Abstract
PURPOSE
To examine retinal structure and changes in photoreceptor intensity post-dark adaptation in patients with complete congenital stationary night blindness and Oguchi disease.
DESIGN
Prospective observational case series.
METHODS
We recruited three patients with complete congenital stationary night blindness caused by mutations in GRM6, two brothers with Oguchi disease caused by mutations in GRK1, and one normal control. Retinal thickness was measured from optical coherence tomography (OCT) images. Integrity of the rod and cone mosaic was assessed using adaptive optics scanning light ophthalmoscopy. We imaged five of the patients following a period of dark adaptation, and examined layer reflectivity on OCT in a patient with Oguchi disease under light- and dark-adapted conditions.
RESULTS
Retinal thickness was reduced in the parafoveal region in patients with GRM6 mutations, as a result of decreased thickness of the inner retinal layers. All patients had normal photoreceptor density at all locations analyzed. Upon removal from dark adaptation, the intensity of the rods (but not cones) in the patients with Oguchi disease gradually and significantly increased. In one Oguchi patient, the outer segment layer contrast on OCT was fourfold higher under dark-adapted versus light-adapted conditions.
CONCLUSIONS
The selective thinning of the inner retinal layers in patients with GRM6 mutations suggests either reduced bipolar/ganglion cell numbers or altered synaptic structure in the inner retina. Our finding that rods, but not cones, change intensity after dark adaptation suggests that fundus changes in Oguchi disease are due to changes within the rods as opposed to changes at a different retinal locus.
Congenital stationary night blindness represents a heterogeneous group of inherited retinal disorders characterized by impaired night vision, and can be inherited in an autosomal dominant, autosomal recessive or X-linked fashion.1 Patients with congenital stationary night blindness have an abnormal rod electroretinogram (ERG) and an abnormal dark-adaptation curve. Additional ocular manifestations are variable but can include reduced visual acuity, refractive error (commonly myopia but occasionally hyperopia), nystagmus, strabismus, and altered fundus appearance.2 Within congenital stationary night blindness with a normal-appearing fundus, X-linked and autosomal recessive congenital stationary night blindness may be further subdivided into complete and incomplete forms.2 Both forms are associated with an electronegative or ‘negative’ ERG in response to a bright white flash in the dark-adapted eye, such that there is a reduced b-wave / a-wave ratio (i.e.“Schubert-Bornschein ERG”).3, 4 The discrimination between the complete and incomplete form is made according to whether there is a rod-specific ERG response to a dim light under dark adaptation.2, 5
In patients with complete congenital stationary night blindness there is no detectable b-wave in the rod-specific ERG. Long flash photopic ERG demonstrates attenuated b-wave amplitude with normal d-wave amplitude, consistent with involvement of the cone ON pathway as well.6, 7 In patients with incomplete congenital stationary night blindness, there is a detectable rod-specific ERG, though the b-wave is reduced from normal. In addition, the long flash photopic ERG demonstrates attenuated d-wave amplitude, consistent with impaired function of both the ON and OFF pathways.6, 7 ERG evidence of inner retinal rod system dysfunction may also occur in autosomal dominant congenital stationary night blindness, but in association with normal cone ERGs. In other cases of autosomal dominant congenital stationary night blindness, ERG rod responses are attenuated with normal cone responses, but the standard bright flash response does not have a negative waveform (i.e.“Riggs ERG”).
Mutations in the NYX, GRM6, TRPM1, and GPR179 genes are associated with complete congenital stationary night blindness, being expressed in ON bipolar cells and encoding proteins that are part of the mGluR6 signaling cascade.8–16 Incomplete congenital stationary night blindness is associated with mutations in genes involved in glutamate release from the photoreceptors (CACNA1F, CABP4, and CACNA2D4).17–21 Mutations in genes encoding three components of the rod-specific phototransduction cascade have all been reported in association with autosomal dominant congenital stationary night blindness(RHO, GNAT1, and PDE6B).22, 23 Recent estimates suggest that approximately 20% of congenital stationary night blindness patients have an unknown genetic basis for the disease.21, 24
Oguchi disease is a distinctive form of autosomal recessive congenital stationary night blindness with abnormal-appearing fundus first described in the Japanese population.25 Patients with Oguchi disease have a distinct dark adaptation curve compared to the other forms of congenital stationary night blindness; following a bleach, rod sensitivity recovers to normal after a prolonged period (>2 hours) of dark adaptation.22 Patients with Oguchi disease have no rod a-wave or b-wave, except after prolonged dark adaptation when a normal single-flash response can be obtained. The fundus has a diffuse or patchy radiant appearance, described as a golden-yellow tapetal-like metallic sheen, with normal fundus coloration restored after prolonged dark adaptation (Mizuo-Nakamura phenomenon).26 Two genes have been implicated in Oguchi disease, both of which are involved in rod phototransduction – SAG (encoding arrestin) and GRK1 (encoding rhodopsin kinase).27, 28
Significant progress in our understanding of how the various mutations implicated in the subtypes of congenital stationary night blindness affect retinal structure has come from examination of mouse models. For example, Pardue et al. showed that the nob mouse had normal cytoarchitecture in the presence of disrupted retinal transimission.29 Rhodopsin kinase knockout mice have been shown to undergo light-induced rod degeneration.30 Across mouse models of congenital stationary night blindness, differences in retinal morphology have been noted between mutations in genes expressed presynaptically (incomplete congenital stationary night blindness) and mutations in genes expressed postsynaptically (complete congenital stationary night blindness).31 Despite this progress utilizing mouse models of the disease, structural data in humans are sparse. A histopathological report of a patient with congenital stationary night blindnessof unknown genetic origin demonstrated normal rod and cone structure,32 and a second study of a patient with suspected congenital stationary night blindness showed normal rod outer segment structure.33 A recent investigation using spectral-domain optical coherence tomography (SD-OCT) demonstrated thinned retinas in five patients with incomplete congenital stationary night blindness, three of whom had a mutation in the CACNA1F gene.34 Despite being more rare than congenital stationary night blindness without fundus abnormalities, anatomical studies in patients with Oguchi disease are rather numerous. Usui et al. reported diffuse, fine white particles on helium-neon laser imaging in the light-adapted retina, which disappeared on dark adaptation and reappeared gradually on exposure to light suggesting that accumulation of an abnormal product in the outer retina causes the golden metallic fundus appearance.35 A histopathological report by Kuwabara et al. suggested the existence of an abnormal layer between the retinal pigment epithelium (RPE) and the outer segments of the photoreceptors along with displaced cone nuclei.36 More recently, SD-OCT studies in patients with Oguchi disease have reported a variety of outer retinal findings, including shortened rod outer segments,37 hyperreflective outer segment regions that disappeared after extended dark adaptation,38 thinning of the parafoveal outer nuclear layer (ONL),39 and presumed shortening of photoreceptor outer segments (evidenced by the disappearance of the extrafoveal inner segment/outer segment (IS/OS) line in the partly dark-adapted state).40, 41 While congenital stationary night blindness(including the Oguchi variant) is traditionally believed to be non-progressive, accumulating evidence suggests that not all cases are truly stationary.22, 23, 39 Thus careful reexamination of retinal morphology with advanced retinal imaging tools may disclose previously unrecognized anatomical manifestations of congenital stationary night blindness.
Here we sought to apply non-invasive, high-resolution imaging tools and quantitative analyses to examine retinal structure in patients with complete congenital stationary night blindness caused by mutations in GRM6 or Oguchi disease caused by mutations in GRK1. We also examined the intensity of individual rod and cone photoreceptors under light adapted conditions as well as after a prolonged period of dark adaptation. Our data indicate that defects in the phototransduction process can alter photoreceptor reflectivity and demonstrate altered inner retinal morphology in patients with GRM6 mutations. This quantitative imaging approach should prove useful in examining retinal structure in other diseases.
METHODS
This prospective observational case series included three patients diagnosed with complete congenital stationary night blindness caused by mutations in GRM6, two brothers with Oguchi disease caused by mutations in GRK1, and one visually-normal healthy adult. A brief summary of the patient characteristics is provided in the Table. Color vision was assessed using the 2002 edition AO-HRR pseudo-isochromatic plates (Richmond Products, Inc., Albuquerque, New Mexico, USA). Axial length measurements were obtained using an IOL Master (Carl Zeiss Meditec, Inc, Dublin, California, USA) for calibrating the lateral scale of all retinal images. For all imaging sessions, each patient’s eye was dilated and accommodation suspended using one drop each of phenylephrine (2.5%) and tropicamide (1%).
TABLE.
Genotype and Phenotype Summary of 5 Patients with Complete Congenital Stationary Night Blindness or Oguchi Disease and 1 Normal Control
| Patient | Age(y)/ Gender |
BCVA (OD; OS) |
Refractive error, diopters (OD;OS) |
Phenotype | Affected Gene |
Allele 1 | Allele 2 | Source |
|---|---|---|---|---|---|---|---|---|
| JC_0677 | 23/F | 20/12; 20/12 | −1.50 sphere; −1.50 sphere |
Normal | ND | ND | ND | This Study |
| DH_0491 | 41/M | 20/16; 20/16 | No correction | Oguchi Disease | GRK1 | c. 1129G>C, p.Ala377Pro | c.1139T>A, p.Val380Asp | This Study |
| DH_0819 | 47/M | 20/32; 20/25 | + 0.25 sphere; No correction |
Oguchi Disease | GRK1 | c. 1129G>C, p.Ala377Pro | c.1139T>A, p.Val380Asp | This Study |
| JC_0550 | 30/F | 20/16; 20/16 | −0.75/0.75 × 30; −0.50 sphere |
complete congenital stationary night blindness |
GRM6 | c.172G>A, p.Gly58Arg | IVS2-1G>T (c.722-1G>T) | This Study |
| JC_0682 | 13/F | 20/32; 20/20 | −1.00/−1.25 × 105; −0.50/−2.50 × 50 |
complete congenital stationary night blindness |
GRM6 | c.2267G>A, p.Gly756Asp | c.2267G>A, p.Gly756Asp | Ref. 42 |
| JC_0684 | 32/F | 20/30; 20/30 | −0.50/−1.00 × 80; −0.50/−1.50 × 105 |
complete congenital stationary night blindness |
GRM6 | c.2029C>T, p.Arg677Cys | c.2029C>T, p.Arg677Cys | Ref. 42 |
F = female; M = male; BCVA = best-corrected visual acuity; OD = right eye; OS = left eye; ND = not done.
The methods of molecular genetic analysis and genotypes for two of the patients with complete congenital stationary night blindness (JC_0682 & JC_0684) have been previously reported.42 For the third patient with complete congenital stationary night blindness (JC_0550), genomic DNA was isolated from whole blood using the ArchivePure DNA extraction kit (5 Prime, Gaithersburg, Maryland, USA). The polymerase chain reaction was used to amplify the GRM6 gene using the primers previously described by Dryja et al. (2005).7 For exon 1, exons 2–3, exons 5–6, and exon 8 (both the 5' and 3' regions) TaKara LA enzyme with the 2XGC buffer (TaKaRa Bio Inc, Japan) was used according to the manufacturer's instructions. All reactions were subjected to an initial denaturing step of 94°C for 1 minute, and a final extension step of 72°C for 10 to 15 minutes. Between the initial and final incubations, PCRs for exon1, exons 2–3, and both PCRs for exon 8 were subjected to 30 cycles of denaturing at 94°C for 30–45 seconds, annealing at 58°C for 1 minute, and extension at 72°C for 2 minutes. The PCR for exons 5–6 used the same denaturing, extension and annealing temperatures, but the times were 45 seconds for denaturing, 2 minutes and 30 seconds for annealing, and 3 minutes and 45 seconds for extension. For exons 4, 7, 9 and 10 a hot-start PCR employing the AmpliTaq Gold PCR kit (Applied Biosystems, Foster City, California, USA) was done according to the manufacturer's instructions. All reactions were subjected to an initial denaturing step at 95°C for 5 minutes, and a final extension step at 72°C for 10–15 minutes. Between the initial and final steps, reactions were subject to 30 cycles of denaturing at 94°C for 30 seconds, annealing for 30 seconds at 56°C for exon 9, 58.5°C for exons 4 and 7, and 60°C for exon 10, and extension at 72°C for 45 seconds. PCR products were directly sequenced using the same primers as were used for amplification, and additional primers described by Dryja et al. (2005) for sequencing exons 5–6.7 Sequencing was performed using BigDye Terminator V3.1 (Applied Biosystems, Foster City, California, USA) and AmpliTaq FS (Applied Biosystems, Foster City, California, USA) and sequencing reactions were analyzed on an ABI 7500 capillary sequencer. All sequencing was bi-directional.
DNA was extracted from two patients with Oguchi disease and their mother by following the manufacturer’s specifications for whole blood DNA extraction using Autopure LS instrument (Gentra Systems, Minneapolis, Minnesota, USA). The entire coding region of the GRK1 gene was sequenced in the proband using fluorescent dideoxynucleotides on an ABI 3730 automated sequencer. Mutations were identified by the approximately equal peak intensity of two fluorescent dyes at the mutant base. Mutant nucleotides identified in the proband (DH_0491) were then evaluated in his sibling (DH_0819) and mother. All sequencing was bi-directional.
SPECTRAL-DOMAIN OPTICAL COHERENCE TOMOGRAPHY
Volumetric images of the macula and optic disc were acquired using the Cirrus HD-OCT (Carl Zeiss Meditec, Inc, Dublin, California, USA). Individual volumes were nominally 6 × 6mm and consisted of 128 B-scans (512 A-scans/B-scan). Retinal thickness was calculated using the built-in macular analysis software (software version 5.2), which is automatically determined by measuring the distance between the inner limiting membrane (ILM) and RPE boundaries. Thickness of the ganglion cell layer plus inner plexiform layer (GCL+IPL) was calculated using the built in analysis software (software version 6.0), which was determined by measuring the distance between the retinal nerve fiber layer/GCL interface and the IPL/inner nuclear layer interface.
We also acquired additional high density SD-OCT line scans through the fovea, with a sampling of 1000 A-scans/B-scan and 100 repeated B-scans (Bioptigen, Inc., Durham, North Carolina, USA). Up to 40 B-scans from these 100-scan sequences were registered and averaged to reduce speckle noise in the image as previously described.43 A total of six layers were manually segmented in the averaged images (ILM; outer plexiform layer, OPL; external limiting membrane, ELM; inner segment ellipsoid, ISe; RPE1 and RPE2). Layer naming is based on the recent work of Spaide and Curcio.44 The innermost peak of the outer photoreceptor complex is invariantly attributed to the ELM. The 2nd layer has been often referred to as the IS/OS layer, though here we adopt the recent nomenclature of Spaide and Curcio, who through a detailed analysis attribute this band to the ellipsoid portion of the inner segment (ISe).44 The 3rd layer is attributed to the RPE contact cylinder while the 4th band is attributed to the apical portion of the RPE. As the RPE contributes to both of these bands, we simply refer to the 3rd and 4th layers as RPE1 and RPE2, respectively. Importantly, the distance between RPE1 and RPE2 is not the thickness of the RPE cell, but provides a convenient way to communicate the presence of multiple bands associated with the RPE.
Total retinal thickness (ILM-RPE2 distance), inner retinal thickness (ILM-OPL distance) and ONL thickness (OPL-ELM distance) was calculated using a custom Matlab (Mathworks, Natick, Massachusettes, USA) program and compared to previously reported values for 93 controls (42 male, 51 female), with an average age of 25.7 ± 8.2 years (range 11–40 years).45 For one of the patients (DH_0491), we acquired SD-OCT images under light- and dark-adapted states. Longitudinal reflectivity profiles were generated as previously described,46, 47 in order to assess the relative intensity of the different retinal layers under these conditions.
ADAPTIVE OPTICS SCANNING LIGHT OPHTHALMOSCOPE
Images of the photoreceptor mosaic were obtained using a previously described adaptive optics scanning light ophthalmoscope.48–50 The wavelength of the super luminescent diodes used for retinal imaging were 680nm and 775nm, subtending a field of view of 0.96° × 0.96°. Separate image sequences of 100–200 frames each were acquired at various parafoveal and perifoveal locations. Parafoveal images were usually acquired by instructing the patient to fixate on one of the corners of the raster scan square, while the perifoveal images were acquired using an external fixation target. To increase the percentage of recorded frames with useable data, the image acquisition software had an “active blink removal” algorithm, which discarded frames with a mean intensity below a specified threshold value.
To correct for intraframe distortions within the frames of the raw image sequence due to the sinusoidal motion of the resonant optical scanner, we estimated the distortion from images of a stationary Ronchi ruling, and then re-sampled each frame of the raw image sequence over a grid of equally spaced pixels. After desinusoiding, a reference frame was manually selected from within each image sequence for subsequent registration using custom software. Registration of frames within a given image sequence was performed by dividing the frame of interest into strips, aligning each strip to the location in the reference frame that maximizes the normalized cross correlation between them.51 The discrete set of locations for the image strips was used to generate a continuous transformation that would register the recorded frame to match the reference frame. Once all the frames were registered, the 50 frames with the highest normalized cross correlation to the reference frame were averaged, in order to generate a final registered image with an increased signal to noise ratio for subsequent analysis.
Cell density was calculated over a 55 × 55µm sampling window using a previously described semi-automated algorithm implemented in Matlab (Mathworks, Natick, Massachusettes, USA),52, 53 with an additional modification that during the user review of the automated cell identifications, a cell removal tool was also available. Intrasession repeatability of the algorithm on parafoveal cone images was reported to be 2.7%.53 Since the rods greatly outnumber the cones in the perifoveal images, we utilized the algorithm to identify the rods by removing any cones during the user review step. Estimates of cone density for these images were obtained using manual identification of the large, coarsely spaced cones.
ANALYZING PHOTORECEPTOR REFLECTIVITY
Five of the six patients (JC_0677, DH_0491, DH_0819, JC_0682, and JC_0684) underwent a second imaging session using the adaptive optics scanning light ophthalmoscope. First, the eye of interest was aligned and optimal focus established for a single parafoveal and a single perifoveal location (~10° eccentricity). The eye was then patched and the patient placed in a dark room for 2 hours. A second drop each of phenylephrine (2.5%) and tropicamide (1%) was administered approximately 15 minutes before the end of dark adaptation, this was done with care not to compromise the dark-adapted state of the retina.
Immediately upon removal from dark adaptation, we began imaging with the 775nm light source. The only additional illumination in the room was that from two computer monitors. Numerous image sequences were acquired at the predetermined parafoveal and perifoveal locations throughout a period of at least 45 minutes. Midway through this period, the room lights were turned on so as to fully return the eye to a light-adapted state. While the light levels and adaption of the eye were not precisely controlled during the 45-minute imaging session, it was the same for the five patients, allowing us to make relative comparisons between the patients.
The reflectivity of individual cells was analyzed as previously described, and the method is summarized next.48 The raw video sequences were processed as described above, and for a given imaging location, the average images from each time point were registered to each other using an affine transformation (i2kRetina, Dual Align, LLC, Clifton Park, New York, USA). This aligned image stack was then cropped to a common area, a reference frame was selected, and the image stack then went through strip registration,51 as described above. Finally, the image series was normalized to the temporal mean of the nonzero portions of the stack. This aligned image stack was then averaged so as to generate a single reference image for each location. These images were then used to determine preliminary cone and rod coordinate locations. The position of perifoveal rods was determined by manual selection, while the position of parafoveal cones was identified using a modified version of previously described semi-automated algorithm, which also allowed manual addition/subtraction of cones missed or selected in error.53 From these preliminary coordinates, the final coordinates were determined using custom Matlab (Mathworks, Natick, Massachusetts, USA) software that identified the local maximum within a 3x3 pixel (~1.25 × 1.25µm) region around the initial cone (or rod) coordinate. Perifoveal cone photoreceptors show a multimodal intensity pattern that varies in both intensity and structure over time, a phenomenon not yet fully understood. In addition, the small number of cones (<50) present in the perifoveal images would make any global conclusion about their reflectance behavior over time difficult. As such, we did not analyze the reflectivity of the perifoveal cones.
The final coordinates were adjusted for each frame within the aligned image stack, in order to compensate for small errors in image registration. This was done by first projecting a mask for each cell through the aligned image stack. A square 3x3 pixel and circular 5 pixel diameter mask was used for rods and cones, respectively. For each frame, each cells’ mask was repositioned to a local maximum, which never occurred greater than 1 pixel away from the original final coordinate. As the time stamp for each image was recorded, we thus could generate plots of intensity as a function of time for the parafoveal cones and perifoveal rods in each of the five patients.
RESULTS
Clinical Findings and Genetic Analysis Results
A summary of the clinical characteristics and molecular diagnosis for each patient is provided in the Table. Two of the complete congenital stationary night blindness patients (JC_0682 and JC_0684) were previously described, with their initial diagnosis based on nyctalopia and electrophysiological findings.42 Both had homozygous mutations in GRM6, p.Gly756Asp (JC_0682) and p.Arg677Cys (JC_0684).42 The third complete congenital stationary night blindness patient (JC_0550) was mis-diagnosed with retinitis pigmentosa at age 7, and presented to us with a history of poor night vision as far as she can recall and no difficulty with her peripheral vision. Full-field ERG showed selective b-wave reduction under scotopic conditions using a bright flash stimulus. The single flash photopic response showed evidence of an a-wave and absence of a b-wave, consistent with the loss of the ON-bipolar cell response. Isolated rod response was not detectable. She was found to be heterozygous for a mutation in GRM6 (p.Gly58Arg), which has been previously reported to be pathogenic.9 She was also found to be heterozygous for a mutation at the intron 2 / exon 3 junction within GRM6 that would be expected to result in a splicing defect (G>T at the 3’ end of intron 2).
The two brothers with Oguchi disease were found to harbor novel compound heterozygous mutations in GRK1, p.Ala377Pro and p.Val380Asp. Sequencing of their mother’s DNA revealed her to carry only the p.Ala377Pro variation, indicating that the Oguchi patients’ variations lie on different alleles. Neither of these variations has been observed in Caucasian (n=81) Hispanic (n=89) or African American (n=78) controls (unpublished observations). Also, neither of these alleles was observed among the more than 3000 Caucasian exomes and more than 1800 African American exomes that are visible on the Exome Variant Server (http://evs.gs.washington.edu/EVS/) of the NHBLI Exome Sequencing Project. DH_0491 originally presented with nyctalopia since early childhood, though he reported his night vision improved after 45–60 minutes of sustained dark adaptation. Maculae and peripheral fundi were normal with the exception of a diffuse hyperreflective sheen from the retinal surface upon indirect ophthalmoscopic illumination. After 105 minutes of dark adaptation, the hyperreflective sheen was noted to be absent ophthalmoscopically, but reappeared after only 5–10 seconds of illumination (consistent with the Mizuo-Nakamura phenomenon). Fundus examination of his brother (DH_0819) revealed fundus discoloration typical of Oguchi disease.
Reduced Retinal Thickness in Complete Congenital Stationary Night Blindness and Normal Retinal Thickness in Oguchi Disease
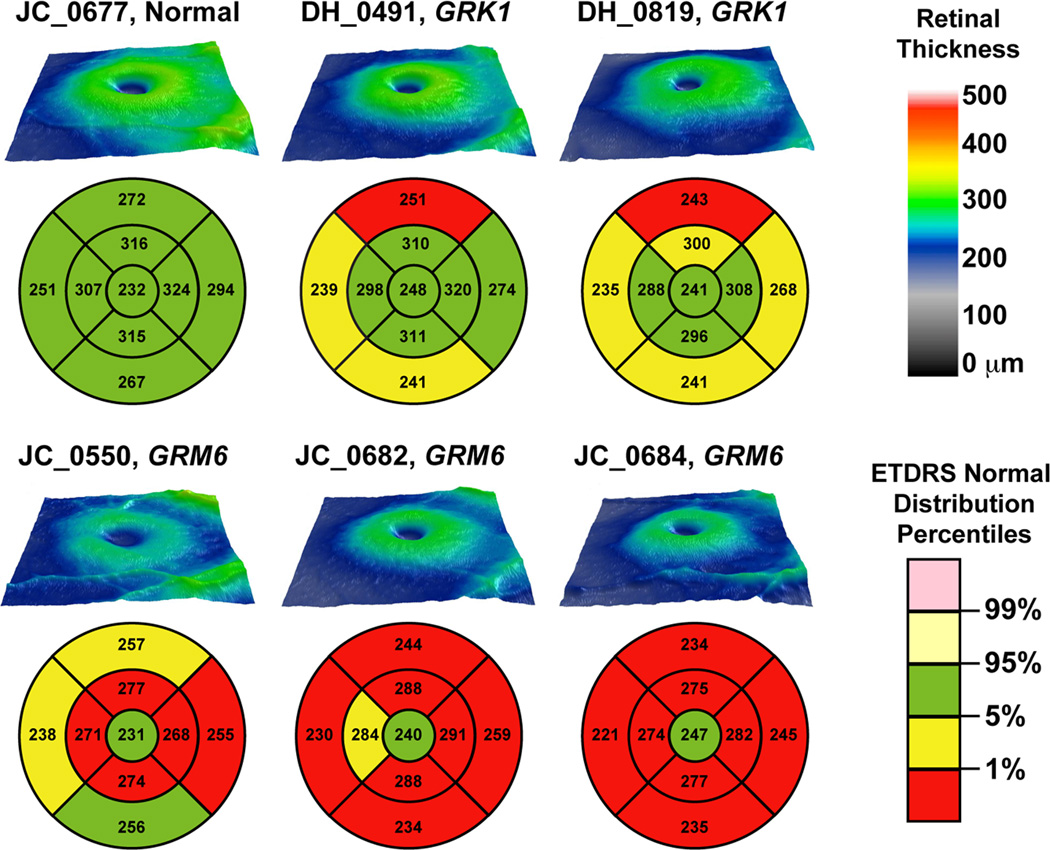
Topographical retinal thickness maps obtained from the Cirrus HD-OCT showed reduced retinal thickness (at least 5/9 Early Treatment Diabetic Retinopathy Study (ETDRS) segments with retinal thickness <1% of normal distribution percentiles) in the extrafoveal region in the three complete congenital stationary night blindness patients with GRM6 mutations (Figure 1). The two Oguchi disease patients with GRK1 mutations had predominantly normal retinal thickness, with each patient having a single ETDRS segment (superior retina, outer ring) with a thickness that was below 1% of the normal distribution percentiles. Visual inspection of the entire volume scans disclosed no anomalies of any of the retinal layers, and detailed examination of high-resolution horizontal line scans demonstrates the presence of all retinal layers (Figure 2).
Figure 1.
Topographical retinal thickness maps for 3 patients with complete congenital stationary night blindness caused by GRM6 mutations, 2 brothers with Oguchi disease caused by GRK1 mutations, and 1 normal control. Early treatment diabetic retinopathy study (ETDRS) grids are plotted below each topographical thickness map and show that retinal thickness was significantly reduced in the 3 complete congenital stationary night blindness patients, and to a much lesser extent in the Oguchi patients.
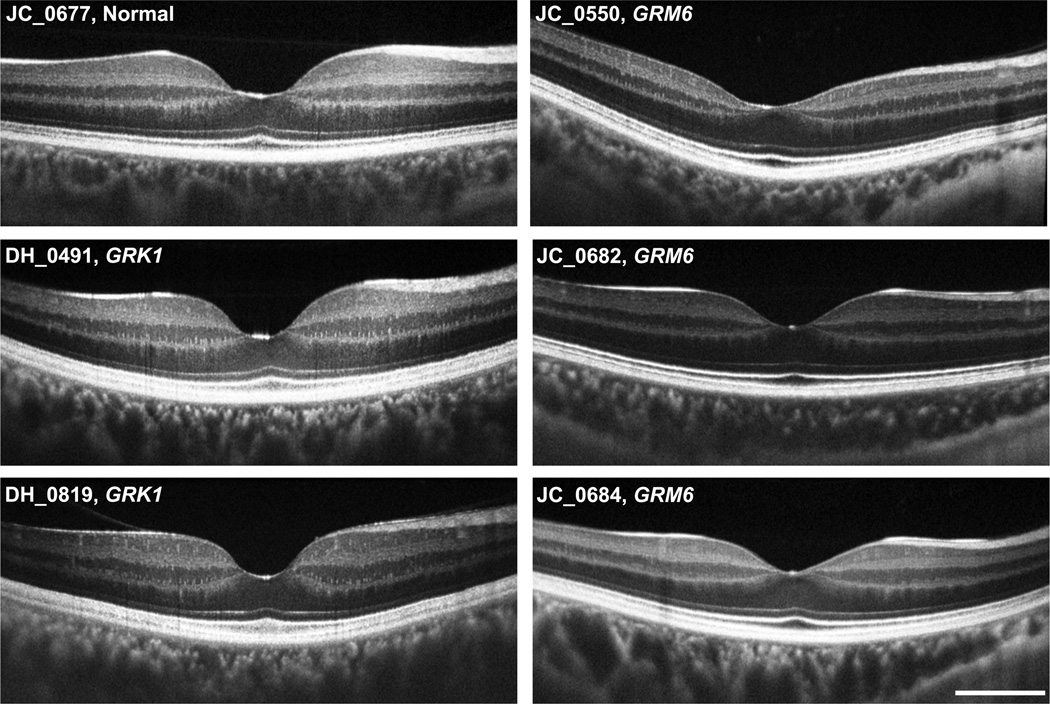
Figure 2.
High-resolution optical coherence tomography line scans (Bioptigen, Inc., Durham, North Carolina, USA) acquired along the horizontal meridian for 3 patients with complete congenital stationary night blindness caused by GRM6 mutations, 2 brothers with Oguchi disease caused by GRK1 mutations, and 1 normal control. All retinal layers can be identified and appear to be continuous. Scale bar is 1mm.
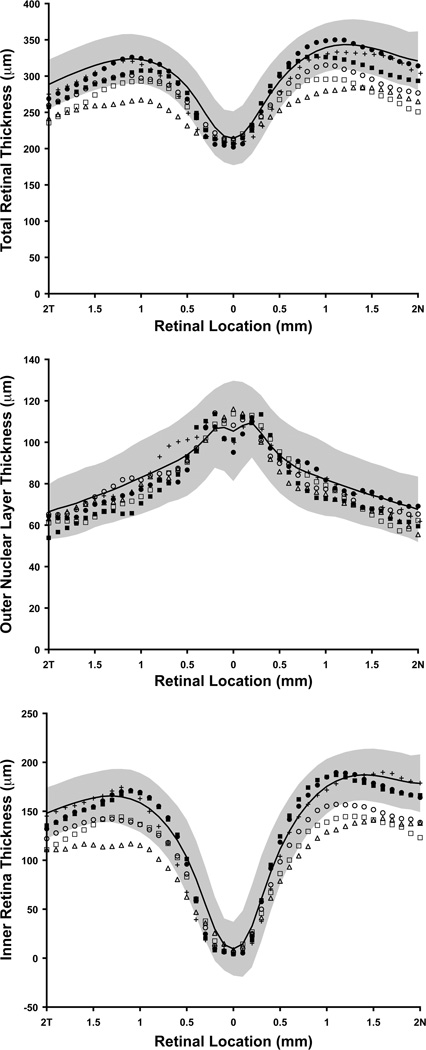
In order to investigate the reduction in retinal thickness in more detail, we examined the thickness of the inner retina and the outer nuclear layer (ONL) from the high-resolution horizontal line scans in Figure 2, and compared it to previously reported normative data from 93 individuals.45 As shown in Figure 3, all five patients had normal foveal thickness, though the complete congenital stationary night blindness patients with GRM6 mutations had reduced total retinal thickness (below 2 standard deviations (SD) from the mean) outside the fovea, consistent with the results from the Cirrus volumetric data. The three patients with GRM6 mutations had normal ONL thickness but reduced inner retinal layer thickness (Figure 3). The GCL+IPL analysis on the Cirrus macular volumes revealed significant thinning in the three patients with GRM6 mutations (the average GCL+IPL thickness was 64 µm for JC_0550, 65 µm for JC_0682, and 59 µm for JC_0684). These data indicate that the thinning of the GRM6 retina is due to inner retinal defects as opposed to photoreceptor loss, and involves the ganglion cell layer. Both patients with Oguchi disease caused by GRK1 mutations had normal ONL and inner retinal layer thickness (Figure 3), and the average GCL+IPL thickness was 70 µm and 76 µm for DH_0491 and DH_0819, respectively.
Figure 3.
Graphs showing total retinal thickness (top), outer nuclear layer (ONL) thickness (middle), and inner retina thickness (bottom), derived from high-resolution optical coherence tomography line scans (Bioptigen, Inc., Durham, North Carolina, USA) acquired along the horizontal meridian. Solid black line represents mean thickness values for 93 normal controls, with shaded region representing ±2 standard deviations from the mean. Filled symbols are the patients with Oguchi disease caused by GRK1 mutations (DH_0491, filled circles; DH_0819, filled squares). Open symbols are the patients with complete congenital stationary night blindness caused by GRM6 mutations (JC_0550, open triangles; JC_0682, open circles; JC_0684, open squares). The crosses are thickness values for the normal control recruited for the current study (JC_0677). Only the congenital stationary night blindness patients with GRM6 mutations had reduced retinal thickness, and this can be attributed to thinning of the inner retina, as opposed to the ONL.
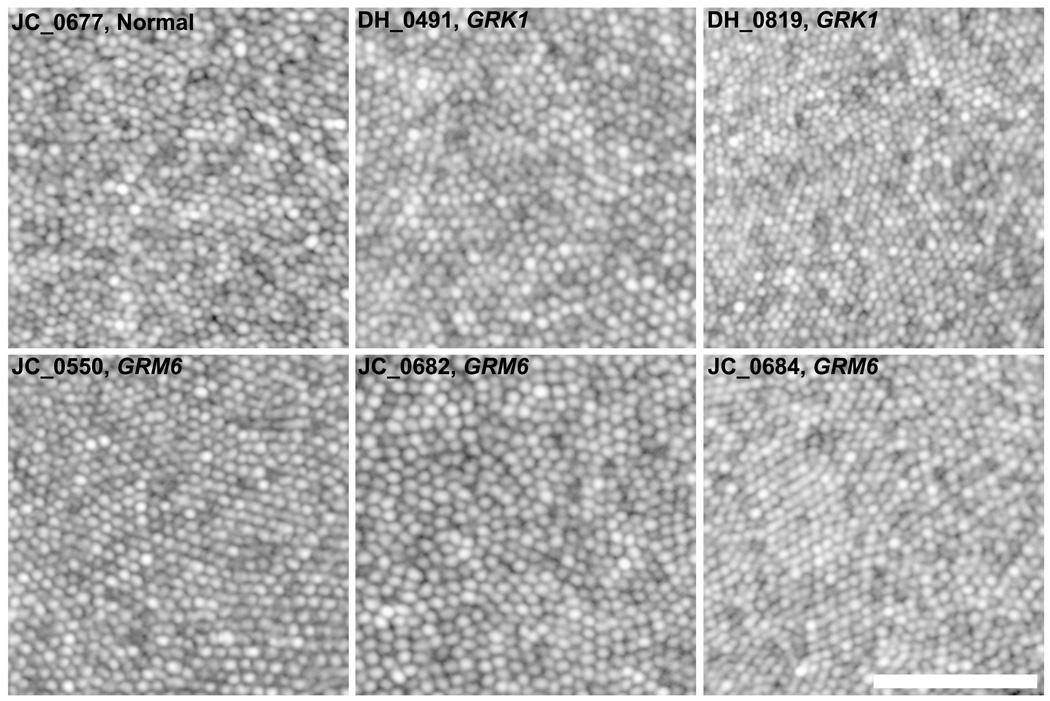
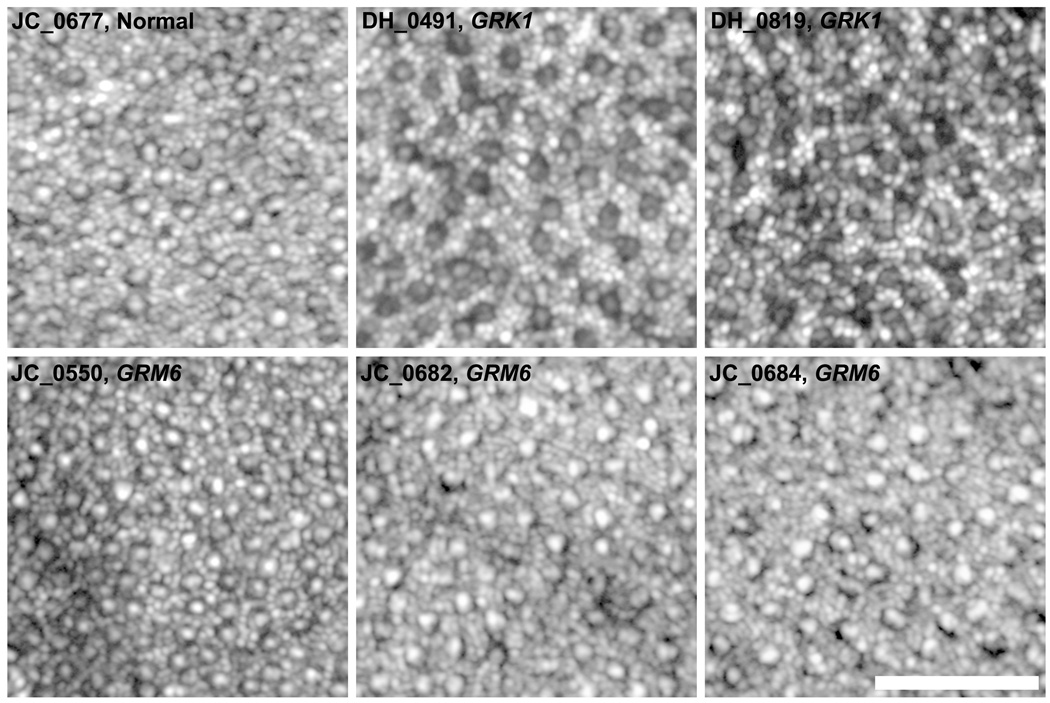
Integrity of the Photoreceptor Mosaic in Complete Congenital Stationary Night Blindness and Oguchi Disease Assessed with Adaptive Optics Retinal Imaging
All patients had a complete contiguous cone mosaic across the foveal region, with representative parafoveal images shown in Figure 4. Parafoveal (~0.6° from fixation) cone density values were 73,058 cones/mm2 (DH_0491), 83,966 cones/mm2 (DH_819), 85,289 cones/mm2 (JC_0682), 88,264 cones/mm2 (JC_0550), 80,331 cones/mm2 (JC_0684), 83,967 cones/mm2 (JC_0677). These cone density measurements were within previously measured normative values from our lab (mean±SD = 72,528±8,539 cones/mm2).53 Perifoveal photoreceptor mosaic images showed normal rod and cone photoreceptor mosaics for all patients (Figure 5). Rod density values measured at ~10° from fixation were 95,868 rods/mm2 (DH_0491), 79,669 rods/mm2 (DH_0819), 98,512 rods/mm2 (JC_0682), 82,644 rods/mm2 (JC_0550), 89,917 rods/mm2 (JC_0684), 100,826 rods/mm2 (JC_0677). Cone density values measured at ~10° from fixation were 6,612 cones/mm2 (DH_0491), 7,933 cones/mm2 (DH_819), 8,595 cones/mm2 (JC_0682), 11,900 cones/mm2 (JC_0550), 6,281 cones/mm2 (JC_0684), 7,934 cones/mm2 (JC_0677). These values were also consistent with normal values previously reported from histology and imaging studies.50, 54
Figure 4.
Images of the parafoveal cone mosaic obtained with an adaptive optics scanning light ophthalmoscope for 3 patients with complete congenital stationary night blindnes caused by GRM6 mutations, 2 brothers with Oguchi disease caused by GRK1 mutations, and 1 normal control. Images are displayed with logarithmic gray scale to facilitate visualization of the entire mosaic. All patients had complete, contiguous parafoveal cone mosaics. Scale bar is 50µm.
Figure 5.
Images of the perifoveal photoreceptor mosaic obtained with an adaptive optics scanning light ophthalmoscope for 3 patients with complete congenital stationary night blindness caused by GRM6 mutations, 2 brothers with Oguchi disease caused by GRK1 mutations, and 1 normal control. Images are displayed with logarithmic gray scale to facilitate visualization of the rod and cone mosaics. All patients had a complete rod mosaic with a coarser cone mosaic interleaved amongst the rods. Scale bar is 50µm.
Photoreceptor Intensity Changes Following Dark Adaptation
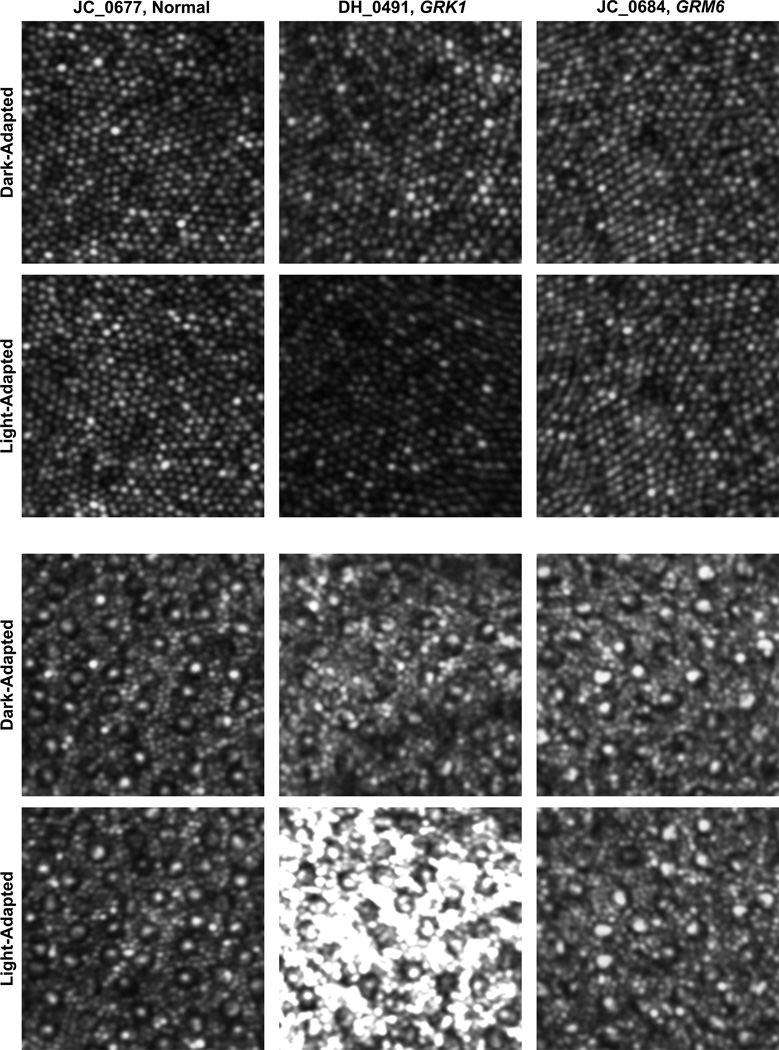
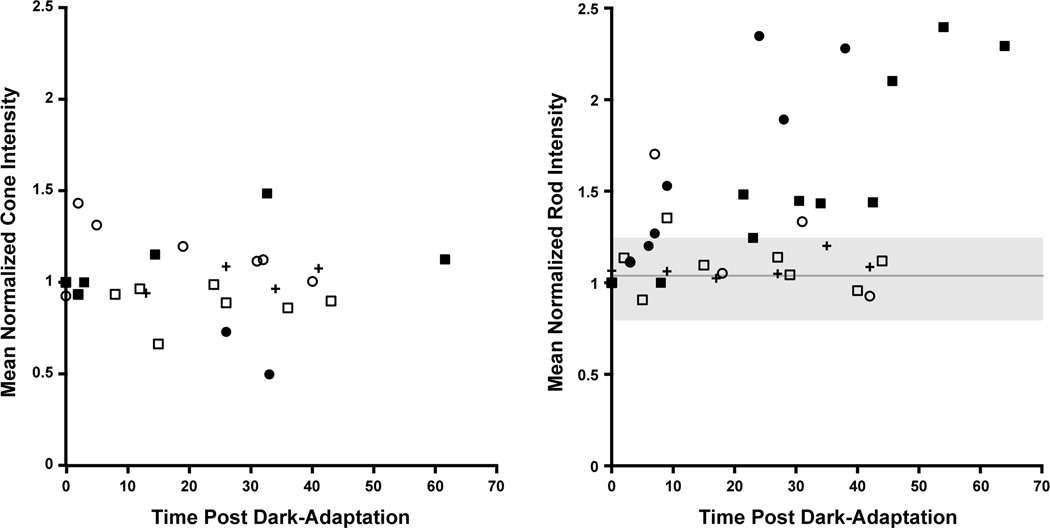
As one of the hallmarks of Oguchi disease is a pronounced difference in retinal appearance under light-versus dark-adapted conditions, we sought to analyze photoreceptor reflectance under these same conditions. Figure 6 shows representative images from three of our patients (JC_0677, DH_0491, and JC_0684). The cell-to-cell variability in intensity is seen within each retina under both conditions for both the cone (Figure 6, top) and rod (Figure 6, bottom) mosaics. The mean parafoveal cone and perifoveal rod intensity did not significantly change for the normal (JC_0677) or the patient with complete congenital stationary night blindness due to GRM6 mutations (JC_0684). However, for the patient with Oguchi disease caused by GRK1 mutations (DH_0491), we observed a pronounced increase in the relative intensity of the rods in the perifoveal image in the light-adapted state compared to the dark-adapted state. Time-lapse movies of the cone and rod mosaics for 2 of the other patients show a similar finding, with the cones and rods of a patient with complete congenital stationary night blindness (JC_0682) remaining stable and the rods (but not cones) of a patient with Oguchi disease (DH_0819) increasing upon removal from dark adaptation (Supplemental Material at AJO.com). Mean cone and rod intensity as a function of time post dark-adaptation is quantified in Figure 7. While the mean cone and rod intensity varies across the time points, the mean cone intensity did not display any systematic trends across the patient groups. In contrast, both patients with Oguchi disease (DH_0491 and DH_0819) showed a systematic increase in the mean rod intensity over time, with over a 2-fold increase in intensity over the course of the recording session. Taken together, these data indicate that changes within the rod photoreceptors are the likely source of the more global fundus changes seen in Oguchi disease.
Figure 6.
Imaging the photoreceptor mosaic under different adaptation states in 1 patient with complete congenital stationary night blindness caused by GRM6 mutations, 1 patient with Oguchi disease caused by GRK1 mutations, and 1 normal control. Images are from the parafoveal location (top set) or the perifoveal location (bottom set). The top row in each set shows images immediately after removal from a period of 2 hours of dark adaptation. The bottom row in each set shows images of the exact same patches of retina after about 30–40 minutes into the imaging session, we thus consider these images to be collected under a light adapted state, at least relative to those in the top rows. Each patch of retina is 82 × 82µm, no scale bar is given so as not to obscure any of the cones or rods. The perifoveal rod images of the Oguchi disease were the only ones to show an increase in intensity in the light-versus dark-adapted conditions.
Figure 7.
Plot of photoreceptor intensity as a function of time after removal from dark adaptation for 2 patients with complete congenital stationary night blindness caused by GRM6 mutations, 2 brothers with Oguchi disease caused by GRK1 mutations, and 1 normal control. Mean intensity for parafoveal cones (left) and perifoveal rods (right) is shown for the five patients. An average of 1,589 cones and 1,047 rods were analyzed per subject. As seen on the left, mean cone intensity was variable, but no clear trends were visible across the subjects. The mean ± SD of the cone intensity values across all subjects are plotted on the right as the solid gray line and shaded gray region, respectively. Generally, the mean rod intensity varies within this region for the two complete congenital stationary night blindness patients and the normal control, while a systematic increase in the mean rod intensity is observed for the two brothers with Oguchi disease. Filled symbols are the brothers with Oguchi disease (DH_0491, filled circles; DH_0819, filled squares), open symbols are the complete congenital stationary night blindness patients (JC_0682, open circles; JC_0684, open squares), and the crosses are the normal control (JC_0677).
Appearance of the Outer Retinal Layers on Optical Coherence Tomography Images in Oguchi Disease
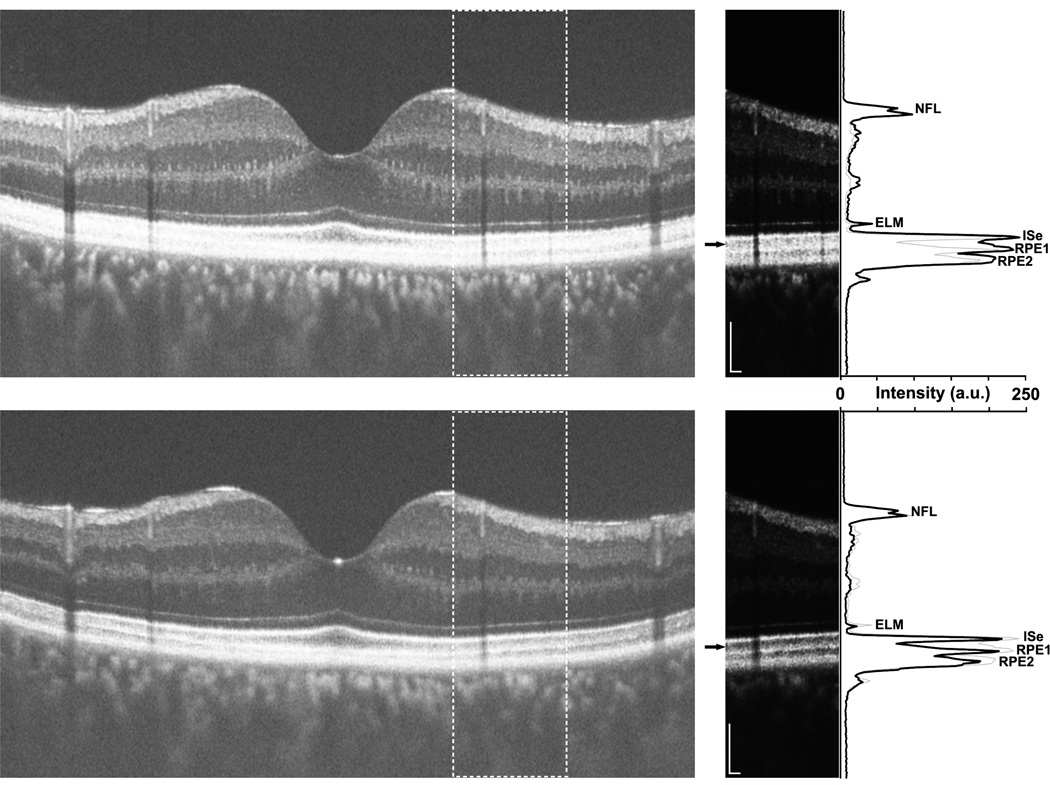
Previous OCT studies in Oguchi disease have reported a variety of outer retinal changes, including an absent IS/OS (referred to here as ISe) band, outer segment shortening, and disappearance of the hyporeflective outer segment band.37, 38, 40, 41 To further explore this issue, we obtained vertical line scans through the fovea in the left eye of one of our patients with Oguchi disease (DH_0491) under both light-adapted and dark-adapted conditions. (Figure 8) SD-OCT images are typically displayed using a logarithmic gray scale, which often makes discrimination between two closely spaced, highly reflective layers difficult. As such, our analysis was performed on linearly scaled images (Figure 8). In both imaging conditions, the four primary outer retinal layers (ELM, ISe, RPE1, RPE2) are easily seen. However, in the dark-adapted images, the Michelson contrast of the outer segment layer (the hyporeflective band between the ISe and RPE1) is higher (48%) compared to that in the light-adapted images (12%). The reduced contrast in the light-adapted images is likely due to increased scattering of the ISe and/or RPE1 layers. Also apparent from this analysis is that the outer segment length (approximated by the distance between ISe and RPE1) was not different between the two adaptation conditions (~ 23µm).
Figure 8.
High-resolution optical coherence tomography line scans (Bioptigen, Inc., Durham, North Carolina, USA) acquired along the vertical meridian of a patient with Oguchi disease caused by GRK1 mutations (DH_0491). Images were acquired under light-adapted (top) or dark-adapted (bottom) conditions. The horizontal line scans on the left are an average of 40 individual B-scans, and are displayed in the conventional logarithmic gray scale. The outer retinal layers are difficult to discriminate in the light-adapted image (top). The inset panels on the right are cropped sections (indicated by the dashed boxes in the larger line scans), displayed in a linear gray scale. Small black arrows indicate the portion of the scan corresponding to the photoreceptor outer segments, clearly visible in both light-adapted (top) and dark-adapted (bottom) conditions. Longitudinal reflectivity profile taken through the center of each inset panel is plotted along the right side of the figure as a solid black line, and demonstrate the presence of the four primary outer retinal layers (external limiting membrane, ELM; inner segment ellipsoid, ISe; and 2 layers corresponding to the retinal pigment epithelium (RPE), RPE1 and RPE2) under both adaptation conditions. The thin gray line on each plot is the longitudinal reflectivity profile from the other imaging condition, allowing comparison of layer position and contrast under the two adaptation conditions. The Michelson contrast of the outer segment layer (the hyporeflective band between the ISe and RPE1) is higher (48%) compared to that in the light-adapted images (12%). Also apparent from this analysis is that the outer segment length (approximated by the distance between ISe and RPE1) was not different between the two adaptation conditions. Scale bar is 100µm.
DISCUSSION
This study demonstrates the utility of applying advanced retinal imaging techniques to the study of inherited retinal disease. In particular, our results indicate that it is important to carefully refine the anatomical phenotype associated with the various forms of inherited retinal disease, as the phenotypic variability is likely to relate to the underlying genotype. In our patients (complete congenital stationary night blindness caused by GRM6 mutations and Oguchi disease caused by GRK1 mutations) we observed normal topography of the rod and cone mosaics, consistent with the idea that these diseases are due to functional defects in retinal neurotransmission and not morphological defects in the photoreceptors themselves. However, a temporal analysis of the rod photoreceptors revealed a notable change in rod intensity under relative light versus dark adaptation states in the two brothers with Oguchi disease caused by GRK1 mutations. Such changes were not observed in the parafoveal cones of these patients, nor in the rods or parafoveal cones of the patients with GRM6 mutations or the normal control.
In in vivo images of the photoreceptor mosaic, it has been shown that there is variability in intensity between different rods and cones. In addition, individual rods and cones vary in intensity over time, on scales ranging from seconds to hours.48, 55–57 Changes in intensity occur both in the absence and presence of a light stimulus, and it has been suggested that these changes reflect physiological activity within these photoreceptors. Our findings in Oguchi disease suggest that changes within the rod photoreceptors themselves, rather than in extracellular structures, underlie the global fundus changes seen in this disorder. This is certainly consistent with conclusions from electrophysiological studies on these patients, that confine the physiological deficit to the rods themselves.22 Previous studies have suggested that the intensity of individual photoreceptors in adaptive optics images can be affected by the length of the outer segment and changes in refractive index of the outer segment.55, 56, 58 As the specific mutations in GRK1 reported here are likely to affect the recovery phase kinetics of the rods, this could result in accumulation of some metabolite that ultimately causes a change in refractive index of the rod outer segments. Such a change could affect the intensity of these cells in the adaptive optics images. GRK1 is expressed in cones as well as rods,59, 60 thus it is surprising that cone intensity did not also change in the Oguchi patients. However as cones also express GRK7,59, 60 and recover much faster than rods (even without GRK1), it may be that this is enough to prevent any measureable change in refractive index of the cones (though deactivation kinetics may still be impacted by the deficient GRK1 activity). Further information about the mechanism of this change will be obtained by examining rod and cone reflectivity in patients with other defects in deactivation, such as RGS9/R9AP retinopathy (Bradyopsia). The ability to monitor the intensity of individual cone and rod photoreceptors provides a powerful tool with which to examine the optical behavior of the photoreceptors, which may provide a surrogate marker for cellular activity. This may be especially useful when using controlled stimulus delivery with near-infrared monitoring of photoreceptor reflectivity.61–63 Future experiments will require finer control of the adaptive state of the retina in order to correlate these changes in cell intensity with clinical measures of photoreceptor function.
Previous studies have found altered appearance of the outer retinal bands on SD-OCT in patients with Oguchi disease. One study concluded that the rod outer segments were shortened in Oguchi disease,37 two others observed high-intensity areas in the outer segment (the normally hyporeflective space between the 2nd and 3rd bands of the outer photoreceptor complex).38, 41 Another study reported disappearance of the IS/OS (referred to here as ISe) line under light-adapted conditions with re-emergence after a period of dark-adaptation.40 Our data did not show a difference in the outer segment length, nor did we observe a disappearance of the ISe band. Rather, we observed a fourfold reduction in contrast of the hyporeflective outer segment band under light adaptation. Analyzing traditional logarithmic gray scale SD-OCT images can be difficult, especially when trying to make inferences about layers that are closely spaced. Moreover, we feel that in cases like this, quantitative analyses of layer reflectivity and separation should take priority over qualitative descriptions of these outer retinal layers. While there may indeed be real differences in outer retinal structure in patients with Oguchi disease caused by different mutations, it is currently not possible to make any firm conclusions in the face of previous superficial analyses of OCT images in patients with Oguchi disease.
Our SD-OCT results also showed significant retinal thinning in the complete congenital stationary night blindness patients with GRM6 mutations, which we were able to attribute to isolated changes in the inner retina (including the ganglion cell layer), not the outer retina. This is in contrast to previous findings by Chen et al., who observed significant thinning of the photoreceptor layer (distance between OPL and Bruch’s membrane) in incomplete congenital stationary night blindness patients with CACNA1F mutations.34 Interestingly, they also observed thinning of the RGC+IPL layer.34 Differences in retinal lamination between different subtypes of congenital stationary night blindness would likely be due to differences in the cell-specific expression of the causative genes, and might be expected based on previous work on mouse models of congenital stationary night blindness.31 However it is important to note that in the Chen et al. study, the 5 incomplete congenital stationary night blindness patients with CACNA1F mutations were compared to only 4 myopic controls,34 whereas here we utilized 93 controls. As such, some of the previously reported thinning may not be real, and it would be important to utilize a single, large control group in subsequent studies of other patients with congenital stationary night blindness to better understand any significant differences in retinal structure across the different subtypes of congenital stationary night blindness.
In conclusion, our data support the emerging view that while congenital stationary night blindness may be considered a primarily stationary disorder, this does not mean that there are no structural changes in the retina in these patients. Whether such micro-architectural alterations are stable or change over time, or whether they might predispose these patients to other retinal changes remains a topic worthy of investigation, provided rigorous analytical procedures are employed.
Supplementary Material
ACKNOWLEDGEMENT
Publication of this article was supported in part by grants P30EY001931 (J.C., A.D., D.P.H.), P30EY001792 (J.J.M., G.A.F.), P30EY001730 (M.N.), T32EY014537 (A.M.D.), R01EY017607 (J.C.), UL1RR031973 (J.C.), and R00EY01951 (J.J.M) from the National Eye Institute, National Institutes of Health, Bethesda, Maryland. This publication was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1RR031973, and was conducted in part in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-016511 from the National Center for Research Resources, National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Additional support came from Foundation Fighting Blindness, Owings Mills, Maryland; the British Retinitis Pigmentosa Society; the Thomas M. Aaberg, Sr., Retina Research Fund; Moorfields Eye Hospital Special Trustees; Fight for Sight; The Gene and Ruth Posner Foundation; the RD and Linda Peters Foundation; the Pangere Corporation; Grant Healthcare Foundation; unrestricted departmental grants from Research to Prevent Blindness; and the National Institute for Health Research UK to the Biomedical Research Centre for Ophthalmology based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology. Joseph Carroll is the recipient of a Career Development Award from Research to Prevent Blindness. Alfredo Dubra holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund, and is the recipient of a Career Development Award from Research to Prevent Blindness. Edwin M. Stone is an Investigator of the Howard Hughes Medical Institute. Michel Michaelides is supported by a Foundation Fighting Blindness Career Development Award. Pooja Godara is supported by a research award from the VitreoRetinal Surgery Foundation.
Biography

Pooja Godara received her medical degree in 2004 from Kasturba Medical College, India. She completed a residency in ophthalmology at University College of Medical Sciences, Delhi, India. She is currently an imaging fellow at the Medical College of Wisconsin, sponsored by the VitreoRetinal Surgery Foundation. After this fellowship, she will begin a residency in ophthalmology at the University of Alabama-Birmingham.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors indicated no conflicts of interest.
Involved in Concept and design (P.G., R.F.C., P.I.S., M.A.G., A.R.W., A.T.M., G.A.F., D.P.H., M.M., J.C.); Analysis and interpretation (P.G., R.F.C., M.A.D., M.R.S., J.J.M., A.M.D., M.N., A.D., E.M.S., J.C.); Writing the article (P.G., R.F.C., M.N., E.M.S., J.C.); Critical revision of the article (P.G., R.F.C., P.I.S., M.A.D., M.R.S., M.A.G., J.J.M., A.R.W., A.T.M., A.M.D., M.N., A.D., G.A.F., D.P.H., M.M., J.C.); Provision of materials, patients, or resources (P.I.S., M.A.G., J.J.M., A.R.W., A.T.M., M.N., E.M.S., G.A.F., D.P.H., M.M., J.C.).
This was a prospective study design, and Institutional Review Board approval no. CHW 07/77; GC 409 was obtained from the Medical College of Wisconsin / Children’s Hospital of Wisconsin, and all research procedures adhered to the tenets of the Declaration of Helsinki. Informed consent to participate in this research was obtained from all subjects in the study, and the Health Insurance Portability and Accountability Act compliance was maintained.
REFERENCES
- 1.Carr RE. Congenital stationary night blindness. Trans Am Ophthalmol Soc. 1974;72:448–487. [PMC free article] [PubMed] [Google Scholar]
- 2.Miyake Y, Yagasaki K, Horiguchi M, Kawase Y, Kanda T. Congenital stationary night blindness with negative electroretinogram. A new classification. Arch Ophthalmol. 1986;104(7):1013–1020. doi: 10.1001/archopht.1986.01050190071042. [DOI] [PubMed] [Google Scholar]
- 3.Karpe G. The basis of clinical electroretinography. Acta Ophthalmol. 1945;24(suppl):1–118. [Google Scholar]
- 4.Schubert G, Bornschein H. Analysis of the human electroretinogram. Ophthalmologica. 1952;123(6):396–413. doi: 10.1159/000301211. [DOI] [PubMed] [Google Scholar]
- 5.Audo I, Robson AG, Holder GE, Moore AT. The negative ERG: Clinical phenotypes and disease mechanisms of inner retinal dysfunction. Surv Ophthalmol. 2008;53(1):16–40. doi: 10.1016/j.survophthal.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Miyake Y, Yagasaki K, Horiguchi M, Kawase Y. On- and off-responses in photopic electroretinogram in complete and incomplete types of congenital stationary night blindness. Jpn J Ophthalmol. 1987;31(1):81–87. [PubMed] [Google Scholar]
- 7.Langrová H, Gamer D, Friedburg C, Besch D, Zrenner E, Apfelstedt-Sylla E. Abnormalities of the long flash ERG in congenital stationary night blindness of the Schubert-Bornschein type. Vision Res. 2002;42(11):1475–1483. doi: 10.1016/s0042-6989(02)00068-8. [DOI] [PubMed] [Google Scholar]
- 8.Bech-Hansen NT, Naylor MJ, Maybaum TA, et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet. 2000;26(3):319–323. doi: 10.1038/81619. [DOI] [PubMed] [Google Scholar]
- 9.Zeitz C, van Genderen M, Neidhardt J, et al. Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15-Hz flicker electroretinogram. Invest Ophthalmol Vis Sci. 2005;46(11):4328–4335. doi: 10.1167/iovs.05-0526. [DOI] [PubMed] [Google Scholar]
- 10.Dryja TP, McGee TL, Berson EL, et al. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci USA. 2005;102(13):4884–4889. doi: 10.1073/pnas.0501233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Audo I, Kohl S, Leroy BP, et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2009;85(5):720–729. doi: 10.1016/j.ajhg.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Sergouniotis PI, Michaelides M, et al. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet. 2009;85(5):711–719. doi: 10.1016/j.ajhg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Genderen MM, Bijveld MM, Claassen YB, et al. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet. 2009;85(5):730–736. doi: 10.1016/j.ajhg.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura M, Sanuki R, Yasuma TR, et al. TRPM1 mutations are associated with the complete form of congenital stationary night blindness. Mol Vis. 2010;16:425–437. [PMC free article] [PubMed] [Google Scholar]
- 15.Peachey NS, Ray TA, Florijn R, et al. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2012;90(2):331–339. doi: 10.1016/j.ajhg.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Audo I, Bujakowska K, Orhan E, et al. Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2012;90(2):321–330. doi: 10.1016/j.ajhg.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bech-Hansen NT, Naylor MJ, Maybaum TA, et al. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19(3):264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 18.Boycott KM, Pearce WG, Bech-Hansen NT. Clinical variability among patients with incomplete X-linked congenital stationary night blindness and a founder mutation in CACNA1F. Can J Ophthalmol. 2000;35(4):204–213. doi: 10.1016/s0008-4182(00)80031-9. [DOI] [PubMed] [Google Scholar]
- 19.Zeitz C, Kloeckener-Gruissem B, Forster U, et al. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet. 2006;79(4):657–667. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wycisk KA, Zeitz C, Feil S, et al. Mutation in the auxiliary calcium-channel subunit CACNA2D4 causes autosomal recessive cone dystrophy. Am J Hum Genet. 2006;79(5):973–977. doi: 10.1086/508944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeitz C, Labs S, Lorenz B, et al. Genotyping microarray for CSNB-associated genes. Invest Ophthalmol Vis Sci. 2009;50(12):5919–5926. doi: 10.1167/iovs.09-3548. [DOI] [PubMed] [Google Scholar]
- 22.Dryja TP. Molecular genetics of Oguchi disease, fundus albipunctatus, and other forms of stationary night blindness: LVII Edward Jackson memorial lecture. Am J Ophthalmol. 2000;130(5):547–563. doi: 10.1016/s0002-9394(00)00737-6. [DOI] [PubMed] [Google Scholar]
- 23.Zeitz C. Molecular genetics and protein function involved in nocturnal vision. Expert Rev Ophthalmol. 2007;2(3):467–485. [Google Scholar]
- 24.Lodha N, Loucks CM, Beaulieu C, Parboosingh JS, Bech-Hansen NT. Congenital stationary night blindness: mutation update and clinical variability. Adv Exp Med Biol. 2012;723(5):371–379. doi: 10.1007/978-1-4614-0631-0_48. [DOI] [PubMed] [Google Scholar]
- 25.Oguchi C. Über eine Abart von Hemeralopie. Acta Soc Ophthalmol Jpn. 1907;11:123–134. [Google Scholar]
- 26.Mizuo G. On a new discovery in the dark adaptation of Oguchi's disease. Acta Soc Ophthalmol Jpn. 1913;17:1854–1859. [Google Scholar]
- 27.Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15(2):175–178. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- 28.Fuchs S, Nakazawa M, Maw MA, Tamai M, Oguchi Y, Ga lA. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10(3):360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 29.Pardue MT, McCall MA, LaVail MM, Gregg RG, Peachey NS. A naturally occurring mouse model of X-linked congenital stationary night blindness. Invest Ophthalmol Vis Sci. 1998;39(12):2443–2449. [PubMed] [Google Scholar]
- 30.Chen CK, Burns ME, Spencer M, et al. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci USA. 1999;96(7):3718–3722. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCall MA, Gregg RG. Comparisons of structural and functional abnormalities in mouse b-wave mutants. J Physiol. 2008;586(18):4385–4392. doi: 10.1113/jphysiol.2008.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaghefi HA, Green WR, Kelley JS, Sloan LL, Hoover RE, Patz A. Correlation of clinicopathologic findings in a patient: congenital night blindness, branch retinal vein occlusion, cilioretinal artery, drusen of the optic nerve head, and intraretinal pigmented lesion. Arch Ophthalmol. 1978;96(11):2097–2104. doi: 10.1001/archopht.1978.03910060477019. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe I, Taniguchi Y, Morioka K, Kato M. Congenital stationary night blindness with myopia: a clinico-pathologic study. Doc Ophthalmol. 1986;63(1):55–62. doi: 10.1007/BF00153012. [DOI] [PubMed] [Google Scholar]
- 34.Chen RWS, Greenberg JP, Lazow MA, et al. Autofluorescence imaging and spectral-domain optical coherence tomography in incomplete congenital stationary night blindness and comparison with retinitis pigmentosa. Am J Ophthalmol. 2011;153(1):143–154. doi: 10.1016/j.ajo.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usui T, Ichibe M, Ueki S, et al. Mizuo phenomenon observed by scanning laser ophthalmoscopy in a patient with Oguchi disease. Am J Ophthalmol. 2000;130(3):359–361. doi: 10.1016/s0002-9394(00)00532-8. [DOI] [PubMed] [Google Scholar]
- 36.Kuwabara Y, Ishihara K, Akiya S. Histopathological and electron microscopic studies on the retina of Oguchi's disease (article in Japanese) Acta Soc Ophthalmol Jpn. 1963;67:1323–1351. [Google Scholar]
- 37.Hashimoto H, Kishi S. Shortening of the rod outer segment in Oguchi disease. Graefes Arch Clin Exp Ophthalmol. 2009;247(11):1561–1563. doi: 10.1007/s00417-009-1114-6. [DOI] [PubMed] [Google Scholar]
- 38.Takada M, Otani A, Ogino K, Yoshimura N. Spectral-domain optical coherence tomography findings in the Mizuo-Nakamura phenomenon of Oguchi Disease. Retina. 2011;31(3):626–628. doi: 10.1097/IAE.0b013e318206cd52. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi T, Tsuzuranuki S, Kozaki K, Urashima M, Tsuneoka H. Macular dysfunction in Oguchi disease with the frequent mutation 1147delA in the SAG gene. Ophthalmic Res. 2011;46(4):175–180. doi: 10.1159/000325024. [DOI] [PubMed] [Google Scholar]
- 40.Yamada K, Motomura Y, Matsumoto CS, Shinoda K, Nakatsuka K. Optical coherence tomographic evaluation of the outer retinal architecture in Oguchi Disease. Jpn J Ophthalmol. 2009;53(5):449–451. doi: 10.1007/s10384-009-0708-1. [DOI] [PubMed] [Google Scholar]
- 41.Sergouniotis PI, Davidson AE, Sehmi K, Webster AR, Robson AG, Moore AT. Mizuo-Nakamura phenomenon in Oguchi disease due to a homozygous nonsense mutation in the SAG gene. Eye. 2011;25(8):1098–1011. doi: 10.1038/eye.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sergouniotis PI, Robson AG, Li Z, et al. A phenotypic study of congenital stationary night blindness (CSNB) associated with mutations in the GRM6 gene. Acta Ophthalmol. 2012;90(3):192–197. doi: 10.1111/j.1755-3768.2011.02267.x. [DOI] [PubMed] [Google Scholar]
- 43.Tanna H, Dubis AM, Ayub N, et al. Retinal imaging using commercial broadband optical coherence tomography. Br J Ophthalmol. 2010;94(3):372–376. doi: 10.1136/bjo.2009.163501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spaide RF, Curcio CA. Anatomical correlates to the bands seen in the outer retina by optical coherence tomography: literature review and model. Retina. 2011;31(8):1609–1619. doi: 10.1097/IAE.0b013e3182247535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carroll J, Baraas RC, Wagner-Schuman M, et al. Cone photoreceptor mosaic disruption associated with Cys203Arg mutation in the M-cone opsin. Proc Natl Acad Sci USA. 2009;106(49):20948–20953. doi: 10.1073/pnas.0910128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang Y, Cideciyan AV, Papastergiou GI, et al. Relation of optical coherence tomography to microanatomy in normal and rd chickens. Invest Ophthalmol Vis Sci. 1998;39(12):2405–2416. [PubMed] [Google Scholar]
- 47.McAllister JT, Dubis AM, Tait DM, et al. Arrested development: High-resolution imaging of foveal morphology in albinism. Vision Res. 2010;50(8):810–817. doi: 10.1016/j.visres.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cooper RF, Dubis AM, Pavaskar A, Rha J, Dubra A, Carroll J. Spatial and temporal variation of rod photoreceptor reflectance in the human retina. Biomed Opt Express. 2011;2(9):2577–2589. doi: 10.1364/BOE.2.002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dubra A, Sulai Y. Reflective afocal broadband adaptive optics scanning ophthalmoscope. Biomed Opt Express. 2011;2(6):1757–1768. doi: 10.1364/BOE.2.001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dubra A, Sulai Y, Norris JL, et al. Non-invasive imaging of the human rod photoreceptor mosaic using a confocal adaptive optics scanning ophthalmoscope. Biomed Opt Express. 2011;2(7):1864–1876. doi: 10.1364/BOE.2.001864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dubra A, Harvey Z. Biomedical Image Registration. Vol 6204. Heidelberg: Springer; 2010. Registration of 2D images from fast scanning ophthalmic instruments; pp. 60–71. [Google Scholar]
- 52.Li KY, Roorda A. Automated identification of cone photoreceptors in adaptive optics retinal images. J Opt Soc Am A. 2007;24(5):1358–1363. doi: 10.1364/josaa.24.001358. [DOI] [PubMed] [Google Scholar]
- 53.Garrioch R, Langlo C, Dubis AM, Cooper RF, Dubra A, Carroll J. Repeatability of in vivo parafoveal cone density and spacing measurements. Optometry Vision Sci. 2012;89(5):632–643. doi: 10.1097/OPX.0b013e3182540562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J Comp Neurol. 1990;292(4):497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- 55.Pallikaris A, Williams DR, Hofer H. The reflectance of single cones in the living human eye. Invest Ophthalmol Vis Sci. 2003;44(10):4580–4592. doi: 10.1167/iovs.03-0094. [DOI] [PubMed] [Google Scholar]
- 56.Pircher M, Kroisamer JS, Felberer F, Sattmann H, Götzinger E, Hitzenberger CK. Temporal changes of human cone photoreceptors observed in vivo with SLO/OCT. Biomed Opt Express. 2010;2(1):100–112. doi: 10.1364/BOE.2.000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rha J, Jonnal RS, Thorn KE, Qu J, Zhang Y, Miller DT. Adaptive optics floodillumination camera for high speed retinal imaging. Opt Express. 2006;14(10):4552–4569. doi: 10.1364/oe.14.004552. [DOI] [PubMed] [Google Scholar]
- 58.Jonnal RS, Besecker JR, Derby JC, et al. Imaging outer segment renewal in living human cone photoreceptors. Opt Express. 2010;18(5):5257–5270. doi: 10.1364/OE.18.005257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weiss ER, Ducceschi MH, Horner TJ, Li A, Craft CM, Osawa S. Species-specific differences in expression of G-protein-coupled receptor kinase (GRK) 7 and GRK1 in mammalian cone photoreceptor cells: implications for cone cell phototransduction. J Neurosci. 2001;21(23):9175–9184. doi: 10.1523/JNEUROSCI.21-23-09175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maeda T, Imanishi Y, Palczewski K. Rhodopsin phosphorylation: 30 years later. Prog Ret Eye Res. 2003;22(4):417–434. doi: 10.1016/s1350-9462(03)00017-x. [DOI] [PubMed] [Google Scholar]
- 61.Jonnal RS, Rha J, Zhang Y, Cense B, Gao W, Miller DT. In vivo functional imaging of human cone photoreceptors. Opt Express. 2007;14(24):16141–16160. [PMC free article] [PubMed] [Google Scholar]
- 62.Grieve K, Roorda A. Intrinsic signals from human cone photoreceptors. Invest Ophthalmol Vis Sci. 2008;49(2):713–719. doi: 10.1167/iovs.07-0837. [DOI] [PubMed] [Google Scholar]
- 63.Rha J, Schroeder B, Godara P, Carroll J. Variable optical activation of human cone photoreceptors visualized using short coherence light source. Opt Lett. 2009;34(24):3782–3784. doi: 10.1364/OL.34.003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.