Abstract
Multicellular organisms are composed of an interactive network of various tissues that are functionally organized as discrete organs. If aging were slowed in a specific tissue or organ how would that impact longevity at the organismal level? In recent years, molecular genetic approaches in invertebrate model systems have dramatically improved our understanding of the aging process and have provided insight into the preceding question. In this review, we discuss tissue and organ-specific interventions that prolong lifespan in the nematode C. elegans and the fruit fly Drosophila melanogaster. These interventions include reduced Insulin/IGF-1 signaling, knockdown of genes important for mitochondrial electron transport chain function and, finally, up-regulation of the Drosophila PGC-1 homolog. An emerging theme from these studies is that the intestine is an important target organ in mediating lifespan extension at the organismal level.
Keywords: Aging, Genetics, Invertebrate, Longevity, Drosophila, C. elegans
1. Introduction
The identification and characterization of genetic mutants that display extended longevity has revolutionized the way we think about and study the aging process (Kenyon, 2005; Kenyon, 2010). Studies in invertebrate model systems, such as the nematode C. elegans and the fruit fly Drosophila melanogaster, have been at the forefront of these discoveries. While employment of these two invertebrate model systems has mostly focused on producing insights into the aging process at the molecular level, these systems have also allowed for an improved understanding about the relationship between aging at the level of specific tissues or organs and organismal lifespan. Many long-lived mutants display a delay in the onset of age-related changes at the level of individual tissues or organs (Biteau et al., 2010; Garigan et al., 2002; Herndon et al., 2002; McGee et al., 2011; Wessells et al., 2004). In addition, the vast array of genetic and molecular tools available in both C. elegans and Drosophila have allowed investigators to address questions centered on the tissue or organ-specific requirements underlying extended lifespan. In other words, genetic studies can be designed to investigate the impact of exclusive manipulation of the aging rate in one tissue or organ on organismal lifespan. In this review, we discuss the large number of studies demonstrating that manipulation of aging-related genes exclusively within one organ—the intestine—results in an increased lifespan. We discuss these findings in terms of: 1) the importance of the intestine to the health and viability of the aging animal and 2) the role of the intestine as a signaling center influencing the rate of aging in responding cells.
Insulin/IGF-1 signaling pathway
The first pathway shown to modulate lifespan in animals was the insulin/IGF-1 signaling (IIS) pathway (Kenyon, 2011). Pioneering studies from a number of C. elegans researchers showed that mutations in daf-2, which encodes a hormone receptor similar to the insulin and IGF-1 receptors, dramatically improve the lifespan of the animal (Kenyon et al., 1993; Kimura et al., 1997). Other mutations affecting downstream IIS components extend lifespan as well (Johnson, 1990; Morris et al., 1996). Lifespan extensions induced by decreasing the activity of DAF-2, or downstream components of the IIS pathway are dependent upon the activity of the worm FOXO transcription factor DAF-16 (Dorman et al., 1995; Larsen et al., 1995; Lin et al., 1997; Ogg et al., 1997), the heat-shock transcription factor HSF-1 (Hsu et al., 2003); and SKN-1 (Tullet et al., 2008), a Nrf-like xenobiotic-response factor. These transcription factors, in turn, regulate the expression of a large number of downstream genes, many of which are important in mediating long life (Hsu et al., 2003; Murphy et al., 2003).
Studies in the fruit fly Drosophila provided the first evidence that reduced IIS is a ‘public’ or evolutionarily conserved mode of life extension (Partridge and Gems, 2002; Russell and Kahn, 2007). Specific mutant alleles of the Drosophila equivalent of daf-2, InR, and also chico, an insulin receptor substrate, confer extended lifespan (Clancy et al., 2001; Tatar et al., 2001). Other manipulations reported to promote long life include partial ablation of median neurosecretory cells, which produce three of the seven fly insulin-like peptides (Broughton et al., 2005) and a kinase-dead, dominant negative version of InR (InRDN) (Slack et al., 2011). Removal of dFOXO, the fly homolog of DAF-16, almost completely suppresses the lifespan extension conferred by reduced IIS in Drosophila (Slack et al., 2011; Yamamoto and Tatar, 2011).
In C. elegans, the tissue or organ-specific requirements for IIS-mediated longevity have been studied by a number of groups. Altered daf-2 gene activity in neurons can impact lifespan (Apfeld and Kenyon, 1998; Wolkow et al., 2000), however, the overall importance of the nervous system in IIS-mediated longevity is controversial (Libina et al., 2003). To determine whether increased DAF-16 activity in any single tissue was sufficient to extend the lifespan of daf-2 mutants, Libina et al., (2003) expressed a DAF-16::GFP fusion in a tissue-specific fashion in a daf-2 −/− background. As expected, expression of the fusion under the control of the daf-16 promoter in daf-16 −/−; daf-2 −/− animals, almost completely rescued their longevity to daf-16 (+); daf-2 −/− levels. Interestingly, neuronal expression of daf-16 produced only a modest (~10%) positive effect on longevity, and expressing daf-16 specifically in muscles produced no lifespan extension at all. In contrast, expressing daf-16 in the intestine increased lifespan substantially, by 50% 60%. Together, these results indicate that the intestine is an important organ in IIS-mediated lifespan extension in the worm.
As mentioned above, the worm homolog of Nrf2, called SKN-1, a member of the cap-n-collar family that induces expression of genes encoding antioxidant and detoxifying enzymes, is an important downstream component of IIS-mediated longevity (Tullet et al., 2008). Specifically, SKN-1 is required for the stress tolerance and longevity phenotypes of daf-2 mutant worms. This requirement is intriguing as it indicates that the longevity of daf-2 mutants (which also requires DAF-16) is mediated in a parallel and non-redundant fashion by both DAF-16 and SKN-1. Interestingly, SKN-1 is expressed in both the intestine and in the ASI chemosensory neurons of the worm. In both locations it has been shown to play a role in lifespan determination-but, in response to different signals. SKN-1 activity in ASI neurons, but not in the intestine, has been reported to be required for dietary restriction (DR)-mediated lifespan extension- suggesting that the role of SKN-1 in these neurons is to regulate the organism-wide response to nutrition (Bishop and Guarente, 2007). In contrast, it appears that SKN-1 activity in the intestine is important in IIS-mediated longevity (Tullet et al., 2008). In response to reduced IIS, changes in the nuclear localization of SKN-1 and upregulation of SKN-1 target genes were observed exclusively in intestinal cells. Moreover, expression of constitutively nuclear SKN-1 in the intestine is sufficient to extend life span (Tullet et al., 2008). Therefore, the intestine appears to play an important role both for DAF-16- (Libina et al., 2003) and also SKN-1-mediated (Tullet et al., 2008) lifespan extension in C. elegans. Given the importance of these molecules in mediating the pro-longevity effects of reduced IIS, these findings provide a compelling argument for a central role for the intestine in this context.
In Drosophila, the inducible Gene-Switch system (Osterwalder et al., 2001; Roman et al., 2001) has been used to activate dFOXO in different tissues of adult flies. Overexpression of dFOXO with two driver lines, S1106 (Giannakou et al., 2007; Giannakou et al., 2004) and S132 (Hwangbo et al., 2004), has been reported to increase longevity. Initially, these findings were interpreted as demonstrating that activation of dFOXO in adipose tissue was sufficient to extend lifespan. However, subsequent characterization of both S1106 (Alic et al., 2011; Poirier et al., 2008) and S132 (Poirier et al., 2008) have shown that both of these driver lines are also expressed in the intestine. Moreover, recent work with the Drosophila homolog of the human insulin-like growth factor binding protein 7, IMP-L2, supports the idea that the intestine is an important organ in IIS-mediated longevity in the fly (Alic et al., 2011). Increased expression of Imp-L2 results in phenotypic changes consistent with reduced IIS (Alic et al., 2011; Honegger et al., 2008). Furthermore, adult-onset induction Imp-L2 with S1106 results in an increased lifespan (Alic et al., 2011). Interestingly, in response to the inducing agent, an increase in IMP-L2 protein levels was detected in the intestine. However, no increase in IMP-L2 was detected in the fat body of long-lived flies. These data suggest that the intestine may be the most relevant organ in S1106-mediated life extension, including the dFOXO studies.
These studies raise an interesting question: are there specific cell types within the intestine that are important in IIS-mediated life extension? The Drosophila midgut displays functional and morphological similarities with the mammalian small intestine, as well as with other vertebrate barrier epithelia. Tissue homeostasis in the midgut is maintained by multipotent intestinal stem cells (ISCs), which are distributed along the basement membrane (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Division of an ISC gives rise to one daughter cell that retains stem cell fate and another daughter cell that becomes an enteroblast (EB), both expressing a transcription factor called Escargot (esg). In the intestine, the 5961-GeneSwitch (GS) driver line is expressed exclusively in ISCs and EBs (Mathur et al., 2010). At the same time, this driver line shows no detectable expression in muscle tissue or major organs such as the brain and gonad (Biteau et al., 2010). Therefore, 5961GS provides a powerful tool to refine our understanding of the relationship between the intestine, IIS and longevity. Remarkably, recent work has shown that moderate inhibition of IIS in ISCs/EBs is sufficient to extend lifespan (Biteau et al., 2010). More specifically, adult-onset induction of InRDN, DP110DN or RNAi of Akt, with 5961GS results in enhanced longevity (Biteau et al., 2010). To identify candidate downstream mediators of this effect, the authors activated certain FOXO target genes in ISCs/EBS and examined lifespan. Interestingly, up-regulation of two stress protective genes, Hsp68 and Jafrac1, was sufficient to extend lifespan. Therefore, expressing selected FOXO target genes in the ISC/EBs is sufficient to recapitulate the effects of reduced IIS in these cells. Taken together, the data with the S1106 and 5961GS driver lines support a model where reduced IIS in the fly intestine can lead to a longer life.
Mitochondrial electron transport chain
Mitochondria have been implicated in the aging process in a number of ways (Balaban et al., 2005; Guarente, 2008; Wallace, 2005). Interestingly, perturbation of genes important for mitochondrial electron transport chain (ETC) function has been reported to extend lifespan in diverse species including yeast (Kirchman et al., 1999), worms (Dillin et al., 2002; Feng et al., 2001; Lee et al., 2003), flies (Copeland et al., 2009) and mice (Dell’agnello et al., 2007; Lapointe and Hekimi, 2008). However, a detailed understanding of the relationship between respiratory chain activity and lifespan determination in these models is lacking. As in the case of IIS-mediated longevity, ETC-mediated longevity has been most extensively studied in C. elegans. Mutations in clk-1 (Wong et al., 1995), isp-1 (Feng et al., 2001) and nuo-6 (Yang and Hekimi, 2010b) lead to enhanced lifespan. clk-1 encodes an enzyme necessary for the biosynthesis of ubiquinone (Ewbank et al., 1997), an electron transporter of the respiratory chain, and isp-1 and nuo-6 encode subunits of the respiratory complexes. In addition, a number of studies have shown that knockdown of worm genes encoding ETC subunits also prolongs lifespan (Dillin et al., 2002; Lee et al., 2003) and this effect is conserved in Drosophila (Copeland et al., 2009).
The idea that inactivating genes required for respiration could have a positive effect on longevity seems counterintuitive. However, a model has emerged where moderate inhibition of certain ETC genes can promote longevity whereas strong inhibition is detrimental to lifespan (Copeland et al., 2009; Rea et al., 2007). Indeed, certain ETC gene mutations in both worms (Ishii et al., 1998) and flies (Walker et al., 2006) do shorten lifespan. Interestingly, in both of these cases respiratory complex II is impaired. In both Drosophila and C. elegans, there is no clear relationship between extended lifespan and metabolic parameters such as assembly of respiratory complexes, oxygen consumption and ATP levels (Copeland et al., 2009; Lee et al., 2003). These findings indicate that ETC-mediated longevity cannot simply be attributed to reduced energy production leading to a decreased ‘rate of living’. Furthermore, in both flies and worms, ETC-mediated longevity can be uncoupled from increased stress resistance and reproductive trade-offs (Copeland et al., 2009; Durieux et al., 2011; Lee et al., 2003).
What is the mechanism underlying ETC-mediated longevity? While still a work in progress, our understanding of ETC-mediated longevity has increased significantly in recent years. That said, there have been a number of surprises along the way. Initially, it was presumed that lifespan extension mediated by RNAi of ETC subunits was the same as that resulting from ETC gene mutations. However, recent work has shown that, at least in C. elegans, the underlying mechanisms appear to be distinct (Yang and Hekimi, 2010b). Historically, it has been believed that reactive oxygen species (ROS) produced by mitochondria cause age-onset damage and shorten lifespan (Balaban et al., 2005). Interestingly, however, clk-1, isp-1 and nuo-6 mutants have been reported to display increased levels of ROS (Lee et al., 2010; Yang and Hekimi, 2010a). Moreover, this increase in ROS levels appears to be both necessary and sufficient to promote longevity ROS (Lee et al., 2010; Yang and Hekimi, 2010a). In this model, ROS can trigger mechanisms that slow aging, presumably via changes in gene expression. More specifically, it has been shown that clk-1 and isp-1-mediated longevity requires the induction (via ROS) of the hypoxia-inducible factor HIF-1 (Lee et al., 2010). Related work has shown that clk-1 mutants display alterations in gene expression reminiscent of the response triggered by inhibiting respiration in yeast and mammalian cells termed the “retrograde response” (Cristina et al., 2009). In addition, both the clk-1 transcriptional response and longevity phenotype require the activities of two similar genes—fsrt-1 and fstr-2—which encode potential signaling proteins (Cristina et al., 2009). Interestingly, however, fsrt-1/2 are not required for the longevity phenotype produced by inhibiting isp-1. An RNAi screen identified the nuclear homeobox protein CEH-23 as an important downstream component of ETC-mediated longevity (Walter et al., 2011). In this case, CEH-23 is required for extended lifespan resulting from mutations in clk-1 and isp-1.
What are the tissue-specific requirements of ETC-mediated longevity? Can altered ETC gene activity in a specific tissue/organ impact longevity? And if so, is the intestine an important mediating organ in this context also? In Drosophila, our group showed that neuronal knockdown of certain ETC genes can extend lifespan (Copeland et al., 2009). However, we did not test the effects of intestine-specific knockdown of ETC genes on fly longevity. In addition, recent work in C. elegans has revealed that tissue or organ-specific manipulations of ETC gene activity can also impact aging at the level of the organism (Durieux et al., 2011). Remarkably, knock-down of cco-1, a complex IV subunit, using an intestine-specific promoter driving a cco-1 hairpin construct significantly increases worm lifespan. Muscle-specific knockdown of cco-1 produced small negative effects on lifespan in worms. Consistent with our findings in the fly (Copeland et al., 2009), neuronal knock-down of cco-1 resulted in an increase in lifespan. Interestingly, however, the impact on longevity from neuronal ETC knockdown was consistently less robust compared to the intestinal knockdown. Furthermore, knock-down of cco-1 in both the nervous system and the intestine did not extend lifespan to a greater degree than knock-down in either tissue alone.
In the same study (Durieux et al., 2011), it was also shown that the mitochondrial unfolded protein response (UPRMT) is required for ETC-mediated longevity. More specifically, treating long-lived ETC mutant animals with RNAi directed toward components of the UPRmt blocked extended lifespan. This observation lead the authors to speculate that induction of the UPRMT may play a role in the tissue-specific nature of ETC-mediated longevity. In support of this idea, RNAi of cco-1 in neurons was found to activate markers of the UPRMT in the intestine. Remarkably, neuronal RNAi of cco-1 induced a UPRMT reporter to the same extent as animals with intestinal cco-1 RNAi. These data strongly support a model where the intestine is an important organ in ETC-mediated longevity. Intestine-specific perturbation of an ETC gene is sufficient to extend lifespan. Furthermore, although neuronal ETC gene perturbation was also found to promote longevity, the intestine may play a key role in mediating this effect. Additional support for an important role for the intestine in ETC-mediated longevity can be found in other genetic studies of long-lived worms with defects in ETC-related components. As mentioned above, fstr-1 was up-regulated in clk-1 mutants, and this gene, and/or its homolog fstr-2, is required for a robust transcriptional retrograde response (Cristina et al., 2009). Moreover, knocking down fstr-1/2 activity with RNAi suppressed the clk-1 longevity phenotype. Interestingly, fstr-1 is expressed in three neurons located in the head and throughout the intestine, particularly in the anterior intestinal cells (Cristina et al., 2009). These data suggest that fstr-1 acts in the intestine and/or in specific neurons to slow the rate of aging in clk-1 mutants. In addition, the homeobox transcription factor CEH-23 is required for the prolonged lifespan phenotype of a number of different mitochondrial mutants (Walter et al., 2011). Interestingly, the expression of CEH-23::GFP is restricted to a handful of neurons and the intestine of the worm. Moreover, overexpression of ceh-23 using an endogenous promoter is sufficient to increase wild-type lifespan (Walter et al., 2011). Again, these data support the idea that the intestine may play a key role in the downstream gene expression changes that mediate longevity in response to ETC perturbations.
The peroxisome proliferator-activated receptor γ coactivator-1 (PGC-1) coactivators
In mammals, members of the PPARγ coactivator-1 (PGC-1) family of transcriptional coactivators serve as inducible coregulators of nuclear receptors in the control of cellular energy metabolism (Lin et al., 2005; Scarpulla, 2008). The founding member of the family PGC-1α was identified through its functional interaction with PPARγ in brown adipose tissue, a mitochondria-rich tissue specialized for thermogenesis (Puigserver et al., 1998). Up-regulation of PGC-1α induces and coordinates gene expression that stimulates mitochondrial oxidative metabolism (Puigserver and Spiegelman, 2003). Other members of the PGC-1 family, namely PGC-1β and PGC-related coactivator (PRC), are also implicated in modulating mitochondrial function, but their exact role is not as well characterized. PGC-1 family members also play diverse roles in energy metabolism: PGC-1α and PGC-1β have been shown to regulate adaptive thermogenesis, mitochondrial biogenesis, glucose/fatty-acid metabolism, peripheral circadian clock, fiber-type switching in skeletal muscle, and heart development (Liu and Lin, 2011).
In recent years, a number of studies have linked alterations in PGC-1 gene activity to aging and age-related diseases in mammalian species (Anderson and Prolla, 2009; Wenz, 2011). Defects in mitochondrial energy metabolism have been implicated in the pathogenesis of several neurodegenerative disorders including Huntington’s disease (HD) and Parkinson’s Disease (PD) (Lin and Beal, 2006). PGC-1α has been identified as a potential therapeutic target to treat both HD (Cui et al., 2006) and PD (Zheng et al., 2010). More specifically, increased expression of PGC-1α provides neuroprotection in transgenic HD mice (Cui et al., 2006) and separately can block dopaminergic neuron loss in cellular models of PD (Zheng et al., 2010). Mildly increased expression of PGC-1α was reported to have a therapeutic effect on the onset and progression of age-related loss of muscle mass (sarcopenia) (Wenz et al., 2009): muscle-specific expression of PGC-1α preserved mitochondrial function, neuromuscular junctions, and muscle integrity during aging.
The first report of a PGC-1 homolog in an invertebrate species came from an analysis of the transcriptional response to refeeding of dietary protein in Drosophila (Gershman et al., 2007). Strikingly, upon refeeding there was an increase in a large number of transcripts encoding genes with roles in mitochondrial biogenesis, such as mitochondrial DNA binding proteins, and nuclear encoded mitochondrial ribosome proteins. Interestingly, the study identified a single ortholog of a PGC-like peptide in the fly genome (CG9809) whose mRNA increases upon refeeding. Sequence alignments revealed that the N-terminal acidic domain that mediates transcriptional coactivation for mammalian PGC-1s, and the C-terminal arginine–serine-rich and RNA recognition domains are highly conserved in the fly protein. In the same study, they reported that CG9809 gene expression was reduced in Drosophila S2 cells that expressed a constitutively active dFOXO-A3 construct (Gershman et al., 2007).
A loss of function study further examined the role of CG9809 in regulating growth and metabolism (Tiefenbock et al., 2010). Indeed, a P-element insertion allele confers a 25% reduction in wet weight in adults, which correlated with reduced protein, lipid, glycogen and trehalose levels per animal. When normalized to body weight, reduced lipid and glycogen levels were observed in adult males. These phenotypes prompted the authors to name the mutant fly spargel-German for ‘asparagus’. Importantly, the authors demonstrated that Drosophila PGC-1 (dPGC-1/Spargel) is required for the normal expression of genes encoding mitochondrial proteins in the larval fat body. At the same time, dPGC-1 does not appear to be limiting for mitochondrial morphology or abundance in the larval fat body under normal conditions. A primary interest of our group is the relationship between mitochondrial energy metabolism and the aging process (Bahadorani et al., 2010a; Bahadorani et al., 2010b; Cho et al., 2011; Copeland et al., 2009; Walker et al., 2006). We reasoned that the fly PGC-1 homolog was an attractive target to examine in the context of lifespan determination. In the first place we set out to determine whether overexpression of dPGC-1 was sufficient to stimulate mitochondrial gene expression and/or activity. First, we examined three independent mitochondrial markers, the amount of mtDNA, citrate synthase activity and the abundance of HSP60, in response to up-regulation of dPGC-1. In doing so, we discovered that overexpression of dPGC-1 with a ubiquitous GAL4 driver (da-GAL4) leads to an increase in all three mitochondrial markers both during development and also in the adult stage (Rera et al., 2011). In addition, we demonstrated that overexpression of dPGC-1 leads to an increase in the abundance of respiratory complexes I, III, IV, and V and an increase in respiratory chain activity. In mammals, PGC-1α regulates glucose homeostasis (Herzig et al., 2001; Yoon et al., 2001) and triglyceride (TAG) metabolism (Zhang et al., 2004). Remarkably, we discovered that up-regulation of dPGC-1 confers ~20% reduction in TAG levels and a significant increase in both the amount of stored glycogen and free glucose.
Having established that dPGC-1 plays an important role in energy homeostasis in the fly-consistent with findings in mammals-we set out to examine the impact of dPGC-1 on longevity. In this context, we had no pre-conceived ideas regarding which (if any) tissue may be important in extending lifespan upon up-regulation of dPGC-1. Instead, we took an unbiased approach where we used a panel of GAL4 driver lines (with distinct expression patterns) to overexpress dPGC-1 and studied the impact on longevity. Interestingly, we failed to observe life extension upon ubiquitous, neuronal or muscle-specific overexpression of dPGC-1. Instead, we observed robust life extension with two different driver lines: S1106 and TIGS-2. These drivers are expressed in adipose tissue and the digestive tract (S1106) and the digestive tract alone (TIGS-2). As both driver lines were expressed in the intestine and the TIGS-2-mediated longevity effect was particularly robust we concluded that the intestine is an important tissue in dPGC-1-mediated life extension (Rera et al., 2011).
Given these findings, we were interested to refine the cell-type specific requirements of dPGC-1-mediated longevity. As discussed earlier, tissue homeostasis in the midgut is maintained by multipotent intestinal stem cells (ISCs), which are distributed along the basement membrane (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Division of an ISC gives rise to one daughter cell that retains stem cell fate and another daughter cell that becomes an enteroblast (EB), both expressing a transcription factor called Escargot (esg). Therefore, we examined the effects of up-regulation of dPGC-1 via esgGAL4 and 5961GS-both of which are expressed in the ISC/EBs within the intestinal epithelium. Overexpression of dPGC-1 with the constitutive esgGAL4 driver resulted in extended lifespan compared to isogenic control flies. To validate and extend this finding, we took advantage of the inducible nature the 5961GS driver to determine whether adult-only induction of dPGC-1 is sufficient to promote longevity. Indeed, 5961GS > dPGC-1 flies were significantly longer lived when exposed to the inducing agent exclusively in the adult stage. This data provides our strongest support for the idea that up-regulation of dPGC-1 within stem and progenitor cells within the intestine is sufficient to extend lifespan at the organismal level: we and others (Biteau et al., 2010) have failed to detect 5961GS induced expression in tissues or organs beyond the intestine.
To better understand these findings, we were interested to determine whether dPGC-1 activity was altered in the intestines of flies as a function of age. Remarkably, we found that dPGC-1 mRNA levels were reduced by ~60% in the intestines of aged flies. Furthermore, we discovered that aging results in a progressive loss of mitochondrial membrane potential in the mid-gut region of the intestine. This provides a potential explanation for our observation that up-regulation of dPGC-1 in the intestine leads to health benefits. Indeed, we observed that esgGAL4 > dPGC-1 flies display an increase in both mitochondrial complex I and complex II activities in the aged intestine and a maintenance of mitochondrial membrane potential, compared to controls. Our data provide an intriguing link between mitochondrial metabolism, stem cell behavior and aging-while highlighting the importance of the intestine in dPGC-1-mediated longevity (Rera et al., 2011).
Understanding the importance of the intestine in tissue-specific life extension
As discussed above, an emerging theme in invertebrate aging studies is that the intestine is an important target organ with respect to genetic interventions that extend lifespan. These findings are not limited to a single model organism, or a specific mode of life extension. Given the evolutionary distance between nematodes and arthropods, this provides a compelling argument for the generality of these findings. Furthermore, although (at least in the worm) the mechanisms of ETC- and IIS-mediated lifespan extension are clearly distinct (Dillin et al., 2002; Lee et al., 2003) the intestine has been shown to be an important target organ in both cases. Thus, the significance of intestine-specific manipulations is a unifying theme in multiple independent interventions that prolong lifespan in two of the best-studied model organisms in aging research.
Why would the intestine play such a prominent role in organismal aging? Two plausible explanations have emerged (Figure 1): 1) maintaining healthy intestinal structure/function is a critical determinant of lifespan, 2) the intestine is an important signaling center influencing the rate of aging in responding cells and tissues. Next, we will discuss the evidence supporting each of these ideas.
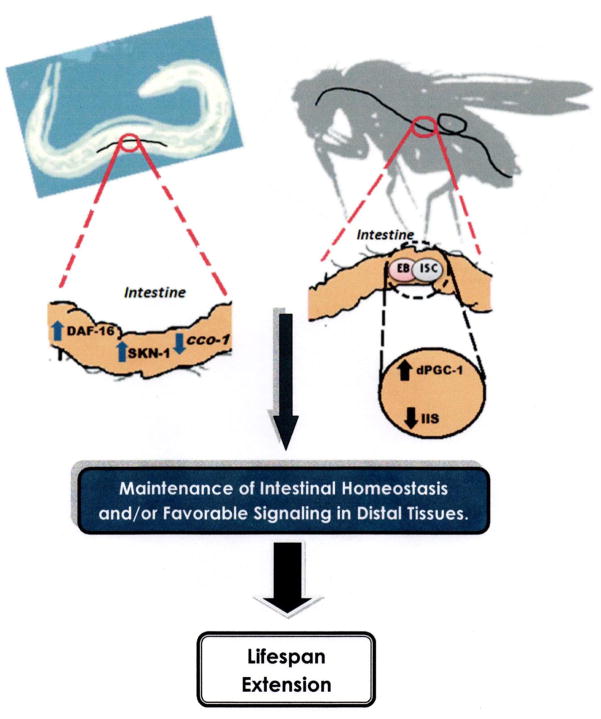
Figure 1. Intestine-specific alterations in gene expression can extend lifespan in C. elegans and D. melanogaster.
In the worm, the intestine is an important target organ for insulin/IGF-1 signaling (IIS)-mediated longevity. Overexpression of either DAF-16 or SKN-1 in the intestine leads to an increase in longevity. Furthermore, knock-down of cco-1, a mitochondrial complex IV subunit, using an intestine-specific promoter driving a cco-1 hairpin construct significantly increases worm lifespan. In the fly, a moderate reduction in IIS in intestinal stem (ISC) and progenitor (enteroblasts; EB) cells can extends lifespan. At the same time, up-regulation of dPGC-1, the Drosophila PGC-1 homolog, in ISCs/EBs also promotes longevity. Taken together, these findings point to the intestine as an important target organ for interventions that delay aging in invertebrate model systems. An emerging theme from these studies is that maintaining intestinal homeostasis may be important for the health and vitality of aging animals.
The intestine as a signaling center influencing aging in other tissues
Understanding how physiological changes in different tissues/organs are coordinated is a challenging and fundamental question in biology. In the context of aging research, progress in this area has been made using invertebrate model systems. Earlier in this review, we discussed the findings in C. elegans demonstrating that intestine-specific expression of daf-16 in daf-16 −/−; daf-2 −/− animals extended lifespan by 50–60% (compared to <20% for neuron-specific daf-16 expression) (Libina et al., 2003). To better understand these findings, the investigators established an in vivo assay for DAF-16 activity: Psod-3::gfp transgenic animals (sod-3 is a DAF-16 target). As expected, the Psod-3::gfp was highly expressed in many cell types (including neurons) in daf-2 −/− compared to wild-type animals. Interestingly, when wild-type worms were subjected to daf-2 RNAi, Psod-3::gfp was upregulated in nonneuronal tissues, but not in neurons. Yet these animals lived twice as long as normal (comparable to daf-2 −/−). Furthermore, when daf-2 −/− animals were subjected to daf-16 RNAi, Psod-3::GFP was downregulated in most tissues, but not downregulated in neurons. However, these animals lived only slightly longer than daf-16 −/−; daf-2 −/− double mutants, consistent with a modest longevity role for neuronal daf-16. Next, the Psod-3::gfp was used to determine whether increased DAF-16 activity in one tissue could lead to the up-regulation of sod-3 in other cells. Remarkably, overexpressing daf-16 in the intestines of wild-type animals increased sod-3::gfp expression not only in the intestine, but also in other tissues, including the epidermis, head and body muscles, though not neurons. Neuronal daf-16 increased sod-3::gfp expression not only in neurons, but also to a small but significant extent in the epidermis, body and head muscles. In contrast, muscle daf-16 did not increase sod-3::gfp expression in the epidermis or neurons. Together, these data demonstrate that in C. elegans the intestine is an important signaling center influencing IIS activity and, therefore, the rate of aging in distal tissues. When considering these findings, however, it is important to remember that the intestine is the major site of fat storage in C. elegans, and thus behaves as the animal’s adipose tissue.
As discussed above, recent work has shown that there exist cell non-autonomous cues from ETC gene knock-down also (Durieux et al., 2011). In this case, it was shown that neuronal ETC knock-down induced the UPRMT in intestinal cells. However, technical limitations prevented ascertaining whether intestine-specific ETC perturbations altered gene expression/physiology in other tissues. It has not escaped our attention that these findings are consistent with a model whereby neuronal knock-down of ETC genes may lead to an increase or maintenance of mitochondrial activity in the intestine (due to the induction of the UPRMT). If so, there may be mechanistic overlap with our findings on dPGC-1-mediated longevity in the fly (Rera et al., 2011).
Maintaining healthy intestinal homeostasis is an important determinant of lifespan
With hindsight, it is not surprising that these findings point to the intestine as an important organ with respect to healthy aging. Not only is the intestine essential for the uptake of nutrients that are a vital source of energy, but it is also an important barrier that protects us from toxins and pathogens in the environment. As well as a potential signaling center, another way of thinking about these findings is that they may be telling us that intestinal homeostasis is of upmost importance to the health and viability of aging animals. But, what do we know of intestinal aging in flies and worms? Despite being used as a model system in aging research for more than 20 years only a handful of studies investigating the pathobiology of aging in C. elegans have been carried out( Garigan et al., 2002; Herndon et al., 2002). However, just last year, a detailed analysis of the aging pathology of the C. elegans intestine was reported (McGee et al., 2011). These pathological changes include an increase in the variability of the shape and size of the intestinal lumen, a loss or foreshortening of microvilli in regions of the intestine, and a loss of intestinal nuclei with age. It is highly likely that these pathological changes will compromise the function of the organ and, hence, the health of the animal. For example, the loss of intestinal nuclei will affect the ability of the intestine to generate new mRNA and proteins. Furthermore, it has been reported, in a beautiful movie on David Gems’s lab website (http://www.ucl.ac.uk/~ucbtdag/), that death ‘begins’ in the worm intestine. Importantly, a systemic reduction of IIS (daf-2 −/−) is sufficient to delay markers of intestinal aging in the worm (McGee et al., 2011). Even after 45 days, at which point wild-type worms are all dead, daf-2 −/− animals do not display loss of intestinal nuclei. However, it is not clear if daf-2 mutants undergo loss of intestinal nuclei (or other markers of intestinal aging) at the very end of their lifespan.
Given the findings above, we think that it is interesting to consider the earlier work showing that DAF-16/SKN-1 activation in the intestine is sufficient to extend longevity, in the context of age-related pathological changes in the intestine. It will be very interesting and informative to examine age-related changes in the intestine in response to intestine-specific alterations in IIS/DAF-16/SKN-1. It is possible that these manipulations may delay (or prevent) markers of intestinal aging. If so, the resulting increase in lifespan that results from intestine-specific alterations in DAF-16/SKN-1 may result from maintenance of healthy intestinal structure/function during aging.
Just a few years ago, little was known with respect to age-related changes in the Drosophila intestine. However, in recent years, progress has been made in this area such that we can now define a number of hallmarks of intestinal aging in Drosophila (Biteau et al., 2008; Choi et al., 2008; Park et al., 2009). As discussed earlier, the fly midgut is composed of a limited number of cell types: large and polyploid EnteroCytes (ECs), the main absorptive cells in the epithelium; several types of small diploid EnteroEndocrine cells (EEs) and the common progenitors of these cells, the intestinal stem cells (ISCs: marked by expression of the transcription factor escargot, esg) and their diploid daughter cells, EnteroBlasts (EBs). Initially, it was reported that there was a dramatic increase in stem cell markers (e.g., esg+) in the guts of old flies (Choi et al., 2008). This was proposed to reflect an increase in the number of stem cells as well as of EBs and a decline in the number of differentiated ECs. Subsequently, it was shown that the increased number of esg+ cells in old guts does not simply reflect an increase in the number of ISCs but is caused by retention of stem cell markers in cells that have EC-like morphology (Biteau et al., 2008). These cells fail to express EC markers and accumulate basally in the epithelium. The observed age-related loss of epithelial homeostasis is thus caused by a combination of increased ISC proliferation and misdifferentiation (Biteau et al., 2008).
As is the case in C. elegans (McGee et al., 2011), a systemic reduction in IIS can delay intestinal aging in Drosophila (Biteau et al., 2010). Long-lived InR or chico mutant flies display a delay in the accumulation of markers of intestinal aging, including markers of ISC proliferation. As discussed earlier, a moderate reduction in IIS in ISCs and EBs is sufficient to extend lifespan (Biteau et al., 2010). Furthermore, overexpression of stress protective genes (regulated by FOXO and hence presumably activated under conditions of reduced IIS), Hsp68 and Jafrac1, is also sufficient to extend lifespan. Importantly, a moderate reduction in IIS in the ISC/EB lineage leads to improved intestinal homeostasis in aged flies (Biteau et al., 2010). These data are consistent with a model whereby a moderate decrease in ISC proliferation is associated with a long lifespan. Additional support from this model comes from our own work with dPGC-1 (Rera et al., 2011): up-regulation of dPGC-1 in the digestive tract, including restricted expression in ISC/EBs, results in extended lifespan. Furthermore, up-regulation of dPGC-1 abrogates the precocious activation of ISC proliferation and delays the accumulation of mis-differentiated cells in the intestinal epithelium (Rera et al., 2011). In other words, these intestine-specific manipulations that promote longevity in Drosophila delay the onset of markers of intestinal aging.
In summary, aging is not kind to the intestines of worms or flies: a number of age-related cellular and structural changes have been documented to occur in the intestines of both organisms. It is tempting to speculate that these cellular changes may manifest functional outcomes in aging animals. An area of obvious interest is the relationship between nutrient uptake, intestinal aging and longevity. It is possible that alterations in nutrient uptake resulting from intestinal degeneration may impact the health of the aging animal. If so, then we would predict that there would be a decline in nutrient stores in aging animals and that manipulations that delay intestinal aging would result in improved maintenance of metabolic health. Indeed, overexpression of stress-protective FOXO target genes, via esgGAL4, results in increased nutrient stores in aged flies (Biteau et al., 2010). This rescue of metabolic homeostasis correlates with increased starvation tolerance, further supporting the idea that maintenance of intestinal homeostasis is critical for the metabolic health of aging flies.
In recent years, a more expansive role for the gastrointestinal tract is emerging as an important source of both neural and systemic signals regulating appetite and body weight (Sandoval et al., 2008; Strader and Woods, 2005). In mammals, specific neuronal populations and brain/gut hormones are responsible for sensing and responding to homeostatic metabolic changes. Deregulation of these sensors/effectors has been shown to contribute significantly to diabetes or metabolic syndrome (Sandoval et al., 2008; Strader and Woods, 2005). Recently, it was shown that distinct domains of the Drosophila intestine are innervated by both efferent and sensory neurons (Cognigni et al., 2011). Moreover, this study uncovered a central role for the Drosophila intestine in the integration of neural, nutritional, and reproductive information to adjust nutrient intake and utilization. Hence, altering or improving gut homeostasis during aging could modify the ability of this organ to sense and respond to environmental conditions-with important consequences to the health of the aging fly.
Although the major function of the intestinal epithelium is to absorb nutrients, we should remind ourselves that the epithelium also serves as an important barrier between the ‘outside’ and ‘inside’ world. Any compromise in intestinal barrier function could expose the ‘inside’ world to potentially harmful toxins, irritants, bacteria and other pathogens that also exist in the gut lumen. But, before we consider such potential consequences, we must first ask a simple question: is there a loss of intestinal barrier function in aged invertebrate model organisms? We were interested in this question with respect to our work on the Drosophila PGC-1 homolog (dPGC-1). To develop an assay of intestinal barrier function, we examined flies of different ages that had consumed a non-absorbable blue food dye (Rera et al., 2011). Remarkably, when we did so, we observed a striking difference in the localization of the dye in young vs old flies after feeding. In young flies, as expected, the blue dye was found exclusively within the proboscis and digestive tract post-feeding. However, when we examined old flies we observed a significant fraction of the population where the dye was found throughout the body. We interpret the “Smurf” phenotype, i.e., the leakage of dye into the haemolymph and consequently all tissues, to reflect a defect(s) in intestinal integrity. Importantly, we observed that esgGAL4-mediated activation of dPGC-1 retards the age-related onset of the “Smurf” phenotype. Therefore, an increase in dPGC-1 expression within the digestive tract results in improved intestinal integrity in aged flies, which is consistent with the delay in cellular markers of intestinal aging that we previously discussed. Our findings suggest that age-related alterations in intestinal barrier function may be important for organismal aging. In this context, it is interesting to consider the observation that IIS-mediated longevity, in worms, flies and mice, has been linked to an up-regulation of broad spectrum cellular detoxification (that is, the phase 1, phase 2 xenobiotic or drug detoxification system) (McElwee et al., 2007).
We acknowledge that the role of intestinal flora is also emerging as potentially relevant to intestinal homeostasis and longevity. Indeed, in Drosophila, age-related intestinal defects are significantly dampened (as indicated by the mitotic index) under axenic conditions (Buchon et al., 2009) and in C. elegans a strong inverse correlation between intestinal bacterial load and lifespan has recently been reported (Portal-Celhay et al., 2012). Furthermore, it has been shown that the Drosophila microbiome modulates host development and energy homeostasis via the IIS pathway (Shin et al., 2011). A promising area of future work, therefore, would be to examine the relationship between intestinal integrity, the gut flora and aging.
Is the intestine significant in mammalian lifespan extension?
Thus far we have discussed data to substantiate the notion that the intestine may be important in mediating the lifespan-extending effect of diverse genetic interventions in C. elegans and Drosophila. Is such importance merely limited to these invertebrates, or may the intestine play a significant role in mammalian longevity as well? As one of the ultimate goals of aging research is to develop therapeutic interventions for aging humans, the elucidation of this question is of obvious value.
The fact that there is a significant evolutionary distance between nematodes and arthropods—yet both demonstrate lifespan-extension that may be mediated by intestinal changes—is evidence that the potential intestinal-mediation of lifespan extension may be a public mechanism applicable to mammals as well. As with nematodes and flies, age-related changes in the intestines of mammals have also been observed—changes that may manifest themselves as functionally detrimental to organismal physiology and survival. For example, rodents exhibit an increase in intestinal barrier permeability (Katz et al., 1987) and a reduction in intestinal sugar and amino acid transport with age (Esposito et al., 1985; Navab and Winter, 1988). To our knowledge, studies of genetic interventions that extend lifespan in mammalian models have not reported associated or required changes in intestinal biology. However, mice subjected to caloric restriction, a dietary intervention that robustly extends lifespan in a number of diverse invertebrates and mammals, were observed to maintain an intestinal absorption capacity for sugars and amino acids nearly twice as high as that of control mice, thereby associating enhanced and/or maintained intestinal function with lifespan extension (Casirola et al., 1997). Whether or not such an association is causal remains to be deciphered.
Conclusions
It is an exciting time to be studying the basic biology of aging. Invertebrate model systems continue to lead the way when it comes to uncovering the deep secrets of aging. One of the most remarkable findings of recent years has been that organ-specific changes in gene expression can extend lifespan. Many of these studies, discussed herein, have focused on the intestine. We have discussed the intestine both as a signaling center and also as a central player in maintaining metabolic homeostasis. Trying to unravel the precise mechanisms by which intestinal homeostasis influences aging at the level of the organism will require considerable time, energy and creativity. In the meantime, are we tempted to speculate? Why is the intestine so important in this context? What is our ‘gut feeling’? Here, we borrow from the immortal words of Carl Sagan: “I try not to think with my gut. If I’m serious about understanding the world, thinking with anything besides my brain, as tempting as that might be, is likely to get me into trouble. Really, it’s okay to reserve judgment until the evidence is in.”
Highlights.
Intestine-specific alterations in Insulin/IGF-1 signaling can result in longer lifespan in worms and flies
Knock-down of a mitochondrial electron transport chain subunit in the intestine of worms extends lifespan
Intestine-specific up-regulation of the Drosophila PGC-1 homolog extends lifespan
In the worm, the intestine acts as a signaling center influencing the rate of aging in responding cells.
Maintaining intestinal homeostasis is important for healthy aging
Acknowledgments
We apologize to our colleagues whose work we were unable to discuss due to space limitations. D.W.W is supported by the National Institute on Aging (NIH RO1 AG037514) and the Ellison Medical Foundation. D.W.W is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alic N, Hoddinott MP, Vinti G, Partridge L. Lifespan extension by increased expression of the Drosophila homologue of the IGFBP7 tumour suppressor. Aging cell. 2011;10:137–147. doi: 10.1111/j.1474-9726.2010.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Prolla T. PGC-1alpha in aging and anti-aging interventions. Biochimica et biophysica acta. 2009;1790:1059–1066. doi: 10.1016/j.bbagen.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Bahadorani S, Cho J, Lo T, Contreras H, Lawal HO, Krantz DE, Bradley TJ, Walker DW. Neuronal expression of a single-subunit yeast NADH-ubiquinone oxidoreductase (Ndi1) extends Drosophila lifespan. Aging cell. 2010a;9:191–202. doi: 10.1111/j.1474-9726.2010.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadorani S, Hur JH, Lo T, Jr, Vu K, Walker DW. Perturbation of mitochondrial complex V alters the response to dietary restriction in Drosophila. Aging cell. 2010b;9:100–103. doi: 10.1111/j.1474-9726.2009.00537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell stem cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS genetics. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, Partridge L. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes & development. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casirola DM, Lan Y, Ferraris RP. Effects of changes in calorie intake on intestinal nutrient uptake and transporter mRNA levels in aged mice. The journals of gerontology Series A, Biological sciences and medical sciences. 1997;52:B300–310. doi: 10.1093/gerona/52a.6.b300. [DOI] [PubMed] [Google Scholar]
- Cho J, Hur JH, Walker DW. The role of mitochondria in Drosophila aging. Experimental gerontology. 2011;46:331–334. doi: 10.1016/j.exger.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA. Age-related changes in Drosophila midgut are associated with PVF2, a PDGF/VEGF-like growth factor. Aging cell. 2008;7:318–334. doi: 10.1111/j.1474-9726.2008.00380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Cognigni P, Bailey AP, Miguel-Aliaga I. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell metabolism. 2011;13:92–104. doi: 10.1016/j.cmet.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Current biology: CB. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS genetics. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst CN, Tanese N, Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Dell’agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Human molecular genetics. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Faelli A, Tosco M, Orsenigo MN, Battistessa R. Age-related changes in rat intestinal transport of D-glucose, sodium, and water. The American journal of physiology. 1985;249:G328–334. doi: 10.1152/ajpgi.1985.249.3.G328. [DOI] [PubMed] [Google Scholar]
- Ewbank JJ, Barnes TM, Lakowski B, Lussier M, Bussey H, Hekimi S. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science. 1997;275:980–983. doi: 10.1126/science.275.5302.980. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiological genomics. 2007;29:24–34. doi: 10.1152/physiolgenomics.00061.2006. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jacobson J, Vinti G, Leevers SJ, Partridge L. Dynamics of the action of dFOXO on adult mortality in Drosophila. Aging cell. 2007;6:429–438. doi: 10.1111/j.1474-9726.2007.00290.x. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–176. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, Sakano Y, Paupard MC, Hall DH, Driscoll M. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- Honegger B, Galic M, Kohler K, Wittwer F, Brogiolo W, Hafen E, Stocker H. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. Journal of biology. 2008;7:10. doi: 10.1186/jbiol72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Johnson TE. Increased life-span of age-1 mutants in Caenorhabditis elegans and lower Gompertz rate of aging. Science. 1990;249:908–912. doi: 10.1126/science.2392681. [DOI] [PubMed] [Google Scholar]
- Katz D, Hollander D, Said HM, Dadufalza V. Aging-associated increase in intestinal permeability to polyethylene glycol 900. Digestive diseases and sciences. 1987;32:285–288. doi: 10.1007/BF01297055. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. The Journal of biological chemistry. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current biology : CB. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Liu C, Lin JD. PGC-1 coactivators in the control of energy metabolism. Acta biochimica et biophysica Sinica. 2011;43:248–257. doi: 10.1093/abbs/gmr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Bost A, Driver I, Ohlstein B. A transient niche regulates the specification of Drosophila intestinal stem cells. Science. 2010;327:210–213. doi: 10.1126/science.1181958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, Gems D. Evolutionary conservation of regulated longevity assurance mechanisms. Genome biology. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Weber D, Day N, Vitelli C, Crippen D, Herndon LA, Hall DH, Melov S. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging cell. 2011;10:699–710. doi: 10.1111/j.1474-9726.2011.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Navab F, Winter CG. Effect of aging on intestinal absorption of aromatic amino acids in vitro in the rat. The American journal of physiology. 1988;254:G631–636. doi: 10.1152/ajpgi.1988.254.4.G630. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Kim YS, Yoo MA. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging (Albany NY) 2009;1:637–651. doi: 10.18632/aging.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D. Mechanisms of ageing: public or private? Nature reviews. Genetics. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- Poirier L, Shane A, Zheng J, Seroude L. Characterization of the Drosophila gene-switch system in aging studies: a cautionary tale. Aging cell. 2008;7:758–770. doi: 10.1111/j.1474-9726.2008.00421.x. [DOI] [PubMed] [Google Scholar]
- Portal-Celhay C, Bradley ER, Blaser MJ. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC microbiology. 2012;12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocrine reviews. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS biology. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler CL, Ulgherait M, Hur JH, Ansari WS, Lo T, Jr, Jones DL, Walker DW. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell metabolism. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR. Endocrine regulation of ageing. Nature reviews Molecular cell biology. 2007;8:681–691. doi: 10.1038/nrm2234. [DOI] [PubMed] [Google Scholar]
- Sandoval D, Cota D, Seeley RJ. The integrative role of CNS fuel-sensing mechanisms in energy balance and glucose regulation. Annual review of physiology. 2008;70:513–535. doi: 10.1146/annurev.physiol.70.120806.095256. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiological reviews. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- Slack C, Giannakou ME, Foley A, Goss M, Partridge L. dFOXO-independent effects of reduced insulin-like signaling in Drosophila. Aging cell. 2011;10:735–748. doi: 10.1111/j.1474-9726.2011.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader AD, Woods SC. Gastrointestinal hormones and food intake. Gastroenterology. 2005;128:175–191. doi: 10.1053/j.gastro.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tiefenbock SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. The EMBO journal. 2010;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DW, Hajek P, Muffat J, Knoepfle D, Cornelison S, Attardi G, Benzer S. Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:16382–16387. doi: 10.1073/pnas.0607918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annual review of genetics. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS biology. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T. Mitochondria and PGC-1alpha in Aging and Age-Associated Diseases. Journal of aging research. 2011;2011:810619. doi: 10.4061/2011/810619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nature genetics. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans lifespan by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Tatar M. Insulin receptor substrate chico acts with the transcription factor FOXO to extend Drosophila lifespan. Aging cell. 2011;10:729–732. doi: 10.1111/j.1474-9726.2011.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS biology. 2010a;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging cell. 2010b;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, Grunblatt E, Moran LB, Mandel SA, Riederer P, Miller RM, Federoff HJ, Wullner U, Papapetropoulos S, Youdim MB, Cantuti-Castelvetri I, Young AB, Vance JM, Davis RL, Hedreen JC, Adler CH, Beach TG, Graeber MB, Middleton FA, Rochet JC, Scherzer CR. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Science translational medicine. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]