Abstract
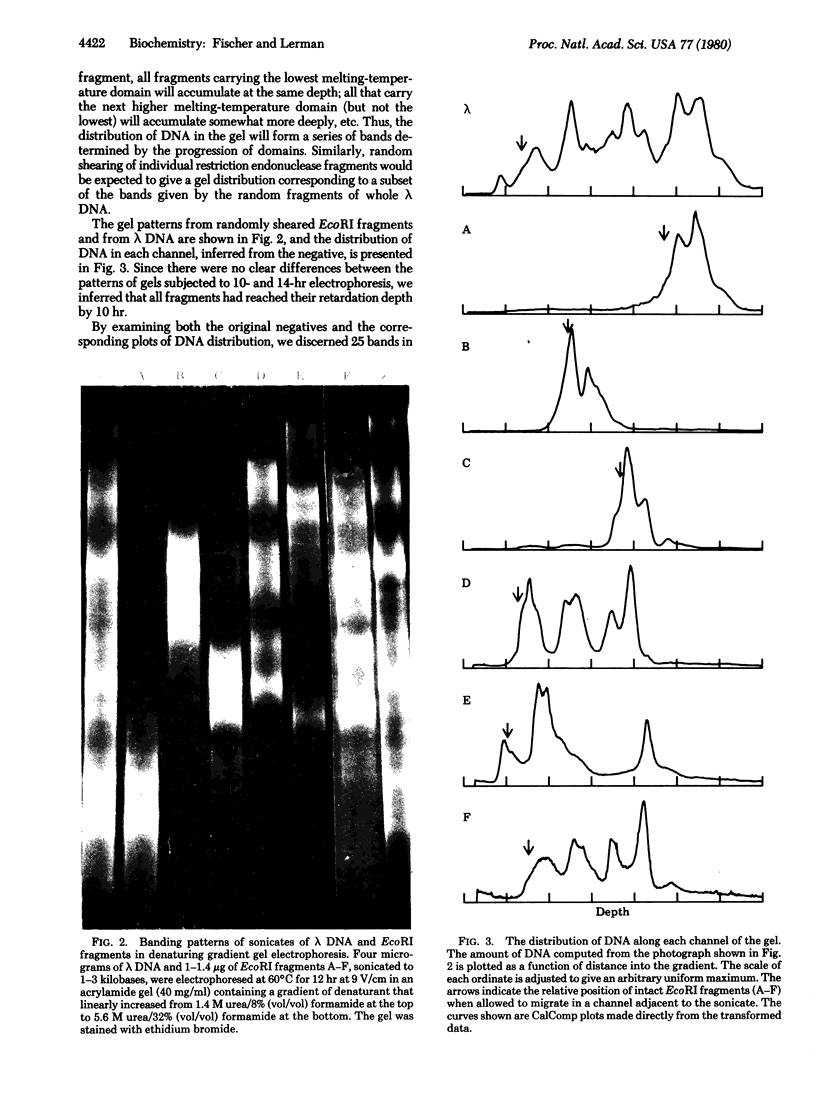
The separation of DNA fragments by electrophoresis at high temperature in a denaturing gradient is independent of the length of the fragments. We have suggested that the basis of fragment separation is that each DNA molecule undergoes partial melting as it encounters a concentration of denaturants sufficient to melt its least stable sequence, while other sequences remain double stranded; in the partially melted configuration, DNA can continue migration only slowly. This model is consistent with the observation that fragments of lambda phage DNA cleaved by different restriction endonucleases reach the same final depth in the gel if they contain the same least-stable sequence. A unique set of bands is produced from the electrophoresis of randomly fragmented DNA; this would be expected if there were a limited number of melting centers occupying discrete genetic loci. An intact DNA molecule penetrates about as deeply into the gel as the uppermost band after fragmentation; this would be expected only if the least-stable sequence controls the final depth of the whole molecule.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama C., Gotoh O., Wada A. Spectral analysis on the melting fine structure of is lambda DNA and T2 DNA. Biopolymers. 1977 Feb;16(2):427–435. doi: 10.1002/bip.1977.360160215. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Haydock P. V. Effect of sodium ion on the high-resolution melting of lambda DNA. Biopolymers. 1979 Dec;18(12):3089–3109. doi: 10.1002/bip.1979.360181214. [DOI] [PubMed] [Google Scholar]
- Falkow S., Cowie D. B. Intramolecular heterogeneity of the deoxyribonucleic acid of temperate bacteriophages. J Bacteriol. 1968 Sep;96(3):777–784. doi: 10.1128/jb.96.3.777-784.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Length-independent separation of DNA restriction fragments in two-dimensional gel electrophoresis. Cell. 1979 Jan;16(1):191–200. doi: 10.1016/0092-8674(79)90200-9. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. Two-dimensional electrophoretic separation of restriction enzyme fragments of DNA. Methods Enzymol. 1979;68:183–191. doi: 10.1016/0076-6879(79)68013-8. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Husimi Y., Yabuki S., Wada A. Hyperfine structure in melting profile of bacteriophage lambda DNA. Biopolymers. 1976 Apr;15(4):655–670. doi: 10.1002/bip.1976.360150406. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Wada A., Yabuki S. Salt-concentration dependence of melting profiles of lambda phage DNAs: evidence for long-range interactions and pronounced end effects. Biopolymers. 1979 Apr;18(4):805–824. doi: 10.1002/bip.1979.360180406. [DOI] [PubMed] [Google Scholar]
- Hutton J. R. Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res. 1977 Oct;4(10):3537–3555. doi: 10.1093/nar/4.10.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B. A denaturation map of the lambda phage DNA molecule determined by electron microscopy. J Mol Biol. 1966 Jul;18(3):464–476. doi: 10.1016/s0022-2836(66)80037-2. [DOI] [PubMed] [Google Scholar]
- Inman R. B. Denaturation maps of the left and right sides of the lambda DNA molecule determined by electron microscopy. J Mol Biol. 1967 Aug 28;28(1):103–116. doi: 10.1016/s0022-2836(67)80081-0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki S., Gotoh O., Wada A. Fine structures in denaturation curves of bacteriophage lambda DNA. Their relation to the intramolecular heterogeneity in base compositon. Biochim Biophys Acta. 1975 Jul 7;395(3):258–273. doi: 10.1016/0005-2787(75)90196-3. [DOI] [PubMed] [Google Scholar]