Abstract
Oropharyngeal squamous cell carcinoma (SCC) is strongly associated with human papillomavirus (HPV) infection, which is distinctively different from most other head and neck cancers. However, a robust quantitative reverse transcription PCR (RT-qPCR) method for comprehensive expression profiling of HPV genes in routinely fixed tissues has not been reported. To address this issue, we have established a new real-time RT-PCR method for the expression profiling of the E6 and E7 oncogenes from 13 high-risk HPV types. This method was validated in cervical cancer and by comparison with another HPV RNA detection method (in situ hybridization) in oropharyngeal tumors. In addition, the expression profiles of selected HPV-related human genes were also analyzed. HPV E6 and E7 expression profiles were then analyzed in 150 archived oropharyngeal SCC samples and compared with other variables and with patient outcomes. Our study showed that RT-qPCR and RNA in situ hybridization were 100% concordant in determining HPV status. HPV transcriptional activity was found in most oropharyngeal SCC (81.3%), a prevalence that is higher than in previous studies. Besides HPV16, three other HPV types were also detected, including 33, 35 and 18. Furthermore, HPV and p16 had essentially identical expression signatures, and both HPV and p16 were prognostic biomarkers for the prediction of disease outcome. Thus, p16 mRNA or protein expression signature is a sensitive and specific surrogate marker for HPV transcriptional activity (all genotypes combined).
Keywords: human papillomavirus, oropharyngeal cancer, prognosis, expression signature
Introduction
Unlike most other head and neck cancers, squamous cell carcinoma (SCC) of the oropharynx has a strong association with human papillomavirus (HPV) infection 1. Epidemiologic studies show that HPV-positive oropharyngeal cancer occurs commonly in patients of younger age, with higher numbers of sex partners, more oral sex exposure, and lower smoking rates 1–4. Despite a steady decrease in the number of overall head and neck cancer cases in the past decades, the incidence of oropharyngeal cancer has increased significantly, especially in recent years 5, 6. Thus, there is an urgent need to focus specifically on oropharyngeal cancer to determine the unique characteristics of this cancer type with the goal of developing specific and targeted treatments.
Many studies have shown that HPV-positive oropharyngeal cancer is associated with significantly better patient survival than HPV-negative oropharyngeal cancer 3, 7–9. However, the molecular mechanisms underlying the differences in disease outcome are poorly understood. It is known that HPV-encoded E6 and E7 oncogenes play critical roles in carcinogenic transformation 10, 11. E6 promotes the degradation of p53, a critical protein for tumor suppression mainly through regulation of growth arrest and apoptosis. On the other hand, E7 binds and inactivates the retinoblastoma protein (Rb), and induces the overexpression of p16, which is a cyclin-dependent kinase inhibitor (CDKN2A). Thus, p16 has been suggested as a marker for HPV-driven oncogenic transformation. While the prognostic value of p16 has been suggested to resemble that of HPV in multiple studies 8, 12–14, it is still controversial whether p16 is a specific and sensitive indicator of HPV activity in oropharyngeal cancer. One major unresolved issue contributing to this uncertainty is the lack of robust assays for the comprehensive analysis of the expression signature of HPV.
In this study, we developed and experimentally validated a new bioinformatics assay design algorithm for the expression profiling of E6 and E7 from 13 high-risk HPV types by RT-qPCR. This method was validated in cervical cancer and also by comparison to another HPV RNA based detection method, RNA in situ hybridization, in oropharyngeal cancer cases. The availability of these robust assays allowed us to comprehensively analyze the expression signature of HPV in oropharyngeal cancer and to thoroughly compare it with p16 expression by RT-qPCR and immunohistochemistry.
Materials and Methods
Design of real-time RT-PCR assays for expression profiling of HPV E6 and E7 transcripts
The whole genome sequences of 108 HPV types were downloaded from NCBI GenBank 15, and further processed with BioPerl (http://bioperl.org) to extract all protein-coding gene sequences from the genomes. These gene sequences were used as templates for PCR primer design. Genotype-specific assays were designed for E6 and E7 genes from thirteen high-risk HPV types (16, 18, 31, 33, 35, 39, 45, 52, 56, 58, 59, 66, and 68) that were selected based on literature review 4, 16. The design algorithm was based partly on our previous algorithms that have been experimentally validated for the expression quantitation of human protein-coding genes as well as non-coding RNAs 17–19. Furthermore, our new algorithm also included many primer selection criteria that were specific for HPV quantitation. The workflow of the design algorithm is described in detail below, with designed primer sequences for all the HPV assays listed in Table 1.
Table 1.
Primer sequences for HPV real-time PCR assays
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| HPV16 E6 Pair 1 | ACAAACCGTTGTGTGATTTGTT | CAGTGGCTTTTGACAGTTAATACA |
| HPV16 E6 Pair 2 | GCAAAGACATCTGGACAAAAAG | ACCGACCCCTTATATTATGGAATC |
| HPV16 E7 Pair 1 | GAACCGGACAGAGCCCATTA | ACACTTGCAACAAAAGGTTACA |
| HPV16 E7 Pair 2 | TAACCTTTTGTTGCAAGTGTGA | TTTGTACGCACAACCGAAGC |
| HPV18 E6 | GATCTGTGCACGGAACTGAACA | GGTTATTTCTATGTCTTGCAGTG |
| HPV18 E7 | CAACGTCACACAATGTTGTGTA | TCAATTCTGGCTTCACACTTAC |
| HPV31 E6 | GGTCAGTTAACAGAAACAGAGG | TGGTGTGTCGTCCCTATATACT |
| HPV31 E7 | AACCGGACACATCCAATTACAA | ACACTGACAACAAAAGGTAACG |
| HPV33 E6 | TGCATTTGCAGATTTAACAGTTG | TTCCAAATGGATTTCCCTCTCT |
| HPV33 E7 | CAACCAGCCACAGCTGATTAC | CAAGTGTGACAACAGGTTACAAT |
| HPV35 E6 | AGAATTACAGCGGAGTGAGGTA | CACAAATCATAGCATGCAAAGTC |
| HPV35 E7 | ATGTGAGGCGACACTACGTC | ATGTCAATGTGTGTGCTCTGTA |
| HPV39 E6 | CCACCTTGCAGGACATTACAAT | TAGTGGTCGTCTGCAATAGACA |
| HPV39 E7 | GCGTCACACAATACAGTGTTCG | TGCAGTGTGTTGTTACACTTAC |
| HPV45 E6 | CGCTTTGACGATCCAAAGCAA | CACAAATCTGGTAGCTTGTAGG |
| HPV45 E7 | GGAACCTCAGAATGAATTAGATCC | TGCTCGTAACACAACAGGTCA |
| HPV52 E6 | TGTCAAACGCCATTATGTCCTG | GCTTGTTTGCATTAACATGTCTT |
| HPV52 E7 | TACGGCTATGCATTCATAGCAC | CTGCTGTAGAGTACGAAGGTCC |
| HPV56 E6 | GCATGCACTGAATTAAACTTAGT | CTGCATAAGGAAAATCATCCCTA |
| HPV56 E7 | AACGCTGCAAGACGTTATATTAG | GGTCAATTTCTGTTTGAGGTGTTA |
| HPV58 E6 | CAGAGGAGAAACCACGGACATT | CCAACGCCTGACACAAATCA |
| HPV58 E7 | AACCGGCCACAGCTAATTACTA | TGTTGATACACAAACGAACCGT |
| HPV59 E6 | GATTCCGTGTATGGAGGAACAT | TGTAACGGTGTCTTGGTTTCAG |
| HPV59 E7 | TGGAACCACAAAATTATGAGGAAG | TAATTGCTCGTAGCACACAAGG |
| HPV66 E6 | TGCACCATCTGAGCGAGGTA | TCCTTTTTGCAGTATACACATGA |
| HPV66 E7 | GCTGAACAACATAAGTGTTACCT | AACTGCACCACCAACTCACA |
| HPV68 E6 | GCCTTTGGTGACTTAAATGTAGT | GCATGCAGCTAATGGTACCC |
| HPV68 E7 | CGGAAGCTACAACTGCTGTTTA | CGGACACACAAAATTTAGTGAGT |
First, we compiled a sequence database containing all 108 known HPV genomes, from which 13 high-risk HPV types were selected for the design of the E6 and E7 assays. The sequences from other non-high-risk HPV types were also included in the algorithm as a screening filter so that the assays designed for the oncogenic E6 and E7 did not cross-react with any transcript encoded by other HPV genomes. For each E6 or E7 gene from the 13 HPV types, primer candidates were selected from regions not involved in alternative splicing as revealed by a previous study 20. Furthermore, common base variations in gene sequences were identified and excluded by multi-alignment of all available genome sequences for the same HPV type with ClustalW 21.
To avoid non-specific priming resulting from low sequence complexity, the DUST program was used and any sequence of low complexity as identified by DUST was rejected 22. In addition, the GC content of a primer was confined to 35–65% to ensure uniform primer annealing. To further enhance priming uniformity, the Tm values of all the primers fell in a narrow range (58–62°C) as calculated with the Nearest Neighbor method. The design program also excluded sequence regions with high likelihood of secondary structure formation as site inaccessibility could lead to insufficient primer annealing 18.
One important goal of the assay design was to specifically detect and quantify one HPV type of interest, with no cross-reactivity to over 100 other HPV types, which have similar sequences, or to cross-react with human transcripts. Several specificity filters were implemented in the design program. One main filter was the exclusion of sequences with a stretch of contiguous bases matched perfectly to other unintended HPV or human genes. The screening for contiguous base match was done using a fast-performing algorithm that we developed previously 17. To further reduce primer cross-reactivity, BLAST searches against other HPV genes as well as the human transcriptome were also performed to identify potential cross-reactive primer candidates.
Patients and tumor samples
This study was approved by the Human Research Protection Office of the Washington University School of Medicine. A total of 150 oropharyngeal squamous cell carcinomas from 150 patients were identified retrospectively from a large radiation oncology database of oropharyngeal SCC patients. Among these tumors, 71 were from tonsil, 51 from base of tongue, 9 from soft palate, oropharyngeal walls or vallecula. For the remaining 19 tumors, the exact anatomic subsite within oropharynx was uncertain. Tumor tissues were collected from patients treated at Washington University in St. Louis between 1997 and 2006. All the patients were treated with either definitive radiation therapy or surgery followed by postoperative radiation therapy by a single study radiation oncologist (WLT). All patients were treated without knowledge of (or regard to) their HPV or p16 status. In addition, half of the patients were also treated with chemotherapy. As standard of care, FFPE tumor tissues were collected for pathological analysis either from biopsy or surgical resection prior to radiation or chemotherapy treatment. These FFPE tumors were analyzed with the following procedure: 1) Existing archival slides stained with hematoxylin and eosin (H&E) were reviewed by two study pathologists (RDC and JSL) to confirm the diagnosis and to identify the tumor regions. 2) Macrodissection was performed to remove non-tumor tissues from the corresponding unstained tumor sections. 3) Total RNA was extracted from the identified tumor regions using the miRNeasy FFPE Kit (QIAGEN) according to the manufacturer’s protocol. In this way, we were able to focus on the analysis of the tumor tissues with minimal contamination from adjacent non-tumor tissues.
Expression profiling of HPV and functional-related human genes
The new HPV assays were used to profile the expression of HPV E6 and E7 in oropharyngeal cancer with total RNA extracted from the tumor blocks. All oligo primers in the assays were purchased from Sigma. Reverse transcription (RT) reaction was done with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was carried out to quantify the cDNA product using Power SYBR Green PCR Master Mix (Applied Biosystems) and 500 nM HPV type-specific primers. Each HPV assay (E6 or E7 from each of the 13 HPV types) was individually performed in a separate well on a 384-well PCR plate. The PCR running protocol was 95°C for 10 min, followed by 36 cycles of amplification (95°C for 10 s, 58°C for 15 s and 60°C for 15 s). Besides HPV E6 and E7, five human genes that are functionally related to HPV infection were also profiled by real-time RT-PCR, including p53, Myc, Rb, p16 and p21 with pre-designed assays 18. In addition, the averaged expression level of GAPDH and β-actin was used as the internal reference control for real-time PCR data normalization.
HPV DNA in situ hybridization
DNA In situ hybridization (ISH) was performed on tissue sections using the ISH I View Blue Plus Detection Kit (Ventana Medical Systems) according to the manufacturer’s instructions on a Ventana Benchmark automated stainer. The probe hybridizes with the high risk HPV genotypes including types 16, 18, 33, 35, 45, 51, 52 56, and 66. Cases were read by the study pathologists (RDC and JSL). Any definitive nuclear staining in the tumor cells was considered positive. Cases were classified in a binary manner as either positive or negative.
HPV RNA in situ hybridization
ISH for HPV E6/E7 mRNA was performed on 49 oropharyngeal SCCs for comparison to RT-qPCR using the RNAscope HPV kit (Advanced Cell Diagnostics, Inc., Hayward, CA) according to the manufacturer’s instructions. In brief, FFPE tissue sections were pretreated with heat and protease before hybridization with target probes to the RNA of mixed HPV genotypes (16, 18, 31, 33, 35, 52, and 58) performed on a single slide. A horseradish peroxidase-based signal amplification system was then hybridized to the target probes followed by color development with 3,3′-diaminobenxidine. Positive staining was identified as brown, punctuate dots present in the nucleus and/or cytoplasm. Control probes for the bacterial gene DapB (negative control) and for the housekeeping gene ubiquitin C (positive control) were also performed on each case. The slides and controls were reviewed by both study pathologists (RDC and JSL) and were interpreted as either positive or negative. Cases were excluded when there was lack of positive staining on the ubiquitin C control slide or when there was staining on the DapB control slide that was equal to or higher than the patient slide.
Immunohistochemistry for p16
Immunoperoxidase staining was performed on FFPE tissue sections using a monoclonal antibody to p16 (MTM Laboratories; E6H4; 1:1 dilution). Immunostaining was performed on a Ventana Benchmark automated immunostainer (Ventana Medical Systems) according to standard protocols with appropriate positive controls. The stained slides were read by one study pathologist (JSL) and classified. For analysis, they were classified in a binary manner as positive when greater than 50% of the cells showed nuclear and cytoplasmic staining.
Survival analysis
Statistical data analysis was done using the R package (http://www.r-project.org/). Univariate Cox proportional hazards regression analysis was done to evaluate the prognostic association to disease outcome. The p-values from the Cox analysis were calculated using the Wald test. Multivariate Cox proportional hazards regression analysis was also performed to evaluate the independent prognostic value of gene expression signatures from clinical features. Furthermore, a Kaplan-Meier estimator was used to evaluate the significance of gene signatures for stratifying patients into different risk groups. The statistical significance from the Kaplan-Meier analysis was calculated with the log-rank test. Overall survival (OS) and disease-free survival (DFS) were used as two clinical end points for outcome assessment. OS was defined as the time interval between the date of treatment and the date of death. Disease-free survival was defined as the time interval between the date of treatment and the date of death or first failure.
Results
Validation of the HPV assays with cervical and oropharyngeal cancer samples
We first designed real-time RT-PCR assays for expression profiling of E6 and E7 mRNA from 13 high-risk HPV types (see Methods). The new quantitative HPV assays were tested on total RNA of eight cervical cancer cell lines obtained from the American Type Culture Collection (ATCC). The HPV status of these cell lines had already been determined in previous studies using various biochemical approaches, with three HPV types detected in six of the eight cell lines 23–25. The new RT-qPCR assays for 13 HPV types were used to determine the presence as well as the expression levels of HPV E6 and E7. In complete agreement with previous studies, our results confirmed that HPV16 was detected in Caski and SiHa cells, HPV18 in C-41, HeLa and SW756 cells, and HPV68 in ME-180 cells 23–25. In contrast, none of the other 10 HPV types were detected. We further tested the HPV assays with 80 FFPE cervical tumors that we collected previously 26. About 90% of these cervical tumors were HPV positive, with nine distinct HPV types identified (unpublished data), which is consistent with expected HPV detection rates in cervical cancer.
RNA in situ hybridization (ISH) was also performed on 49 oropharyngeal SCCs in parallel with the RT-qPCR assays in order to validate these assays with a second HPV RNA detection technique. HPV RNA ISH is a highly sensitive new method that is in commercial development but not yet available for clinical use. Due to technical failures from positive or negative controls, RNA ISH data cannot be obtained in 4 of the 49 cases. For the remaining 45 cases with HPV data available from both RT-qPCR and RNA ISH, 39 were HPV positive by both RT-qPCR and RNA ISH. The remaining 6 cases were negative for HPV by both RT-qPCR and RNA ISH. Thus, there was a perfect correlation (100% sensitivity and specificity) between HPV RNA ISH and RT-qPCR. The majority of the cases were HPV type 16. In addition, there was one case of each of the following HPV types: 18, 33 and 35 by RT-qPCR. Since the RNA in situ hybridization probes were pooled in this analysis, specific HPV types were not resolvable. Thus, the profiling data on cervical cancer cells and FFPE cervical and oropharyngeal tissues demonstrate the robust performance of the new HPV RT-PCR assays with high specificity and sensitivity.
Expression correlation of HPV and related human genes
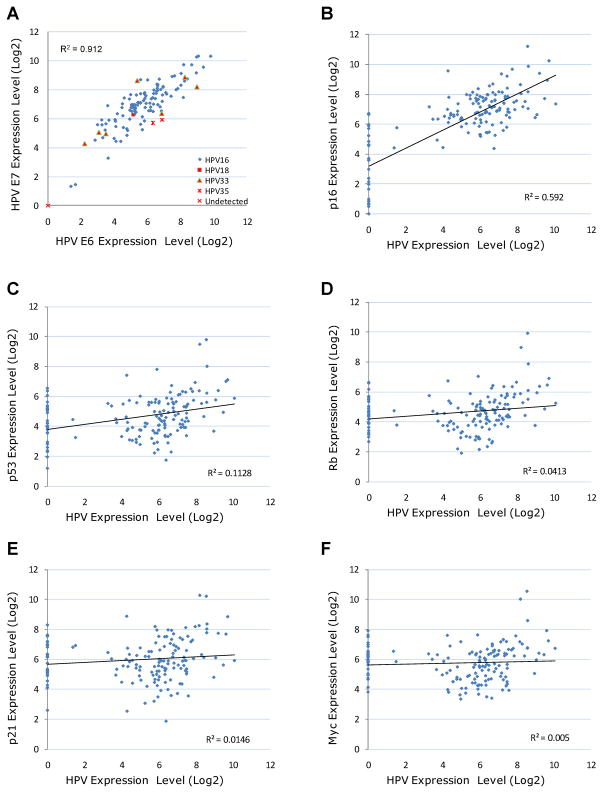
Using the new HPV RT-qPCR assays, we profiled 150 oropharyngeal tumors (Table 2) for E6 and E7 transcriptional activity for the 13 high-risk HPV types. The profiling results indicated that HPV16 was the most prevalent type, with 112 positive cases (74.7%). In addition, seven HPV33-positive, two HPV35-positive and one HPV18-positive cases were also identified. Combined together, 122 of the 150 tumors were HPV positive, representing 81.3% of all cases. Both E6 and E7 transcripts from the same HPV type were detected in each of the 122 HPV-positive cases. On the other hand, in the 28 HPV-negative cases, neither E6 nor E7 transcripts were detected. Overall, the expression profiles of E6 and E7 were highly correlated among all the 150 cases (Pearson correlation coefficient of r= 0.96, Figure 1A).
Table 2.
Patient characteristics
| Age at diagnosis (mean ± SD) | 56.1 ± 9.1 |
| Sex | |
| Male | 136 (91%) |
| Female | 14 (9%) |
| Race | |
| White | 131 (87%) |
| Other | 19 (13%) |
| Smoking* | |
| Yes | 106 (71%) |
| No | 43 (29%) |
| T stage | |
| T1 | 32 (22%) |
| T2 | 56 (38%) |
| T3 | 24 (16%) |
| T4 | 36 (24%) |
| N stage | |
| N0 | 11 (7%) |
| N1 | 21 (14%) |
| N2 | 108 (72%) |
| N3 | 10 (7%) |
| Chemotherapy | |
| Yes | 81 (56%) |
| No | 64 (44%) |
As any lifetime smoking use versus no history of smoking.
Figure 1.
The expression profiles of HPV E6/E7 and functionally-related human genes in 150 oropharyngeal tumors. (A) The expression profiles of HPV E6 and E7 transcripts were determined by real-time RT-PCR. (B–F) The average expression of E6 and E7 transcripts was used to represent the expression of HPV in each tumor. Each data point in the graph represents one tumor sample, with the x-axis representing normalized HPV expression and the y-axis representing normalized expression of a human gene.
Besides HPV E6 and E7, we also profiled the transcriptional expression of multiple human genes that are functionally related to HPV infection, including p53, Myc, Rb, p16 and p21. The expression correlations of HPV and these human genes are presented in Figure 1. Interestingly, the expression profiles of HPV and p16 were highly correlated, with a Pearson correlation coefficient of r = 0.77. In addition, the expression profiles of HPV and p53 were also moderately correlated (r = 0.33). In contrast, Myc, Rb and p21 were not closely correlated with HPV expression (r ≤ 0.20). However, when only HPV-positive tumors were evaluated, all five human gene transcripts were more closely correlated to HPV expression, with r=0.41, 0.40, 0.35, and 0.27 for Rb, p53, Myc and p21, respectively. Overall, the profiling results suggested that p16 was specifically activated by HPV in oropharyngeal cancer, while the transcriptional activities of p53, Rb, Myc and p21 were also likely regulated by other mechanisms in addition to HPV infection.
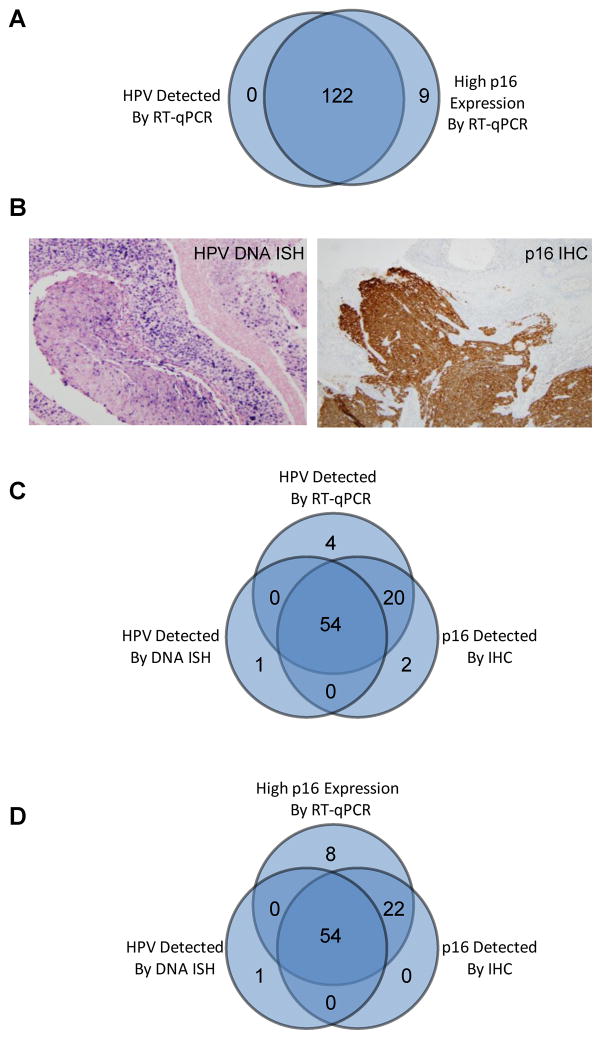
All 122 HPV-positive tumors that were identified by the new HPV E6/E7 RT-qPCR assays were also identified to have high p16 expression by RT-qPCR. On the other hand, there were only nine tumors that had high p16 expression by RT-qPCR, but negative for HPV by RT-qPCR (Figure 2A). Thus, the expression profiles of HPV and p16 transcripts were strongly correlated as revealed by the new RT-qPCR assays. The expression correlation of HPV and p16 was further compared to assays utilized in routine clinical practice, DNA in situ hybridization (ISH) for HPV detection and immunohistochemistry (IHC) for p16 protein expression (Figure 2B). Both HPV DNA ISH and p16 IHC data were obtained from 98 oropharyngeal tumors. As shown in Figure 2C, 76 of the 98 tumors were strongly p16 IHC positive. Among these tumors, HPV transcripts were detected in 74 (97%) cases by RT-qPCR. Compared to p16 IHC, HPV DNA ISH identified a smaller set of tumors (n=55), 54 of which (98%) were also confirmed to be HPV-positive by RT-qPCR assays. On the other hand, there were four tumors that were identified as HPV-positive by RT-qPCR, but not by HPV ISH or p16 IHC. In a similar way, we also correlated the p16 RT-qPCR profiles with HPV ISH and p16 IHC. As shown in Figure 2D, all 76 p16 IHC positive tumors as well as 54 of 55 HPV ISH positive tumors were also confirmed to have high p16 expression by RT-qPCR. Thus, the RT-qPCR profiling data for both HPV and p16 were also highly correlated with the clinically utilized p16 IHC for detection of protein expression.
Figure 2.
HPV and p16 have consistent expression profiles in oropharyngeal cancer. (A) Correlation of HPV RT-qPCR profile and p16 RT-qPCR profile. A normalized expression value of 4 or higher was used as the threshold to define high p16 mRNA expression. (B) In situ hybridization for high-risk HPV DNA and p16 immunohistochemistry. A tumor that is strongly positive for HPV DNA by in situ hybridization showing granular, blue staining in tumor cell nuclei; a tumor that is strongly and diffusely positive for p16 with strong nuclear and cytoplasmic staining in the tumor cells. (C) Correlation of HPV RT-qPCR profile, HPV DNA ISH profile and p16 IHC profile. (D) Correlation of p16 RT-qPCR profile, HPV DNA ISH profile and p16 IHC profile.
HPV and p16 as prognostic biomarkers for oropharyngeal cancer
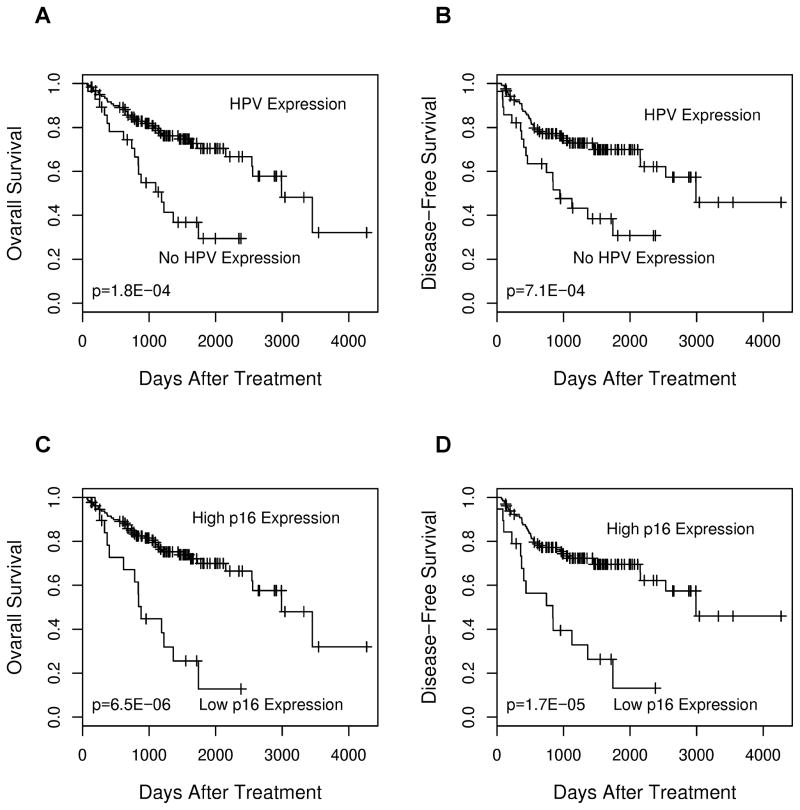
The prognostic significance of HPV and functionally-related human genes was determined by univariate Cox regression analysis of the RT-qPCR expression profiles. The results are summarized in Table 3. Only the expression profiles of HPV and p16 were prognostic of overall patient survival (p=0.005 and 0.0007, respectively, by the Wald test), while the other genes were not. Kaplan-Meier survival analysis was also performed to further evaluate the prognostic significance of HPV and p16 expression. The 150 patients were stratified into two groups based on HPV status (present vs. absent) or p16 status (high vs. low or no expression). As shown in Figure 3, both HPV and p16 expression status was prognostic of disease outcome, using either overall survival or disease-free survival as the endpoint.
Table 3.
Cox regression analysis for association with overall survival
| Gene Feature | Univariate Cox Analysis | Multivariate Cox Analysis |
|---|---|---|
| HPV E6/E7 | 0.005 | 0.019 |
| Myc | 0.12 | — |
| Rb | 0.40 | — |
| p16 | 0.0007 | 0.005 |
| p21 | 0.078 | — |
| p53 | 0.86 | — |
The numbers in the table represent p-values calculated with the Wald test. In the multivariate Cox analysis, the cases were controlled for patient age, sex, race, smoking history, tumor stage and chemotherapy status.
Figure 3.
Kaplan-Meier survival analysis to evaluate the prognostic value of HPV and p16 expression status in 150 tumors as revealed by RT-qPCR. (A) HPV expression and overall survival. (B) HPV expression and disease-free survival. (C) p16 expression and overall survival. A normalized expression value of 4 or higher was used as the threshold to define high p16 mRNA expression. (D) p16 expression and disease-free survival. The p-values were calculated with the log-rank test.
We further assessed whether HPV and p16 expression signatures have independent prognostic value in the context of common clinicopathologic features, including patient age, sex, race, smoking history, tumor stage and chemotherapy status. Multivariate Cox regression analysis was performed to control for these clinical features. As shown in Table 3, HPV and p16 expression signatures retained their prognostic significance, independent of the other variables.
Discussion
The transcriptional activity of HPV is closely related to viral function in carcinogenesis. Thus, in this study, we focused our analysis on the transcriptional expression profiles of both E6 and E7, which are the most important HPV oncogenes. At present, there is still a lack of robust assays for the profiling of E6 and E7 transcripts from many HPV types out of FFPE. HPV type 16 is the most prevalent type associated with oropharyngeal cancer, representing the majority of all HPV positive cases. However, there are many other high-risk HPV types that are found less commonly. One major challenge in HPV assay development is to design specific assays that can differentiate highly homologous sequences among various HPV types, and have no cross-reactivity to any transcript in the human transcriptome. This design challenge is further exacerbated for E6 and E7, because their mRNA transcripts are very short with frequent base variations. To address these challenges, we have developed and also experimentally validated a robust bioinformatics algorithm for HPV RT-qPCR assay design.
As part of the validation process of the RT-qPCR assays, we compared RT-qPCR to slide based RNA in situ hybridization in a subset of the oropharyngeal SCCs. HPV RNA in situ hybridization is another novel method for detection of transcriptionally-active HPV in FFPE that is currently in commercial development and which is starting to be validated in oropharyngeal SCC. In this study, there was perfect correlation between RT-qPCR and RNA in situ hybridization in 45 oropharyngeal SCCs tested by both methods. As far as we are aware, this represents the first study to compare two methods for detecting transcriptionally active HPV in oropharyngeal SCC. Our results indicate that both RT-qPCR and RNA in situ hybridization are highly sensitive and specific methods. In contrast to RT-qPCR, which is considered the gold standard for the detection of transcriptionally-active HPV, RNA in situ hybridization is not quantitative. Further, accurate interpretation of RNA ISH requires positive and negative controls for each case to stain appropriately. These may fail in a minority of cases, as was seen in this study. However, since it is slide based it is perfectly suitable for small tissue samples that may yield insufficient RNA for RT-qPCR.
Using the RT-qPCR assays to quantitate 13 high-risk HPV types in oropharyngeal SCCs (150 cases), our study revealed that the vast majority (81.3%) were HPV positive, which is more prevalent than previously reported 1, 3, 8. Interestingly, only four HPV types were detected from the 150 oropharyngeal tumors. In contrast, when using the same assays to profile 80 cervical tumors, nine HPV types were detected (unpublished data). Why only a small subset of oncogenic HPV viruses appear to efficiently infect the oropharynx and lead to cancer development isn’t clear and may be a subject for further study. The infection rate in normal oropharynx is low. In contrast, the vast majority of oropharyngeal cancers have HPV infection. As demonstrated by real-time RT-PCR, HPV actively transcribes their genes in these cancer tissues. Thus, HPV is viewed as a driving force for oropharyngeal carcinogenesis, and targeted therapy against HPV activities could potentially be effective for most oropharyngeal cancers.
One important finding from our work is that p16 can consistently reflect the transcriptional activity of HPV in oropharyngeal cancer. Previous work demonstrated that both HPV and p16 are prognostic markers; however, it has been under heated debate whether p16 expression specifically reflects HPV activity in oropharyngeal cancer. The controversy mainly arises from: 1) the limited scope of previous HPV analyses, as many studies only focused on HPV type 16; and 2) the relative low sensitivity of HPV DNA in situ hybridization, a common detection method used in previous studies. With our new RT-qPCR assays for the detection and quantitation of many HPV types, we have established a new gold standard for evaluating the expression correlation of p16 to HPV infection. We found that all 122 HPV-positive tumors had high p16 expression and almost all the tumors with high p16 expression (93%) were also HPV positive. There were only nine cases with high p16 expression but no HPV detected. For these cases, it could be that p16 was activated by a mechanism not related to HPV infection, or that other HPV types (not among the 13 high-risk types we analyzed) led to the activation of p16 27. In any case, the discrepancy between p16 and HPV detection with our RT-qPCR assays was very small (6% of all oropharyngeal tumors). Thus, based on the profiling data, it is reasonable to conclude that p16 can be used as a highly sensitive and specific surrogate marker for HPV activity in oropharyngeal cancer. Importantly, we also showed that p16 protein expression by IHC identified essentially the same patients as identified by either HPV RT-qPCR or p16 RT-qPCR, although with a detection sensitivity slightly lower than p16 RT-qPCR (Figure 2C&D). Given its convenience as well as cost-effectiveness, p16 IHC could have important prognostic value in clinical practice for routine care of oropharyngeal cancer patients.
In summary, we have developed a robust RT-qPCR method for HPV detection and quantitation which shows high rates of HPV in oropharyngeal SCC. These RT-qPCR assays correlate highly with slide based HPV RNA in situ hybridization. There also is a very high correlation between HPV detection by RT-qPCR and p16 detection by either RT-qPCR of IHC such that the two test methods are essentially equivalent for the identification of those with improved clinical outcomes.
Novelty & Impact Statements.
A new real-time RT-PCR method has been established for the expression profiling of 13 high-risk HPV types. This new method detects transcriptionally active HPV and highly correlates with RNA in situ hybridization. The study further shows that p16 mRNA or protein expression signature is a sensitive and specific surrogate marker for HPV transcriptional activity (all genotypes combined) in oropharyngeal cancer, and p16 and HPV signatures have equivalent performance for oropharyngeal cancer prognosis.
Acknowledgments
This research was supported by the National Institutes of Health (grant R01GM089784). We would like to thank Jianping Li and Allissa Nielsen for their technical assistance.
Footnotes
H.W., X.M. and Y.L. are employees of Advanced Cell Diagnostics and hold stock options from the company. The remaining co-authors have no financial or other conflicts of interest to disclose.
References
- 1.D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, Westra WH, Gillison ML. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–1956. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 2.D’Souza G, Agrawal Y, Halpern J, Bodison S, Gillison ML. Oral sexual behaviors associated with prevalent oral human papillomavirus infection. J Infect Dis. 2009;199:1263–1269. doi: 10.1086/597755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillison ML, D’Souza G, Westra W, Sugar E, Xiao W, Begum S, Viscidi R. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernster JA, Sciotto CG, O’Brien MM, Finch JL, Robinson LJ, Willson T, Mathews M. Rising incidence of oropharyngeal cancer and the role of oncogenic human papilloma virus. Laryngoscope. 2007;117:2115–2128. doi: 10.1097/MLG.0b013e31813e5fbb. [DOI] [PubMed] [Google Scholar]
- 6.Syrjanen S. Human papillomavirus (HPV) in head and neck cancer. J Clin Virol. 2005;32 (Suppl 1):S59–66. doi: 10.1016/j.jcv.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 8.Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, O’Sullivan B, Waldron J, Cummings B, Kim J, Ringash J, Dawson LA, et al. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27:6213–6221. doi: 10.1200/JCO.2009.23.1670. [DOI] [PubMed] [Google Scholar]
- 9.Licitra L, Perrone F, Bossi P, Suardi S, Mariani L, Artusi R, Oggionni M, Rossini C, Cantu G, Squadrelli M, Quattrone P, Locati LD, et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol. 2006;24:5630–5636. doi: 10.1200/JCO.2005.04.6136. [DOI] [PubMed] [Google Scholar]
- 10.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–1017. doi: 10.2741/2739. [DOI] [PubMed] [Google Scholar]
- 11.Narisawa-Saito M, Kiyono T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Sci. 2007;98:1505–1511. doi: 10.1111/j.1349-7006.2007.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reimers N, Kasper HU, Weissenborn SJ, Stutzer H, Preuss SF, Hoffmann TK, Speel EJ, Dienes HP, Pfister HJ, Guntinas-Lichius O, Klussmann JP. Combined analysis of HPV-DNA, p16 and EGFR expression to predict prognosis in oropharyngeal cancer. Int J Cancer. 2007;120:1731–1738. doi: 10.1002/ijc.22355. [DOI] [PubMed] [Google Scholar]
- 13.Kumar B, Cordell KG, Lee JS, Worden FP, Prince ME, Tran HH, Wolf GT, Urba SG, Chepeha DB, Teknos TN, Eisbruch A, Tsien CI, et al. EGFR, p16, HPV Titer, Bcl-xL and p53, sex, and smoking as indicators of response to therapy and survival in oropharyngeal cancer. J Clin Oncol. 2008;26:3128–3137. doi: 10.1200/JCO.2007.12.7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JS, Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, El-Mofty SK. p16 positive oropharyngeal squamous cell carcinoma:an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol. 2010;34:1088–1096. doi: 10.1097/PAS.0b013e3181e84652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14:467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X. A PCR-based platform for microRNA expression profiling studies. RNA. 2009;15:716–723. doi: 10.1261/rna.1460509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sotlar K, Stubner A, Diemer D, Menton S, Menton M, Dietz K, Wallwiener D, Kandolf R, Bultmann B. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J Med Virol. 2004;74:107–116. doi: 10.1002/jmv.20153. [DOI] [PubMed] [Google Scholar]
- 21.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 22.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput Appl Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 23.Yee C, Krishnan-Hewlett I, Baker CC, Schlegel R, Howley PM. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter S, Delius H, Kahn T, Hofmann B, zur Hausen H, Schwarz E. Characterization of a novel human papillomavirus DNA in the cervical carcinoma cell line ME180. J Virol. 1991;65:5564–5568. doi: 10.1128/jvi.65.10.5564-5568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durst M, Croce CM, Gissmann L, Schwarz E, Huebner K. Papillomavirus sequences integrate near cellular oncogenes in some cervical carcinomas. Proc Natl Acad Sci U S A. 1987;84:1070–1074. doi: 10.1073/pnas.84.4.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu X, Schwarz JK, Lewis JS, Jr, Huettner PC, Rader JS, Deasy JO, Grigsby PW, Wang X. A microRNA expression signature for cervical cancer prognosis. Cancer Res. 2010;70:1441–1448. doi: 10.1158/0008-5472.CAN-09-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L, Graubard BI, Chaturvedi AK. Prevalence of oral HPV infection in the United States, 2009–2010. JAMA. 2012;307:693–703. doi: 10.1001/jama.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]