Abstract
The isolation and characterization of a viable mutant of simian virus 40 (SV40) called "in1449" are described. the mutant DNA is found to have a 157-nucleotide-long insertion at map position 0.649 within the 5' untranslated sequence of the early region of SV40. The complete nucleotide sequence of the insert is presented. Sequence comparisons show that the insert is not of SV40 origin. The insert is presumably of monkey origin since in1449 was produced within monkey kidney cells. The sequence of the in1449 insert matches remarkably well with sequences of a certain predominant family of interspersed repeated sequences in human DNA (called the Alu family) and cloned members thereof. This high degree of sequence homology suggests that the in1449 insert is derived from a member of a family of interspersed repeated sequences in monkey DNA related to the human Alu family. The in1449 insert (and the Alu family members) contain certain oligonucleotide sequences that also are found conserved in the replication origins of papovaviruses and certain other oligonucleotides found in repetitive double-stranded regions of mammalian heterogeneous nuclear RNAs. Sequences around the two recombinant joints in in1449 exhibit a definite pattern of homology. An octanucleotide present in the SV40 part of the first recombinant joint is exactly repeated 15 nucleotides away within the insert; another octanucleotide present within the insert at the second joint is exactly repeated 21 nucleotides away in the viral DNA. The viral DNA sequences flanking the insert in in1449 also exhibit some homology.
Full text
PDF
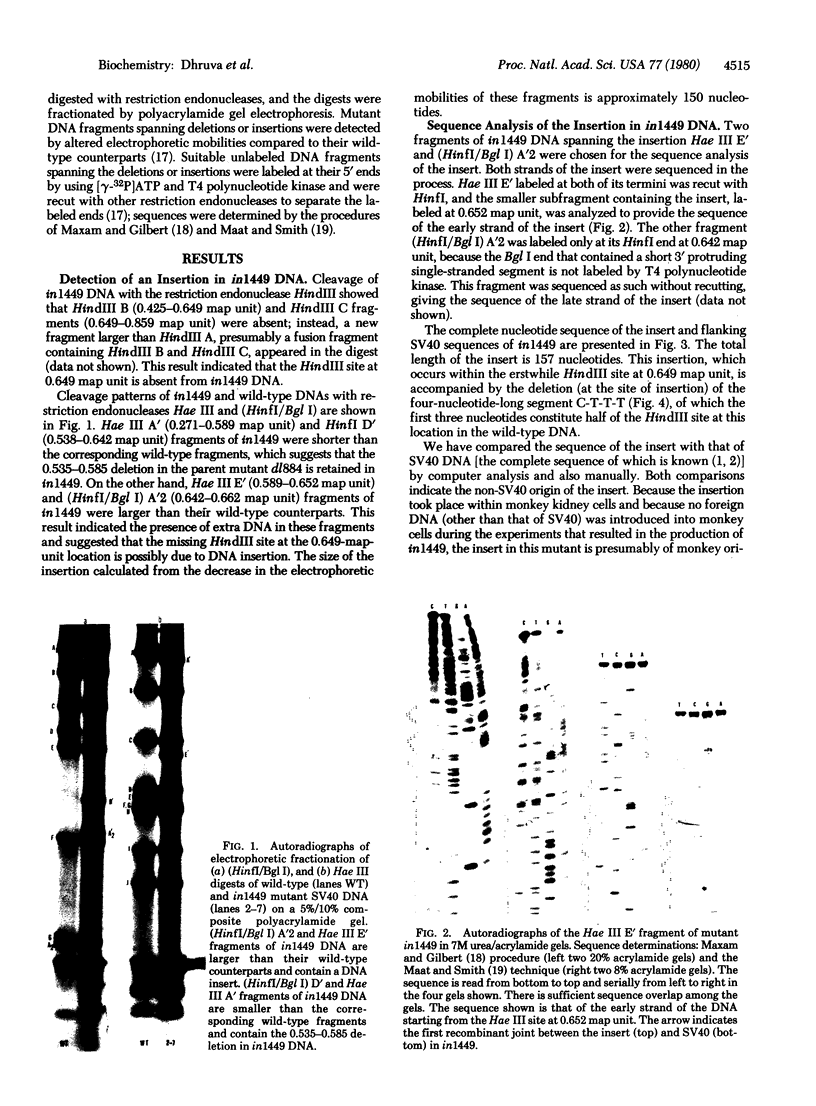



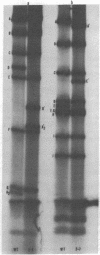
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Botchan M., Topp W., Sambrook J. The arrangement of simian virus 40 sequences in the DNA of transformed cells. Cell. 1976 Oct;9(2):269–287. doi: 10.1016/0092-8674(76)90118-5. [DOI] [PubMed] [Google Scholar]
- Dhar R., Lai C. J., Khoury G. Nucleotide sequence of the DNA replication origin for human papovavirus BKV: sequence and structural homology with SV40. Cell. 1978 Feb;13(2):345–358. doi: 10.1016/0092-8674(78)90203-9. [DOI] [PubMed] [Google Scholar]
- Duncan C., Biro P. A., Choudary P. V., Elder J. T., Wang R. R., Forget B. G., de Riel J. K., Weissman S. M. RNA polymerase III transcriptional units are interspersed among human non-alpha-globin genes. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5095–5099. doi: 10.1073/pnas.76.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Friedmann T., LaPorte P., Esty A. Nucleotide sequence studies of polyoma DNA. The Hpa II 3/5 junction to the Hpa II 4/Hae III 18 junction, encoding the origin of DNA replication and the 5' end of the early region. J Biol Chem. 1978 Sep 25;253(18):6561–6567. [PubMed] [Google Scholar]
- Grindley N. D. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell. 1978 Mar;13(3):419–426. doi: 10.1016/0092-8674(78)90316-1. [DOI] [PubMed] [Google Scholar]
- Gutai M. W., Nathans D. Evolutionary variants of simian virus 40: Cellular DNA sequences and sequences at recombinant joints of substituted variants. J Mol Biol. 1978 Dec 5;126(2):275–288. doi: 10.1016/0022-2836(78)90363-7. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Houck C. M., Rinehart F. P., Schmid C. W. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979 Aug 15;132(3):289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R. Inverted repeated DNA from Chinese hamster ovary cells studied with cloned DNA fragments. Proc Natl Acad Sci U S A. 1978 Jun;75(6):2679–2683. doi: 10.1073/pnas.75.6.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Ketner G., Kelly T. J., Jr Integrated simian virus 40 sequences in transformed cell DNA: analysis using restriction endonucleases. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1102–1106. doi: 10.1073/pnas.73.4.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. N., Brockman W. W., Nathans D. Evolutionary variants of simian virus 40: cloned substituted variants containing multiple initiation sites for DNA replication. Virology. 1975 Jul;66(1):53–69. doi: 10.1016/0042-6822(75)90178-6. [DOI] [PubMed] [Google Scholar]
- Maat J., Smith A. J. A method for sequencing restriction fragments with dideoxynucleoside triphosphates. Nucleic Acids Res. 1978 Dec;5(12):4537–4545. doi: 10.1093/nar/5.12.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan T., Singer M., Rosenberg M. Structure of simian virus 40 recombinants that contain both host and viral DNA sequences. II. The structure of variant 1103 and its comparison to variant CVPS/1P2 (EcoRI res). J Biol Chem. 1979 May 10;254(9):3592–3597. [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Ghosh P. K., Lebowitz P., Piatak M., Weissman S. M. Simian virus 40 early mRNA's. I. Genomic localization of 3' and 5' termini and two major splices in mRNA from transformed and lytically infected cells. J Virol. 1979 Apr;30(1):279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Jelinek W. Determination of nucleotide sequences from double-stranded regions of HeLa cell nuclear RNA. J Mol Biol. 1977 Oct 5;115(4):571–589. doi: 10.1016/0022-2836(77)90103-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg H., Singer M., Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978 Apr 28;200(4340):394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Houck C. M., Deininger P. L., Friedmann T., Schmid C. W. Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature. 1980 Mar 27;284(5754):372–374. doi: 10.1038/284372a0. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Westphal H., Srinivasan P. R., Dulbecco R. The integrated state of viral DNA in SV40-transformed cells. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1288–1295. doi: 10.1073/pnas.60.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. A biochemical method for increasing the size of deletion mutations in simian virus 40 DNA. J Mol Biol. 1977 Jul 5;113(3):503–515. doi: 10.1016/0022-2836(77)90235-2. [DOI] [PubMed] [Google Scholar]
- Subramanian K. N., Dhar R., Weissman S. M. Nucleotide sequence of a fragment of SV40 DNA that contains the origin of DNA replication and specifies the 5' ends of "early" and "late" viral RNA. III. Construction of the total sequence of EcoRII-G fragment of SV40 DNA. J Biol Chem. 1977 Jan 10;252(1):355–367. [PubMed] [Google Scholar]
- Subramanian K. N. Segments of simian virus 40 DNA spanning most of the leader sequence of the major late viral messenger RNA are dispensable. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2556–2560. doi: 10.1073/pnas.76.6.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B., Shenk T. Nucleotide sequence analysis of viable deletion mutants lacking segments of the simian virus 40 genome coding for small t antigen. J Virol. 1979 Jun;30(3):668–673. doi: 10.1128/jvi.30.3.668-673.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolun A., Aleström P., Pettersson U. Sequence of inverted terminal repetitions from different adenoviruses: demonstration of conserved sequences and homology between SA7 termini and SV40 DNA. Cell. 1979 Jul;17(3):705–713. doi: 10.1016/0092-8674(79)90277-0. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Feunteun J., Crawford L. V., Berg P., Fiers W. Nucleotide sequence deletions within the coding region for small-t antigen of simian virus 40. J Virol. 1979 Jun;30(3):674–682. doi: 10.1128/jvi.30.3.674-682.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]