Abstract
General intelligence (g) and virtually all other behavioral traits are heritable. Associations between g and specific single-nucleotide polymorphisms (SNPs) in several candidate genes involved in brain function have been reported. We sought to replicate published associations between 12 specific genetic variants and g using three independent, longitudinal datasets of 5571, 1759, and 2441 well-characterized individuals. Of 32 independent tests across all three datasets, only one was nominally significant at the p < .05 level. By contrast, power analyses showed that we should have expected 10–15 significant associations, given reasonable assumptions for genotype effect sizes. As positive controls, we confirmed accepted genetic associations for Alzheimer disease and body mass index, and we used SNP-based relatedness calculations to replicate estimates that about half of the variance in g is accounted for by common genetic variation among individuals. We conclude that different approaches than candidate genes are needed in the molecular genetics of psychology and social science.
Genetics has great potential to contribute to psychology and the social sciences for at least two reasons. First, as human behavior involves the operation of the brain, understanding the genes whose expression affects the development and physiology of the brain can further our understanding of the causal chains connecting evolution, brain, and behavior. Second, because genetic differences can potentially account for some of the differences among individuals in cognitive function, behavior, and outcomes, any effort to paint a picture of the structure of human differences that does not incorporate genetics will be incomplete and possibly misleading.
Within psychology, the genetics of behavior has been explored since the earliest twin studies (for an overview, see Plomin et al., 2008). Behavior genetic studies have shown that nearly all human behavioral traits are heritable (Turkheimer, 2000). If a trait is heritable in the general population, then—with sufficiently large samples—it should be possible in principle to identify molecular genetic variants that are associated with the trait. General cognitive ability, or g (Spearman, 1904; Neisser et al., 1996; Plomin et al., 2008) is among the most heritable behavioral traits. Estimates of broad heritability as high as 0.80 have been reported for adult IQ measured in modern Western populations (Bouchard, 1998). Although the exact figures have been the topic of much debate, the claim that IQ is at least moderately heritable is widely accepted. IQ may in fact be similar in heritability to the physical trait of height (Weedon & Frayling, 2008). Both height and IQ are genetically “complex” because these traits are influenced by many genes, acting in concert with environmental factors, rather than being determined by single genetic variants. Finding genes associated with g could yield many potential benefits, among them new insights into the biology of cognition and its disorders. Such discoveries might suggest new therapeutic targets or pathways for potential treatments to improve cognition. Uncovering the molecular genetics of other traits and abilities, such as personality, time and risk preferences, and social skills could have similarly beneficial consequences (Benjamin et al., 2007).
By now there is a large literature of candidate gene studies showing associations between many single-nucleotide polymorphisms (SNPs) and g.1 Payton (2009) produced a comprehensive review of these studies. Here we report the results of a series of attempts to replicate as many published SNP-g associations as possible, using data from three independent, large, well-characterized, longitudinal samples. We begin, in Study 1, with the Wisconsin Longitudinal Study (WLS; www.ssc.wisc.edu/wlsresearch), which includes genotypes for 13 of the SNPs reported by Payton (2009) to have published associations with g. These 13 SNPs are located in or near 10 different genes. In followup studies, we test 10 of the original 13 SNPs that were available in two other samples. In Study 2, we use the Framingham Heart Study (FHS; www.framinghamheartstudy.org), and in Study 3, we use data from the Swedish Twin Registry (STR; ki.se/ki/jsp/polopoly.jsp?d=9610&l=en) to examine associations with g. Although we analyzed them separately, the combined sample size of these datasets is almost 10,000 individuals, which gives us considerable statistical power.
If the published SNP-g associations we examined were true positives in the general population, then we would expect many of them to replicate at the 5% significance level in our much larger datasets. However, if the literature on SNP-g associations consists mostly of false positives, then we would expect very few replications in our data. Such a result would not likely be due to differences in the methods used to estimate g in the various datasets under comparison, since g is consistently measured by a wide variety of well-designed tests (Ree & Earles, 1991).
Study 1
Method
The Wisconsin Longitudinal Study (WLS) is based on a one-third sample of all Spring 1957 Wisconsin high school graduates (initial N = 10,317). A randomly selected sibling of a subsample of these graduates was enrolled in 1977 and a randomly selected sibling of each remaining graduate was enrolled in 1993 (N = 5,219). g was measured by the Henmon-Nelson Test of Mental Ability (Lamke & Nelson, 1957) for both graduate and sibling sample members when they were in the 11th grade, and obtained from administrative records. Percentile scores were rescaled to the conventional IQ metric of a mean of 100 and standard deviation of 15.
We studied all 13 SNPs that were both previously associated with g according to Payton’s review (2009) and included among the 90 SNPs genotyped in the WLS. They were: rs429358 and rs7412 in APOE (these SNPs define the e2/e3/e4 haplotype associated with Alzheimer disease), rs6265 in BDNF, rs2061174 in CHRM2, rs8191992 in CHRM2/CHRNA4, rs4680 in COMT, rs17571 in CTSD, rs821616 in DISC1, rs1800497 in DRD2/ANKK1, rs1018381 in DTNBP1, rs760761 in DTNBP1, rs363050 in SNAP25, and rs2760118 in SSADH (aka ALDH5A1).
Of the 6,908 WLS respondents with adequate covariate and genotype data, 5,571 had data for g and for all 13 SNPs previously associated with g. All 13 SNP genotypes were in Hardy-Weinberg equilibrium, and their frequencies matched those reported in the literature for European samples.
As positive controls for global problems in genotyping or data quality, we considered two genotype-phenotype associations that have been established and accepted: APOE and Alzheimer’s disease (AD), and FTO and body mass index (BMI). We tested the two SNPs in the APOE gene that define the common, well-established risk haplotype for AD (e2/e3/e4) for association with parental AD status. As expected, subjects with at least one e4 allele were more likely to report having a parent with AD than were subjects with no e4 alleles (p < .0001). Likewise, the previously reported and replicated association between the number of C alleles of SNP rs1421085 in FTO and body mass index (Tung & Yeo, 2011) was observed here (self-reported BMIs of 27.5, 27.9, and 28.3 for 0, 1, and 2 C alleles, respectively; p < .001).
For each SNP we adopted a standard linear allele dosage model; we regressed Henmon-Nelson IQ on the number minor (less frequent) alleles. However, for the two APOE SNPs, we instead analyzed a dummy variable indicating the presence of at least one e4 allele, since this allele is defined by a haplotype of these two SNPs and is the genotype previously studied in conjunction with g (and AD). All of our analyses controlled for graduate/sibling status, age, gender, and the interactions of these factors, as well as the first three principal components of the genetic data from the full set of 90 genotyped SNPs (to account for possible population stratification). [For additional Methods details, see Supporting Online Material.]
Results
Table 1 displays the results of this analysis. None of the 12 genotypes (11 SNPs and the APOE e4 variable) were significantly associated with g (p ≥ .10 in all cases). We conducted an omnibus F-test for all 11 SNPs and the APOE dummy combined in a single regression, and could not reject the null hypothesis that all of the SNPs jointly have zero effect on g (F = 0.88, p = .56). We calculated the statistical power associated with this omnibus test and found that if, in aggregate, our 12 genotypic predictors jointly explain at least 0.52% of the variance of g, the F-test should reject the null hypothesis more than 99% of the time. The thresholds associated with 80% and 95% rejection are 0.26% and 0.39% of the variance, respectively.
Table 1.
Results of Study 1. Each line gives the results for each SNP of a separate linear regression of g (Henmon-Nelson IQ) on dosage of the minor allele (0, 1, or 2 copies), controlling for age, sex, graduate/sibling status, and the interactions of these factors, as well as the first three principal components of the 90-SNP genotype correlation matrix available in the Wisconsin Longitudinal Study dataset. Sample size varies slightly among SNPs due to missing data. The last two rows show genotypes that were available in the WLS dataset, but not in the FHS dataset (Study 2). The R2 column gives the percentage of variance explained by a univariate regression of g on minor allele dosage for each SNP.
| SNP | CHR | Gene | N | R2 (%) | Beta | Standard Error | t | p | MAF | Minor Allele | Major Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1018381 | 6p | DTNBP1 | 6507 | 0.04 | 0.809 | 0.514 | 1.57 | .12 | .080 | C | T |
| rs17571 | 11p | CTSD | 6464 | 0.01 | 0.310 | 0.481 | 0.64 | .52 | .079 | A | G |
| rs1800497 | 11q | DRD2/ANKK1 | 6469 | 0.00 | 0.007 | 0.356 | 0.02 | .98 | .191 | A | G |
| rs2061174 | 7q | CHRM2 | 6392 | 0.00 | 0.091 | 0.294 | 0.31 | .76 | .328 | G | A |
| rs2760118 | 6p | SSADH (ALDH5A1) | 6479 | 0.01 | −0.114 | 0.340 | −0.34 | .74 | .340 | T | C |
| rs4680 | 22q | COMT | 6420 | 0.02 | −0.350 | 0.270 | −1.30 | .20 | .471 | G | A |
| rs6265 | 11p | BDNF | 6489 | 0.02 | 0.367 | 0.331 | 1.11 | .27 | .190 | T | C |
| rs760761 | 6p | DTNBP1 | 6438 | 0.00 | 0.128 | 0.330 | 0.39 | .70 | .206 | A | G |
| rs8191992 | 7q | CHRNA4/CHRM2 | 6492 | 0.00 | 0.122 | 0.273 | 0.45 | .66 | .474 | T | A |
| rs821616 | 1q | DISC1 | 6478 | 0.04 | −0.483 | 0.293 | −1.65 | .10 | .283 | T | A |
| rs429358, rs7412 | 19q | APOE e4 present/absent | 6390 | 0.00 | 0.041 | 0.426 | 0.10 | .92 | .137 | e4 | e2/e3 |
| rs363050 | 20p | SNAP25 | 6464 | 0.04 | 0.323 | 0.275 | 1.18 | .24 | .427 | G | A |
Note: CHR = Chromosome; MAF = Minor Allele Frequency.
A recent meta-analysis (Barnett et al., 2008) suggests that the well-researched Val158Met polymorphism in COMT (rs4680) may explain around 0.10% of the variance of g. This estimate is likely to still be biased upward, because it assumes no publication bias or winner’s curse is affecting the literature on this association. If we make the reasonable assumption that our SNPs, which are mostly distributed across several chromosomes, are independent, these results imply that the average effect size of the 12 genotypic predictors (which include rs4680) must be even smaller than 0.05% of the variance (because 0.52%/12 = 0.043%), although we cannot rule out the possibility that most are zero and a few exceed 0.10%. These effect sizes are small—e.g., 0.05% of the variance is about 0.45 IQ points for a SNP whose minor allele frequency is close to 50%, as in the case of rs4680—and much lower than the effect sizes reported for the SNPs in the initial publications of their g associations. From these calculations, we conclude that our analysis has a high level of statistical power for effect sizes of meaningful magnitude.
Study 2
Method
In study 2, we attempted to repeat the same analysis as closely as possible with data from the “Initial” and “Offspring” cohorts of the Framingham Heart Study (FHS), which has tracked residents of Framingham, Massachusetts, and their descendants since the 1940s. Dawber et al. (1951) and Feinleib et al. (1975) provide more details on these two cohorts of the FHS. Our dataset included 1759 individuals, of whom 45.4% were male. Participants ranged from 40–100 years in age when they completed a battery of cognitive tests as part of a neuropsychological component of the FHS. These tests included Trails A and B, WRAT-Reading, Boston Naming, WAIS Similarities, Hooper Visual Organization, WMS Visual Reproductions, and WMS Logical Memory (for more information see Seshadri et al., 2007).
To estimate general cognitive ability, we first conducted a principal component analysis on the cognitive test data (controlling for sex, birth year, and cohort); the first component accounted for 45.6% of the variance in test performance, consistent with the normal pattern in studies of general intelligence (Chabris, 2007). For each individual in the full sample, g was then defined as the subject’s score on the first principal component. Finally, the scores were normalized to have mean 100 and variance 15.
Ten of the 13 WLS SNPs were available in a set of genotypes previously imputed. (The two SNPs in APOE, rs7412 and rs429358, and one in SNAP25, rs363050, were not available.) [For additional Methods details, see Supporting Online Material.]
Results
Tests of association with each SNP were conducted using the standard linear allele dosage model as with the WLS data, with the standard errors clustered by extended family. Table 2 displays the results. Nine of the ten SNPs were not significantly associated with g, p ≥ .10 in all cases. We also did an omnibus F-test for all 10 SNPs in a single regression, and could not reject the null hypothesis that all of the SNPs have zero joint effect on g (F = 0.85, p = .58).
Table 2.
Results of Study 2. Each line gives the results for each SNP of a separate linear regression of g (score on the first principal component extracted from a battery of nine cognitive tests) on dosage of the minor allele (0, 1, or 2 copies), controlling for a cubic of age, a cubic of age interacted with sex, the first 10 principal components of the SNP genotype correlation matrix, and study cohort, with clustering by extended families, in the Framingham Heart Study dataset (N = 1759). The R2 column gives the percentage of variance explained by a univariate regression of g on minor allele dosage for each SNP.
| SNP | CHR | Gene | R2 (%) | Beta | Standard Error | t | p | MAF | Minor Allele | Major Allele |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1018381 | 6p | DTNBP1 | 0.02 | 0.607 | 0.928 | 0.655 | .51 | .088 | C | T |
| rs17571 | 11p | CTSD | 0.06 | −0.935 | 1.105 | −0.846 | .40 | .086 | A | G |
| rs1800497 | 11q | DRD2/ANKK1 | 0.14 | −0.914 | 0.632 | −1.448 | .15 | .202 | A | G |
| rs2061174 | 7q | CHRM2 | 0.00 | −0.009 | 0.600 | −0.014 | .10 | .318 | G | A |
| rs2760118 | 6p | SSADH (ALDH5A1) | 0.23 | −1.158 | 0.576 | −2.011 | .04 | .309 | T | C |
| rs4680 | 22q | COMT | 0.02 | −0.260 | 0.539 | −0.481 | .63 | .486 | G | A |
| rs6265 | 11p | BDNF | 0.01 | 0.298 | 0.695 | 0.429 | .67 | .189 | T | C |
| rs760761 | 6p | DTNBP1 | 0.01 | 0.218 | 0.687 | 0.317 | .75 | .191 | A | G |
| rs8191992 | 7q | CHRNA4/CHRM2 | 0.00 | −0.039 | 0.551 | −0.071 | .94 | .440 | T | A |
| rs821616 | 1q | DISC1 | 0.02 | −0.387 | 0.608 | −0.636 | .53 | .287 | T | A |
Note: CHR = Chromosome; MAF = Minor Allele Frequency.
One SNP, rs2760118 in SSADH (also known as ALDH5A1), exhibited a nominally significant association with g (t = 2.01, p = .04), but this association did not survive a Bonferroni correction. The mean g values (transformed to the IQ scale) by genotype for this SNP were 98.3, 99.7, and 100.6 for genotypes TT, TC, and CC respectively. This SSADH polymorphism was first reported to be associated with g by Plomin et al. (2004), with directionality the same as in our FHS data, and some rare SSADH mutations are robustly associated with mental retardation and seizures via a well-known biological pathway involving the metabolism of the inhibitory neurotransmitter GABA (Pearl et al., 2009).
Benjamin et al. (2011) reported that rs2760118 was associated with educational attainment in an Icelandic sample; the association was replicated in a second Icelandic sample and appeared to be partially mediated by an association between SSADH and cognitive function in both samples. However, the same study reported that the association between rs2760118 and education did not replicate in three other datasets (WLS, FHS, and a control group from the NIMH Swedish Schizophrenia Study). It is possible that this SSADH SNP has a true, but small, effect on g that is only observed in some studies and/or under some environmental conditions.
Study 3
Method
To verify that the results of Study 1 and Study 2 were not artifacts of any factors specific to the WLS and FHS datasets, we repeated the analysis in a sample of recently genotyped Swedish twins born between 1936 and 1958. The subjects were all participants in the SALT survey (see Lichtenstein et al., 2002, for a description of the sample); 10,946 of the SALT respondents have been genotyped.
Until recently, Swedish men were required by law to participate in military conscription at or around the age of 18, and a test of cognitive ability was part of the screening process. Since performance on the test influenced a recruit’s ultimate position in the military, incentives to perform well on the test were strong. The recruits studied here took either four or five cognitive tests, depending on their cohort; the tests used included measures of problem solving, concept discrimination, technical comprehension, multiplication, and mechanical or spatial ability. Carlstedt (2000) describes the batteries in more detail and reports evidence that they provide good measures of g. Since there are minor variations across years in the specific questions asked, we conducted a separate principal component analysis of the subtests for each birth year. For each individual in the full sample, g was then defined as the subject’s score on the first principal component. As with the WLS and FHS, we normalized the scores to have mean 100 and standard deviation 15.
Ten of the original 12 WLS genotypes were available in the imputed data, exactly the same SNPs as in the Framingham data. Tests of association with each SNP were conducted using linear regression analysis. The sample is exclusively male, g was estimated separately for each cohort defined by birth year, and there is no meaningful variation in the age at which the men take the test (as conscription nearly always occurs around the age of 18), so age and sex were not included as covariates, but the first ten principal components of genetic data were included. The final sample includes 2,441 individuals for whom genetic and IQ test data is available: 811 twins without a co-twin in the sample, 418 complete MZ pairs, and 397 complete DZ pairs. [For additional Methods details, see Supporting Online Material.]
Results
Tests of association with each SNP were conducted using the same approach as with the WLS and FHS data; Table 3 displays the results. The association that came closest to significance is with SNP rs2760118 in SSADH (t = 1.58, p = .11), the same SNP that was nominally significant in the FHS sample. However, the direction of the association here is the opposite of what was observed in the FHS. In STR the mean IQ scores were 99.2, 100.4, and 100.9 for genotypes CC, TC and TT respectively. The omnibus F-test for all 10 SNPs in a single regression fails to reject the null hypothesis that the SNPs jointly have zero effect on g (F = 0.89, p = .55).
Table 3.
Results of Study 3. Each line gives the results for each SNP of a separate linear regression of g (score on the first principal component extracted from a battery of nine cognitive tests) on dosage of the minor allele (0, 1, or 2 copies), controlling for the first 10 principal components of the SNP genotype correlation matrix, and study cohort, with clustering by family. The sample is comprised exclusively of male Swedish twins born between 1936 and 1958, who all took the tests near the age of 18.
| SNP | CHR | Gene | N | R2 (%) | Beta | Std Error | t | p | MAF | Minor Allele | Major Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs1018381 | 6p | DTNBP1 | 2441 | .103 | −1.350 | 1.120 | −1.21 | .228 | .069 | C | T |
| rs17571 | 11p | CTSD | 2441 | .044 | 0.744 | 0.943 | 0.79 | .430 | .073 | A | G |
| rs1800497 | 11q | DRD2/ANKK1 | 2441 | .007 | −0.345 | 0.698 | −0.49 | .621 | .180 | A | G |
| rs2061174 | 7q | CHRM2 | 2441 | .005 | −0.112 | 0.540 | −0.21 | .835 | .319 | G | A |
| rs2760118 | 6p | SSADH (ALDH5A1) | 2441 | .163 | 0.803 | 0.508 | 1.58 | .114 | .375 | T | C |
| rs4680 | 22q | COMT | 2441 | .020 | −0.233 | 0.498 | −0.47 | .640 | .447 | G | A |
| rs6265 | 11p | BDNF | 2441 | .038 | 0.592 | 0.653 | 0.91 | .365 | .195 | T | C |
| rs760761 | 6p | DTNBP1 | 2441 | .109 | −0.907 | 0.631 | −1.44 | .151 | .221 | A | G |
| rs8191992 | 7q | CHRNA4/CHRM2 | 2441 | .074 | 0.524 | 0.495 | 1.06 | .290 | .456 | T | A |
| rs821616 | 1q | DISC1 | 2441 | .015 | −0.420 | 0.520 | −0.81 | .419 | .318 | T | A |
Note: CHR = Chromosome; MAF = Minor Allele Frequency.
Discussion
We attempted to replicate published associations of 12 specific genotypes with measures of general cognitive ability in three large, well-characterized longitudinal datasets. In the Wisconsin Longitudinal Study, none of the 12 genotypes were significantly associated with g. In the Framingham Heart Study, 9 of the 10 SNPs we were able to test were also not associated with g. The only nominally significant association involved SNP rs27660118. In the Swedish Twin Registry sample, none of the 10 available SNPs were significantly associated with g. The association between rs27660118 and IQ approached significance (before correction for multiple hypothesis testing), but the effect was opposite to that observed in the FHS sample.
There have been previous failures to replicate published candidate gene studies of g (e.g., Houlihan et al., 2009). Our research is distinguished by a large combined sample of almost 10,000 individuals across three independent samples and an attempt to replicate all published associations for which we had available data in all three datasets. The contrast between the outcome expected from the literature and the outcome we actually observed in our investigation is striking. Assuming that the SNPs are independently distributed, under the null hypothesis that every genotype we examined was unrelated to g, the expected number of significant associations at the 5% level is 1.6 (out of our 32 total tests). We observed exactly one nominally significant association, slightly less than would be expected by chance alone.
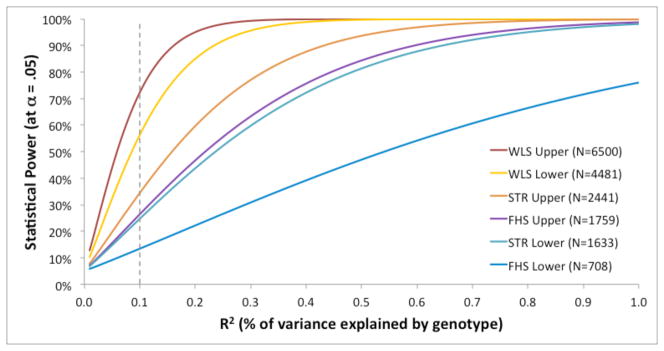
This result is not likely due to lack of statistical power. Figure 1 shows the number of significant associations expected under a range of alternative hypotheses for the size of each genotype’s effect on g, with the effect size ranging from R2 = 0% to 1% of the variance. For example, had all of the associations that we tested been true positives in the population with an effect size of R2 = 0.1%—the effect size that Barnett et al.’s (2008) meta-analysis found for COMT—then the expected number of significant (p < .05) associations would have been approximately 14.7 in the 32 tests we did: the sum of 8.7 out of 12 in the WLS data, 2.6 out of 10 in the FHS data, and 3.4 out of 10 in the STR data.2 Even after accounting conservatively for the genetic relatedness of some participants (siblings in the WLS, family members in the FHS, and twins in the STR), we would still expect 10.6 total associations, or ten times more than we found. And an effect of one tenth of one percent of the phenotypic variance is tiny; as Figure 1 shows, assuming anything larger increases the power of our studies, and thus the divergence between the number of associations expected and the number we observed.
Figure 1.
Statistical power of Studies 1–3 to detect significant associations between SNPs and g, plotted as a function of the percentage of variance in g explained by the SNP (or genotype in the case of APOE e4). Note that the x-axis runs from 0% to 1% out of a total of 100% variance in g, so that 0.1 corresponds to 1/1000 of the total trait variance. Power was estimated for the three studies using the full sample size (“Upper” bound on power for WLS, STR, and FHS) and using the number of unrelated individuals only (“Lower” bound on power for WLS, STR, and FHS), yielding six power curves. Calculations were performed using the tool created by Purcell, Cherny, and Sham (2003) [pngu.mgh.harvard.edu/~purcell/gpc/qtlassoc.html]. Assuming an effect size of 0.1% of variance for each genotype tested (shown by the dashed line), we should have observed between 10.6 and 14.7 significant associations (for the unrelated and full samples, respectively), but we only observed 1.
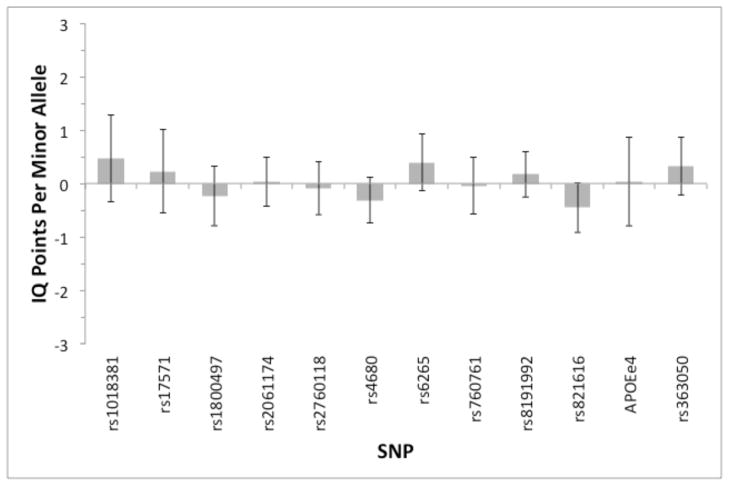
To assess the potential size of any effects on g of the genotypes we examined, we meta-analyzed the results from our three studies. Figure 2 shows that the pooled estimates are sufficiently precise to rule out anything but very small effects. Even the widest 95% confidence interval excludes effect sizes larger than 1.3 IQ points, which is less than one tenth of a standard deviation. Most of the effects are estimated with considerably greater precision.
Figure 2.
Regression coefficients for each genotype (i.e., difference in number of IQ points associated with each copy of the minor allele), pooled across Studies 1–3. To minimize the variance of the estimator, pooling was done by weighting the three estimated regression coefficients for each SNP by the inverse of their estimated variances, with the weights then normalized so that they sum to one. Error bars show 95% confidence intervals. For APOE, the bar shows the number of IQ points associated with possessing at least one e4 allele.
The failure thus far to find genes associated with g does not mean that g has no genetic component. Davies et al. (2011) used data from five different genome-wide association studies (GWAS) and failed to identify any individual markers robustly associated with crystalized or fluid intelligence. They then applied a recently developed method (Yang et al., 2010; Visscher et al., 2010) for testing the cumulative effects of all the genotyped SNPs. In essence, this method calculates the overall genetic similarity between each pair of individuals in a sample and then correlates this genetic similarity with phenotypic similarity across all pairs. Following Yang et al. (2010), we dropped one twin per pair, and then estimated all pairwise genetic relationships in the resulting sample. We then dropped individuals whose relatedness exceeded .025, just as in Davies et al. (2011). Davies et al. reported that the ~550,000 SNPs in their data could jointly explain 40% of the variation in crystalized g (N = 3,254) and 51% of the variation in fluid g (N = 3,181). We applied the same procedure to the STR sample from Study 3 and estimated that the ~630,000 SNPs in our data jointly account for 47% of the variance in g (p < .02), confirming the Davies et al. (2011) findings in an independent sample. These and our other results, together with the failure of whole-genome association studies of g to date, are consistent with general intelligence being a highly polygenic trait on which common genetic variants individually have only small effects.
Conclusion
A consensus is emerging that most published results from candidate gene studies that originally used small samples fail to replicate (Siontis et al., 2010; Ioannidis et al., 2011; cf. Ioannidis, 2005). There are several possible reasons, none of them mutually exclusive, for this state of affairs. Failure to replicate can be attributed to lack of statistical power in the replication sample, but this is unlikely to apply here, because our replication samples are much larger than the samples used in the original studies or in most candidate gene studies. Genetic associations may also fail to replicate when the identified variants are not the ones that cause the trait variation, but are correlated with the true causal variants, with different patterns of linkage disequilibrium in different samples. Patterns of failed replication may also arise due to differing effects of genes on traits across environments.
By far the most plausible explanation in our case, however, is that the original studies we seek to replicate did not have sufficient sample sizes—and not because of any error in design or execution. Expectations that individual SNPs might have large effects on g, which could be detected with small samples, seemed reasonable before genome-wide association studies were possible, and when genotyping was orders of magnitude more expensive than it is now. But if the true effect sizes of common variants are small, as now seems clear, then the early studies whose results we have failed to replicate were inadvertently underpowered. Bayesian calculations imply that results reported from underpowered studies, even if statistically significant, are likely to be false positives (e.g., Ioannidis, 2005; Benjamin, 2010).
The results reported here illustrate for g the problem of “missing heritability” (Manolio et al., 2009), which is the failure—so far—to find specific molecular variants that account for the substantial genetic influences identified by twin and family studies of medical and psychiatric phenotypes. For comparison, height is approximately 90% heritable in Western populations, but so far no common variants contributing more than 0.5cm per allele have been discovered, and the set of 180 height-associated SNPs identified by the most comprehensive meta-analysis only explains about 10% of the population phenotypic variance (Lango Allen et al., 2010). We suspect that our results for g are not an isolated exception, but instead illustrative of a larger pattern in the genetics of cognition and social science (Beauchamp et al., 2011; Benjamin, 2010). There are several possible explanations for the missing heritability. One view is that common variants explain much of the heritable variation but that the individual effects are so small that enormous samples are required to reliably detect them (Visscher, 2008; Visscher et al., 2008). An alternative view is that much of the heritable variation comes from rare, perhaps structural, genetic variants with modest to large effect sizes (Dickson et al., 2010; Yeo et al., 2011).
At the time most of the results we have attempted to replicate were obtained, candidate gene studies of complex traits were commonplace in medical genetics research. Such studies are now rarely published in leading journals. Our results add IQ to the list of phenotypes that must be approached with great caution when evaluating published molecular genetic associations. In our view, excitement over the value of behavioral and molecular genetic studies in the social sciences should be tempered—as it has been in the medical sciences—by an appreciation that for complex phenotypes, individual common genetic variants of the sort assayed by SNP microarrays are likely to have very small effects. Associations of candidate genes with psychological and other social science traits should be viewed as tentative until they have been replicated in multiple large samples. Doing otherwise may hamper scientific progress by proliferating potentially false positive results, which may then influence the research agendas of other scientists who do not appreciate that the associations they take as a starting point for their efforts may not be real. And the dissemination of false results to the public risks creating an incorrect perception about the state of knowledge in the field, especially the existence of genes described as being “for” traits on the basis of unintentionally inflated estimates of effect size and statistical significance.
We think that a profitable way forward for molecular genetic investigations in social science is to follow the lead of medical genetics researchers, who have formed international consortia that include as many large studies with genomic and (harmonized) phenotypic data as possible. A plausible sample size of 100,000 individuals has statistical power of 80% to discover genetic variants accounting for as little as 0.04% of the variance in a trait at a “genome-wide significance level” of p < 5 × 10−8. With sufficient power, it will also be feasible to study gene-gene interactions (e.g., Roetker et al., 2011), which may account for more of the variance in complex phenotypes than individual SNPs considered in isolation.
Finally, we emphasize that the negative results reported here should not detract from research into the behavioral and molecular genetics of g and other social science traits, but rather point the way to study designs that are more likely to yield robust knowledge.
Supplementary Material
Acknowledgments
This research was supported by the NIA (grants P01AG005842 and T32-AG000186-23). The Swedish Twin Registry is supported by the Swedish Department of Higher Education, the European Commission (grant QLG2-CT-2002-01254), the Swedish Research Council, the Swedish Foundation for Strategic Research, the Jan Wallander and Tom Hedelius Foundation, and the Swedish Council for Working Life and Social Research. We thank Paul de Bakker and the Broad Institute for imputing the Framingham Heart Study genotypic data and for making the results available to other FHS researchers. We thank Emil Rehnberg of the Karolinska Institute for conducting the imputation and computing the principal components in the Study 3 dataset. We thank Yeon Sik Cho for research assistance.
Footnotes
Because our goal is to replicate the results of published candidate gene studies of g, we do not consider the results of genome-wide association studies (GWAS), none of which have yet identified replicable SNPs that meet conventional thresholds for significant associations with g (e.g., Butcher et al., 2008; Davies et al., 2011; Seshadri et al., 2007).
For our full samples, power at R2 = 0.1% (the dotted line in Figure 1) is .72 for WLS, .26 for FHS, and .34 for STR. Assuming independence across SNPs—a reasonable assumption since almost all of the SNPs are far apart or on separate chromosomes—the expected number of significant associations in a sample is the power times the number of SNPs tested. (For the smaller samples of unrelated individuals, the power values are .56, .13, and .25 respectively.)
References
- Barnett JH, Scoriels L, Munafò MR. Meta-analysis of the cognitive effects of the catechol-O-methyltransferase gene Val158/108Met polymorphism. Biological Psychiatry. 2008;64:137–144. doi: 10.1016/j.biopsych.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Beauchamp JP, Cesarini D, Johannesson M, van der Loos M, Koellinger P, Groenen PJF, Fowler JH, Rosenquist N, Thurik AR, Christakis NA. Molecular genetics and economics. Journal of Economic Perspectives. 2011;25(4):57–82. doi: 10.1257/jep.25.4.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Chabris CF, Glaeser EL, Gudnason V, Harris T, Laibson DI, Launer L, Purcell S. Genoeconomics. In: Weinstein M, Vaupel JW, Watcher KW, editors. Biosocial surveys. Washington, DC: The National Academies Press; 2007. pp. 304–335. [Google Scholar]
- Benjamin DJ. White paper on genoeconomics. In: Lupia A, editor. Genes, Cognition, and Social Behavior: Next Steps for Foundations and Researchers. University of Michigan; 2010. pp. 66–77. manuscript. [ www.isr.umich.edu/cps/workshop/NSF_Report_Final.pdf] [Google Scholar]
- Benjamin DJ, Cesarini DA, Chabris CF, Glaeser EL, Laibson DI, et al. The Promise and Pitfalls of Genoeconomics. 2011. Manuscript submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard TJ., Jr Genetic and environmental influences on adult intelligence and special mental abilities. Human Biology. 1998;70:257–179. [PubMed] [Google Scholar]
- Butcher LM, Davis OS, Craig IW, Plomin R. Genome-wide quantitative trait locus association scan of general cognitive ability using pooled DNA and 500K single nucleotide polymorphism microarrays. Genes, Brain, and Behavior. 2008;7(4):435–446. doi: 10.1111/j.1601-183X.2007.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlstedt B. PhD thesis. Gothenburg University; 2000. Cognitive abilities: Aspects of structure, process and measurement. [ http://gupea.ub.gu.se/bitstream/2077/9600/3/gupea_2077_9600_3.pdf] [Google Scholar]
- Chabris CF. Cognitive and neurobiological mechanisms of the Law of General Intelligence. In: Roberts MJ, editor. Integrating the mind: Domain specific versus domain general processes in higher cognition. Hove, UK: Psychology Press; 2007. pp. 449–491. [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011 doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: The Framingham Study. American Journal of Public Health. 1951;41:279–286. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson S, Wang K, Krantz I, Hakonarson H, Goldstein D. Rare variants create synthetic genome-wide associations. PLoS Biology. 2010;8(1) doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study: Design and preliminary data. Preventive Medicine. 1975;4:518–552. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genetics in Medicine. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Houlihan LM, Harris SE, Luciano M, Gow AJ, Starr JM, Visscher PM, Deary IJ. Replication study of candidate genes for cognitive abilities: The Lothian Birth Cohort 1936. Genes, Brain and Behavior. 2009;8:238–247. doi: 10.1111/j.1601-183X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nature Genetics. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Why most published research findings are false. PLoS Medicine. 2005;2(8):e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- Lamke TA, Nelson MJ. Henmon-Nelson Tests of Mental Ability. Boston: Houghton Mifflin; 1957. (rev. ed.) [Google Scholar]
- Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- Lichtenstein P, de Faire U, Floderus B, Svartengren M, Svedberg P, Pedersen NL. The Swedish Twin Registry: A unique resource for clinical, epidemiological and genetic studies. Journal of Internal Medicine. 2002;252:184–205. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindor LA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisser U, et al. Intelligence: Knowns and unknowns. American Psychologist. 1996;51(2):77–101. [Google Scholar]
- Payton A. The impact of genetic research on our understanding of normal cognitive ageing: 1995 to 2009. Neuropsychology Review. 2009;19:451–477. doi: 10.1007/s11065-009-9116-z. [DOI] [PubMed] [Google Scholar]
- Pearl PL, Gibson KM, Cortez MA, Wu Y, Snead OC, 3rd, Knerr I, Forester K, Pettiford JM, Jakobs C, Theodore W. Succinic semialdehyde dehydrogenase deficiency: Lessons from mice and men. Journal of Inherited Metabolic Disease. 2009;32(3):343–352. doi: 10.1007/s10545-009-1034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Turic DM, Hill L, Turic DE, Stephens M, Williams J, et al. A functional polymorphism in the succinate-semialdehyde dehydrogenase (aldehyde dehydrogenase 5 family, member A1) gene is associated with cognitive ability. Molecular Psychiatry. 2004;9:582–586. doi: 10.1038/sj.mp.4001441. [DOI] [PubMed] [Google Scholar]
- Plomin R, Kennedy JKJ, Craig IW. The quest for quantitative trait loci associated with intelligence. Intelligence. 2006;34(6):513–526. [Google Scholar]
- Plomin R, McClearn GE, McGuffin P, DeFries J. Behavioral Genetics. 5. New York: Worth; 2008. [Google Scholar]
- Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- Ree MJ, Earles JA. The stability of g across different methods of estimation. Intelligence. 1991;15:271–278. [Google Scholar]
- Roetker NS, Yonker JA, Lee C, Chang V, Basson J, Roan CL, et al. Exploring epistasis in clinically diagnosed depression in the Wisconsin Longitudinal Study: A pilot study utilizing recursive partitioning analysis. 2011. Manuscript submitted for publication. [Google Scholar]
- Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Medical Genetics. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siontis KC, Patsopoulos NA, Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. European Journal of Human Genetics. 2010;18(7):832–837. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman C. “General intelligence,” objectively determined and measured. American Journal of Psychology. 1904;15:201–293. [Google Scholar]
- Tung YC, Yeo GS. From GWAS to biology: Lessons from FTO. Annals of the New York Academy of Sciences. 2011;1220:162–171. doi: 10.1111/j.1749-6632.2010.05903.x. [DOI] [PubMed] [Google Scholar]
- Turkheimer E. Three laws of behavior genetics and what they mean. Current Directions in Psychological Science. 2000;9:160–164. [Google Scholar]
- Weedon MN, Frayling TM. Reaching new heights: Insights into the genetics of human stature. Trends in Genetics. 2008;24(12):595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. Heritability in the genomics era: Concepts and misconceptions. Nature Reviews Genetics. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Visscher PM. Sizing up human height variation. Nature Genetics. 2008;40(5):489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Yang J, Goddard ME. A commentary on “Common SNPs explain a large proportion of the heritability for human height” by Yang et al. (2010) Twin Research and Human Genetics. 2010;13:517–524. doi: 10.1375/twin.13.6.517. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo RA, Gangestad SW, Liu J, Calhoun VD, Hutchison KE. Rare copy number deletions predict individual variation in intelligence. PLoS One. 2011;6(1):e16339. doi: 10.1371/journal.pone.0016339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.