have not observed the full spectrum of ISG induction after ribavarin administration seen following IFN treatment (Figs. 1F and 2B). Second, RNAi against signaling components, which are critical for IFN production and activity, did not affect ribavirin-stimulated gene induction (Fig. 6A–D). Finally, we did not observe gene induction of either the type I or type III IFNs following ribavirin treatment (Supporting Fig. 6A). This may be particularly relevant for viruses such as HCV that have developed numerous strategies to circumvent the IFN system.59 This class of molecules, like ribavirin, may thus be able to potentiate the antiviral effects of IFN through synergistic induction of antiviral genes using different molecular mechanisms.
Although it was originally thought that the development of agents specifically targeting steps in the HCV life cycle would soon make IFN and ribavirin obsolete, emerging data suggest that this may not be the case. Because direct antivirals quickly select for resistant strains, it appears that PEG-IFN will likely be required to achieve an optimal response. Importantly, recent trials with HCV protease inhibitors have shown that ribavirin is also needed to prevent viral relapse.60 Therefore, the search for alternative ribavirin-like molecules in potentiating IFN action and for use in combination with specifically targeted antiviral agents remains a promising approach in the therapy of HCV-infected patients.
Supplementary Material
Acknowledgments
Supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, and in part by a National Institutes of Health K24 mentoring award DK066144 (to M. W. F.).
Abbreviations
- HCV
hepatitis C virus
- IFN
interferon
- IKK
I kappa B kinase
- IRF
interferon regulatory factor
- ISG
interferon-stimulated gene
- mRNA
messenger RNA
- NFκB
nuclear factor κB
- PEG-IFN
pegylated interferon
- poly(I:C)
polyinosinic:polycytidylic acid
- qPCR
quantitative polymerase chain reaction
- RNAi
RNA interference
- siRNA
short interfering RNA
- STAT
signal transducer and activator of transcription
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofurano-syl-1,2,4-triazole-3-carboxamide. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH, Seeff LB. Peginterferon and ribavirin for chronic hepatitis C. N Engl J Med. 2006;355:2444–2451. doi: 10.1056/NEJMct061675. [DOI] [PubMed] [Google Scholar]
- 3.Pawlotsky JM, Dahari H, Neumann AU, Hezode C, Germanidis G, Lonjon I, et al. Antiviral action of ribavirin in chronic hepatitis C. Gastroenterology. 2004;126:703–714. doi: 10.1053/j.gastro.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Jr, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 5.Dixit NM, Layden-Almer JE, Layden TJ, Perelson AS. Modelling how ribavirin improves interferon response rates in hepatitis C virus infection. Nature. 2004;432:922–924. doi: 10.1038/nature03153. [DOI] [PubMed] [Google Scholar]
- 6.Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann WP, Herrmann E, Sarrazin C, Zeuzem S. Ribavirin mode of action in chronic hepatitis C: from clinical use back to molecular mechanisms. Liver Int. 2008;28:1332–1343. doi: 10.1111/j.1478-3231.2008.01896.x. [DOI] [PubMed] [Google Scholar]
- 8.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 9.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maag D, Castro C, Hong Z, Cameron CE. Hepatitis C virus RNA-dependent RNA polymerase (NS5B) as a mediator of the antiviral activity of ribavirin. J Biol Chem. 2001;276:46094–46098. doi: 10.1074/jbc.C100349200. [DOI] [PubMed] [Google Scholar]
- 11.Malinoski F, Stollar V. Inhibitors of IMP dehydrogenase prevent sindbis virus replication and reduce GTP levels in Aedes albopictus cells. Virology. 1981;110:281–289. doi: 10.1016/0042-6822(81)90060-x. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Liu R, Baroudy BM, Malcolm BA, Reyes GR. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology. 2003;310:333–342. doi: 10.1016/s0042-6822(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 13.Tam RC, Pai B, Bard J, Lim C, Averett DR, Phan UT, et al. Ribavirin polarizes human T cell responses towards a Type 1 cytokine profile. J Hepatol. 1999;30:376–382. doi: 10.1016/s0168-8278(99)80093-2. [DOI] [PubMed] [Google Scholar]
- 14.Feld JJ, Nanda S, Huang Y, Chen W, Cam M, Pusek SN, et al. Hepatic gene expression during treatment with peginterferon and ribavirin: identifying molecular pathways for treatment response. Hepatology. 2007;46:1548–1563. doi: 10.1002/hep.21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Jamaluddin M, Wang S, Tian B, Garofalo RP, Casola A, et al. Ribavirin treatment up-regulates antiviral gene expression via the interferon-stimulated response element in respiratory syncytial virus-infected epithelial cells. J Virol. 2003;77:5933–5947. doi: 10.1128/JVI.77.10.5933-5947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feld JJ, Lutchman GA, Heller T, Hara K, Pfeiffer JK, Leff RD, et al. Ribavirin improves early responses to peginterferon through improved interferon signaling. Gastroenterology. 2010;139:154–162. doi: 10.1053/j.gastro.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brok J, Gluud LL, Gluud C. Ribavirin monotherapy for chronic hepatitis C infection: a Cochrane Hepato-Biliary Group systematic review and meta-analysis of randomized trials. Am J Gastroenterol. 2006;101:842–847. doi: 10.1111/j.1572-0241.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, et al. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437–1444. doi: 10.1053/j.gastro.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 23.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, et al. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009;5(1):e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumpter R, Jr, Loo Y-M, Foy E, Li K, Yoneyama M, Fujita T, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 27.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 28.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 29.Hellerbrand C, Jobin C, Iimuro Y, Licato L, Sartor RB, Brenner DA. Inhibition of NFkappaB in activated rat hepatic stellate cells by proteasome inhibitors and an IkappaB super-repressor. Hepatology. 1998;27:1285–1295. doi: 10.1002/hep.510270514. [DOI] [PubMed] [Google Scholar]
- 30.Doyle SL, O’Neill LA. Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Takeuchi O, Akira S. MyD88 as a bottle neck in Toll/IL-1 signaling. Curr Top Microbiol Immunol. 2002;270:155–167. doi: 10.1007/978-3-642-59430-4_10. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 33.Onoguchi K, Yoneyama M, Takemura A, Akira S, Taniguchi T, Namiki H, et al. Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem. 2007;282:7576–7581. doi: 10.1074/jbc.M608618200. [DOI] [PubMed] [Google Scholar]
- 34.Kisseleva T, Brenner DA. Role of hepatic stellate cells in fibrogenesis and the reversal of fibrosis. J Gastroenterol Hepatol. 2007;22(Suppl 1):S73–S78. doi: 10.1111/j.1440-1746.2006.04658.x. [DOI] [PubMed] [Google Scholar]
- 35.Hoofnagle JH, Ghany MG, Kleiner DE, Doo E, Heller T, Promrat K, et al. Maintenance therapy with ribavirin in patients with chronic hepatitis C who fail to respond to combination therapy with interferon alfa and ribavirin. Hepatology. 2003;38:66–74. doi: 10.1053/jhep.2003.50258. [DOI] [PubMed] [Google Scholar]
- 36.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 37.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 38.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor MW, Tsukahara T, Brodsky L, Schaley J, Sanda C, Stephens MJ, et al. Changes in gene expression during pegylated interferon and ribavirin therapy of chronic hepatitis C virus distinguish responders from nonresponders to antiviral therapy. J Virol. 2007;81:3391–3401. doi: 10.1128/JVI.02640-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sen GC, Sarkar SN. The interferon-stimulated genes: targets of direct signaling by interferons, double-stranded RNA, and viruses. Curr Top Microbiol Immunol. 2007;316:233–250. doi: 10.1007/978-3-540-71329-6_12. [DOI] [PubMed] [Google Scholar]
- 41.Lanford RE, Guerra B, Lee H, Averett DR, Pfeiffer B, Chavez D, et al. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(i)-poly(c), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J Virol. 2003;77:1092–1104. doi: 10.1128/JVI.77.2.1092-1104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young KC, Lindsay KL, Lee KJ, Liu WC, He JW, Milstein SL, Lai MM. Identification of a ribavirin-resistant NS5B mutation of hepatitis C virus during ribavirin monotherapy. Hepatology. 2003;38:869–878. doi: 10.1053/jhep.2003.50445. [DOI] [PubMed] [Google Scholar]
- 43.Kim SS, Peng LF, Lin W, Choe WH, Sakamoto N, Schreiber SL, et al. A cell-based, high-throughput screen for small molecule regulators of hepatitis C virus replication. Gastroenterology. 2007;132:311–320. doi: 10.1053/j.gastro.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 44.Fried MW, Jensen DM, Rodriguez-Torres M, Nyberg LM, Di Bisceglie AM, Morgan TR, et al. Improved outcomes in patients with hepatitis C with difficult-to-treat characteristics: randomized study of higher doses of peginterferon alpha-2a and ribavirin. Hepatology. 2008;48:1033–1043. doi: 10.1002/hep.22448. [DOI] [PubMed] [Google Scholar]
- 45.Lindahl K, Stahle L, Bruchfeld A, Schvarcz R. High-dose ribavirin in combination with standard dose peginterferon for treatment of patients with chronic hepatitis C. Hepatology. 2005;41:275–279. doi: 10.1002/hep.20563. [DOI] [PubMed] [Google Scholar]
- 46.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 47.Snoeck E, Wade JR, Duff F, Lamb M, Jorga K. Predicting sustained virological response and anaemia in chronic hepatitis C patients treated with peginterferon alfa-2a (40KD) plus ribavirin. Br J Clin Pharmacol. 2006;62:699–709. doi: 10.1111/j.1365-2125.2006.02741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maynard M, Pradat P, Gagnieu MC, Souvignet C, Trepo C. Prediction of sustained virological response by ribavirin plasma concentration at week 4 of therapy in hepatitis C virus genotype 1 patients. Antivir Ther. 2008;13:607–611. [PubMed] [Google Scholar]
- 49.Jen JF, Glue P, Gupta S, Zambas D, Hajian G. Population pharmacokinetic and pharmacodynamic analysis of ribavirin in patients with chronic hepatitis C. Ther Drug Monit. 2000;22:555–565. doi: 10.1097/00007691-200010000-00010. [DOI] [PubMed] [Google Scholar]
- 50.Toyoda H, Kumada T, Kiriyama S, Sone Y, Tanikawa M, Hisanaga Y, et al. Correlation of serum ribavirin concentration with pretreatment renal function estimates in patients with chronic hepatitis C receiving combination antiviral therapy with peginterferon and ribavirin. J Viral Hepat. 2008;15:651–658. doi: 10.1111/j.1365-2893.2008.01004.x. [DOI] [PubMed] [Google Scholar]
- 51.Glue P. The clinical pharmacology of ribavirin. Semin Liver Dis. 1999;19(Suppl 1):17–24. [PubMed] [Google Scholar]
- 52.Baiocchi L, De Leonardis F, Delle Monache M, Nosotti L, Conti RL, Lenci I, et al. Plasma/erythrocyte ribavirin ×100 ratio as an indicator of sustained virological response in HCV genotype 1 patients with early virological response. Antivir Ther. 2010;15:633–639. doi: 10.3851/IMP1560. [DOI] [PubMed] [Google Scholar]
- 53.Homma M, Matsuzaki Y, Inoue Y, Shibata M, Mitamura K, Tanaka N, et al. Marked elevation of erythrocyte ribavirin levels in interferon and ribavirin-induced anemia. Clin Gastroenterol Hepatol. 2004;2:337–339. doi: 10.1016/s1542-3565(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 54.Ibarra KD, Pfeiffer JK. Reduced ribavirin antiviral efficacy via nucleoside transporter-mediated drug resistance. J Virol. 2009;83:4538–4547. doi: 10.1128/JVI.02280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry SD, Metselaar HJ, Lonsdale RC, Kok A, Haagmans BL, Tilanus HW, et al. Mycophenolic acid inhibits hepatitis C virus replication and acts in synergy with cyclosporin A and interferon-alpha. Gastroenterology. 2006;131:1452–1462. doi: 10.1053/j.gastro.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 57.Gish RG. Treating HCV with ribavirin analogues and ribavirin-like molecules. J Antimicrob Chemother. 2006;57:8–13. doi: 10.1093/jac/dki405. [DOI] [PubMed] [Google Scholar]
- 58.Rustgi VK, Lee WM, Lawitz E, Gordon SC, Afdhal N, Poordad F, et al. Merimepodib, pegylated interferon, and ribavirin in genotype 1 chronic hepatitis C pegylated interferon and ribavirin nonresponders. Hepatology. 2009;50:1719–1726. doi: 10.1002/hep.23204. [DOI] [PubMed] [Google Scholar]
- 59.Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 60.Shiffman ML. What future for ribavirin? Liver Int. 2009;29(Suppl 1):68–73. doi: 10.1111/j.1478-3231.2008.01936.x. [DOI] [PubMed] [Google Scholar]
- 61.Weiskirchen R, Gressner AM. Isolation and culture of hepatic stellate cells. Methods Mol Med. 2005;117:99–113. doi: 10.1385/1-59259-940-0:099. [DOI] [PubMed] [Google Scholar]
- 62.Kato T, Matsumura T, Heller T, Saito S, Sapp RK, Murthy K, et al. Production of infectious hepatitis C virus of various genotypes in cell cultures. J Virol. 2007;81:4405–4411. doi: 10.1128/JVI.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.