Abstract
DB289 is the first oral drug shown in clinical trials to have efficacy in treating African trypanosomiasis (African sleeping sickness). Mild liver toxicity was noted but was not treatment limiting. However, development of DB289 was terminated when several treated subjects developed severe kidney injury, a liability not predicted from preclinical testing. We tested the hypothesis that the kidney safety liability of DB289 would be detected in a mouse diversity panel (MDP) comprised of 34 genetically diverse inbred mouse strains. MDP mice received 10 days of oral treatment with DB289 or vehicle and classical renal biomarkers blood urea nitrogen (BUN) and serum creatinine (sCr), as well as urine biomarkers of kidney injury were measured. While BUN and sCr remained within reference ranges, marked elevations were observed for kidney injury molecule-1 (KIM-1) in the urine of sensitive mouse strains. KIM-1 elevations were not always coincident with elevations in alanine aminotransferase (ALT), suggesting that renal injury was not linked to hepatic injury. Genome-wide association analyses of KIM-1 elevations indicated that genes participating in cholesterol and lipid biosynthesis and transport, oxidative stress, and cytokine release may play a role in DB289 renal injury. Taken together, the data resulting from this study highlight the utility of using an MDP to predict clinically relevant toxicities, to identify relevant toxicity biomarkers that may translate into the clinic, and to identify potential mechanisms underlying toxicities. In addition, the sensitive mouse strains identified in this study may be useful in screening next-in-class compounds for renal injury.
Key Words: pharmacogenetic, mouse diversity panel, DB289, human African trypanosomiasis, sleeping sickness
Human African trypanosomiasis (HAT), also known as sleeping sickness, is a vector-borne parasitic disease caused by Trypanosoma brucei subspecies transmitted to humans by the bite of infected tsetse flies. The disease is primarily found in areas indigenous to the tsetse fly, particularly rural areas of sub-Saharan Africa. Despite improvements in regional HAT control, the disease remains prevalent and new treatments are needed. It has been recently estimated that there are slightly fewer than 10,000 new cases reported each year; as a result, the World Health Organization has identified HAT as a core neglected tropical disease (Molyneux and Malecela, 2011). If left untreated, HAT progresses to a second stage involving the central nervous system, which can be fatal. There are few therapeutic agents available to treat the disease and these have several drawbacks, including the need for parenteral administration, and a high incidence of adverse events that can be severe or fatal. In addition, resistance to available drugs is a growing concern (Delespaux and de Koning, 2007).
To address this unmet need, the Consortium for Parasitic Drug Development (CPDD) developed the first oral drug to treat HAT, DB289 (pafuramidine maleate; 2,5-bis(4-amidinophenyl)-furan-bis-O-methylamidoxime). DB289 is a diamidine prodrug (Thuita et al., 2008; Yeramian et al., 2005) that is converted to the active metabolite DB75 (furamidine; 2,5-bis(4-aminophenyl)-furan), which exhibits a broad spectrum of antiparasitic activity for Trypanosoma subspecies. DB289 efficacy was demonstrated in phase II and phase III trials conducted in the Democratic Republic of Congo, Angola, and Sudan. The major safety concern noted in these treatment trials was a 7% incidence of liver injury; however, the incidence of this adverse event was substantially less than in the comparator group treated with the usual first line HAT therapy, pentamidine (77%).
Despite the promising results of this clinical trial, further development of DB289 was abruptly terminated due to an expanded safety trial conducted in South Africa. In this trial, 100 healthy adult volunteers were treated with 100mg DB289 b.i.d. for 14 days. As expected, generally mild and reversible liver injury was observed. However, 6 of 100 subjects experienced delayed renal insufficiency, and 1 subject required extended dialysis. Whereas preclinical testing in rats and monkeys had demonstrated liver injury as a dose-limiting event, these studies had not suggested the potential for DB289 to cause kidney injury. Identification of an animal model for DB289 kidney injury was important because similar aza analogs have shown promise in ability to treat second-stage HAT and are under consideration for clinical trials.
Since kidney injury due to DB289 had only been reported in the South African population, we hypothesized that genetic variation may underlie susceptibility to this serious adverse event. We reasoned that the introduction of genetic diversity to preclinical screening might have detected the kidney safety liability of DB289. The Mouse Diversity Panel (MDP) is a commercially available panel of well-characterized inbred mouse strains that have a genetic diversity comparable to a heterogeneous human population (McClurg et al., 2007). An advantage of the MDP is that because the genetics of each mouse strain are extensively characterized, it provides the means for detecting genes that contribute to a variable response, such as toxic response. This approach has previously been utilized to identify genetic variants that contribute to susceptibility to toxicity due to acetaminophen (Harrill et al., 2009a,b; Liu et al., 2010) and trichloroethylene (Bradford et al., 2011) exposures. In the case of acetaminophen-induced liver toxicity, the MDP identified that variation in the gene for CD44 was associated with an increased risk of toxicity in mice and this association was subsequently validated in clinical cohorts (Harrill et al., 2009b). In this study, it was hypothesized that the MDP pharmacogenetic approach would detect the kidney injury potential DB289 and would additionally provide mechanistic insight into the toxicity.
MATERIALS AND METHODS
Drugs and internal standards. DB289, DB75, and deuterated standards were acquired from the CPDD and were synthesized according to previously described methods (Boykin et al., 1995; Saulter et al., 2005) in the laboratories of Dr David Boykin (Georgia State University).
Animals. Male and female mice aged 6–8 weeks were obtained from The Jackson Laboratory or Charles River (CD-1) and housed in the Laboratory Animal Resources and Technical Support Facility at The Hamner Institutes. Mice utilized in this study from the Jackson Laboratory comprised 34 inbred strains that are priority strains for the Mouse Phenome Project (Bogue and Grubb, 2004). Mice were allowed to acclimate to the facility for 2 weeks prior to dosing; during this time, animals were randomized to treatment groups by body weight using the Instem Provantis system. Animals were given water ad libitum throughout the study and were fed NIH-07 wafer feed except for the time period 18h prior to necropsy. Animals were maintained on a 12-h light-dark cycle. Care of mice followed institutional guidelines under a protocol approved by the Institutional Animal Care and Use Committee.
DB289 administration. DB289 dosage selection was based on a pilot study in which CD-1 male and female mice were administered 0, 75, 125, or 250 μmol/kg of DB289 daily for 2, 4, or 14 days (i.e., with five mice per group); due to significant morbidity and mortality caused by higher doses, 75 μmol/kg was utilized for the inbred strain panel study in female mice. An initial evaluation of this dose in two inbred mouse strains, BALB/cByJ and C3H/HeJ, demonstrated that DB289 caused a marked decrease in body weight in sensitive mice; therefore, a 10-day treatment regimen was selected. Mice from 34 inbred strains were randomized into treatment groups and dosed intragastrically with 75 μmol/kg DB289 (N = 5 per strain) or vehicle (N = 5 per strain, 0.5% methyl 2-hydroxyethyl cellulose; Sigma-Aldrich) once daily for 10 days with a dosing volume of 10ml/kg for all doses. Dosing was performed at the same time of day throughout the study to avoid diurnal variability (Boorman et al., 2005). Animals were fasted 18h prior to necropsy.
Sample collection. Urine was collected in chilled containers for 18h overnight between study days 3–4, 7–8, and 10–11 using mouse metabolic cages (Hatteras Instruments, Inc., Cary, NC). Total urine volume was measured. On day 11, animals were euthanized by CO2 asphyxia and blood was collected via cardiac puncture. The livers and kidneys were quickly excised and sections of the left liver lobe and left kidney were placed in 10% phosphate-buffered formalin for histological analyses. The remaining tissue was snap-frozen in liquid nitrogen and stored at −80°C. Mice of strain P/J underwent necropsy 24h ahead of schedule, on day 10, due to morbidity associated with treatment.
Clinical Chemistry
Samples were assayed for serum biomarkers by standard enzymatic procedures using a SpectraMax microtiter plate reader (Molecular Devices, LLC). Blood urea nitrogen (BUN) was assayed using the MaxDiscovery Blood Urea Nitrogen Enzymatic Kit (Bioo Scientific Corporation) using the manufacturer’s recommendations. The Quantichrom Creatinine Assay Kit (BioAssay Systems) was utilized to measure creatinine in the serum (sCr) using the manufacturer’s protocol.
Kidney Injury Molecule-1 Measurements
Biomarkers that were assayed in mouse pilot studies included kidney injury molecule-1 (KIM-1; proximal tubule injury; Uscn Life Science, Inc.) β2-microglobulin (glomerular injury; Uscn Life Science, Inc.), Lipocalin-2/NGAL (renal epithelial injury; R&D Systems, Inc.), tissue inhibitor of metalloproteinases 1 (TIMP-1; R&D Systems, Inc.), and cystatin C (glomerular injury; BioVendor, LLC). KIM-1 was quantified in urine collected overnight prior to necropsy using the ELISA Kit for KIM-1 (Uscn Life Science, Inc.) as detailed by the manufacturer. Briefly, samples and standards were added to a microtiter plate precoated with biotin-conjugated polyclonal antibody preparation specific to KIM-1. Avidin conjugated with horseradish peroxidase was then incubated in each well prior to the addition of a substrate solution that elicited a colorimetric change. The reaction was halted upon addition of sulfuric acid solution, and KIM-1 was quantified spectrophotometrically at a wavelength of 450nm. Sample values were calculated using a standard curve and normalized to the urine volume collected over the 18-h collection period.
Measurement of DB75 Tissue Concentrations
Kidney specimens were processed for quantification of DB75, the active metabolite of DB289, by high-performance liquid chromatography/triple quadrupole mass spectrometry (HPLC/MS-MS) using a previously published method (Wang et al., 2010) with modifications. Briefly, specimens were homogenized with two volumes of HPLC-grade water (1g of wet tissue was considered equivalent to 1ml in volume) using a sonic dismembrator (Model 120; Fisher Scientific Co.). Homogenates obtained from DB289-treated animals were diluted 1:10 (vol/vol) with blank rat kidney homogenate. DB75 was extracted by mixing homogenates (25 μl) with 200 μl of 7:1 (vol/vol) methanol:water containing 0.1% (vol/vol) trifluoroacetic acid and 100μM internal standard (DB75-d 8). After centrifugation and evaporation of the resulting supernatants, the remaining pellet was reconstituted with 100 μl of 15% (vol/vol) methanol containing 0.1% (vol/vol) trifluoroacetic acid. DB75 quantification employed an HPLC-MS/MS system consisting of two Shimadzu solvent delivery pumps, a Valco (Valco Instruments Co., Inc.) switching valve, and a thermostated (6°C) LEAP HTC autosampler (LEAP Technologies) connected to an Applied Biosystems API 4000 triple quadrupole mass spectrometer equipped with a heated nebulizer (APCI) ionization source. Reverse-phase gradient chromatography, conducted at a flow rate of 0.75ml/min, was used to elute DB75 and DB75-d 8 from an Aquasil C18 HPLC column (50 × 2.1mm, 5 µm; Thermo Fisher Scientific, Waltham, MA) following a 4-µl injection of reconstituted pellet. Mobile phase (A) consisted of HPLC-grade water with 35mM formic acid and 15mM ammonium acetate; (B) consisted of 80:20 (vol/vol) acetonitrile:water with 35mM formic acid and 15mM ammonium acetate. The initial gradient condition was 100% A, which was held for 0.5min. From 0.5 to 4min, B increased linearly to 90%. The eluent was directed to waste for the first 0.6min of each injection, then directed to the mass spectrometer. At 4min, the eluent was redirected to waste, and the column was washed with 90% B for 1min. The system was re-equilibrated for 1min with 100% A. Total run time was 6min/injection. Automated sample acquisition and data analysis were accomplished using Analyst software (version 1.4.1; Applied Biosystems). DB75 and DB75-d 8 were analyzed in positive ion mode using the following transitions preset in multiple reaction monitoring scans: 305.1 → 288.1 (DB75) and 313.2 → 296.2 (DB75-d 8). Duplicate 11-point calibration curves for DB75 (100–250,000nM), prepared in blank mouse kidney homogenate, were constructed using the peak area ratio of DB75 to DB75-d 8 and fit with a quadratic equation using 1/x weighting. Quality controls and dilution controls (1:10 in blank rat kidney homogenate) were prepared in triplicate. The lower limit of quantification (LLQ) was 100nM. Accuracy and precision were at least 85 and 6%, respectively.
Histology
Formalin-fixed kidney and liver specimens were embedded in paraffin, and 5-µm sections cut in duplicate were applied to each slide. Sections were stained with hematoxylin and eosin. Liver and kidney tissue from the CD-1, BALB/cByJ, and C3H/HeJ pilot studies were assessed by light microscopy by a pathologist blinded to treatment assignment (Experimental Pathology Laboratories, Inc.).
Quantitative Trait Locus Mapping
Genome-wide association (GWA) mapping was performed as described elsewhere using efficient mixed-model association (EMMA; Kang et al., 2008). In this approach, a mixed model was used to incorporate pairwise genetic relatedness between every pair of mouse strains in the statistical model directly. Thus, the number of false positives due to the inherent genetic relatedness of strains is reduced, while simultaneously increasing the power to detect causative QTL. Briefly, GWA was performed in R (http://cran.r-project.org) using a 260,733 single-nucleotide polymorphism (SNP) dataset and the fold change (DB289/vehicle) for urinary KIM-1 or for serum ALT for each strain. Eight strains (NOD/ShiLtJ, NZW/LacJ, SWR/J, C57BR/cdJ, C58/J, P/J, SEA/GnJ, and I/LnJ) had KIM-1 values below the limit of quantitation or mortality that prevented sample collection, and were therefore excluded from association analysis. −Log P values were plotted across the mouse genome using the RGenetics package in Galaxy (http://main.g2.bx.psu.edu/). Genomic intervals with association scores greater than the Bonferroni correction threshold were considered significant (−log10 P ≥ 6.84). Genes within significant intervals were identified with Ensembl and the UCSC Genome Browser using NCBI mouse genome build 37 (http://www.ensembl.org; http://genome.ucsc.edu). Network analysis was performed using the pathway explorer function in Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA). The genetic network was recreated using Microsoft PowerPoint for visual clarity.
Statistical Methods
Phenotypic values were expressed as the mean ± standard error of the mean. Differences between vehicle and DB289-treated mice within a strain were determined via a two-tailed Student’s t-test. Correlation between study endpoints was assessed by linear regression analysis using GraphPad Prism (GraphPad Software, Inc.). Statistical outcomes were considered significant when the p-value was < 0.05.
RESULTS
Determination of Optimal DB289 Dosing Conditions
To determine the optimal dose for the MDP study, a dose range-finding experiment was conducted in CD-1 mice. The highest doses of DB289 were not well tolerated during the 14-day exposure with three (one male and two females) animals at the 125 μmol/kg dose level and nine animals (five males and four females) at the 250 μmol/kg found dead or euthanized early due to moribundity. Dose-dependent hepatocellular necrosis was evident in the liver of CD-1 mice, but was minimal at the 75 μmol/kg dose. Severe liver injury observed histologically, characterized by centrilobular necrosis, inflammation, hemorrhage, and fibrosis, was suspected by the study pathologist to have plausibly contributed to the observed morbidity and mortality.
Minimal to mild degeneration and necrosis was observed in the proximal tubule epithelia of the kidney in the female CD-1 mice treated at all three dose levels (Supplemental tables 1 and 2). This pathology was also observed in males but only at the 125 and 250 μmol/kg dose levels. The observed kidney pathology was not associated with increases in sCr or BUN (Supplemental table 1).
Due to the observed kidney lesions, modest liver injury, and lack of morbidity in CD-1 mice at the lowest dose level, exposure to 75 μmol/kg DB289 for 14 days in female mice was initially selected as the experimental design for the inbred strain study. However, due to weight loss observed in BALB/cByJ mice treated with DB289 in a subsequent study, the dosing duration was shortened to 10 days.
Consistent with observations in female CD-1 mice, sCr and BUN were not elevated in female BALB/cByJ or C3H/HeJ mice tested in the subsequent tolerability study. Urine was assayed for KIM-1, β2-microglobulin, TIMP-1, and cystatin C to determine the optimal kidney injury biomarker to assay. KIM-1 exhibited the largest elevations and dynamic range due to DB289 treatment, consistent with the restriction of the observed histological lesions to the proximal tubule in CD-1 mice (Supplemental table 2).
Effects of DB289 on Animal Health Outcomes in the MDP
DB289 was well tolerated in the majority of mouse strains tested. However, all DB289-treated animals of strains C58/J, MA/MyJ, NOR/LtJ, NZW/LacJ, P/J, PL/J, and SM/J received supportive care due to minor dehydration and weight loss associated with drug treatment. Supportive care consisted of subcutaneous injections of saline and the addition of HydroGel (Clear H2O) to the diet. One C58/J animal was euthanized early and all five P/J mice treated with DB289 underwent necropsy 24h ahead of schedule due to drug-associated morbidity.
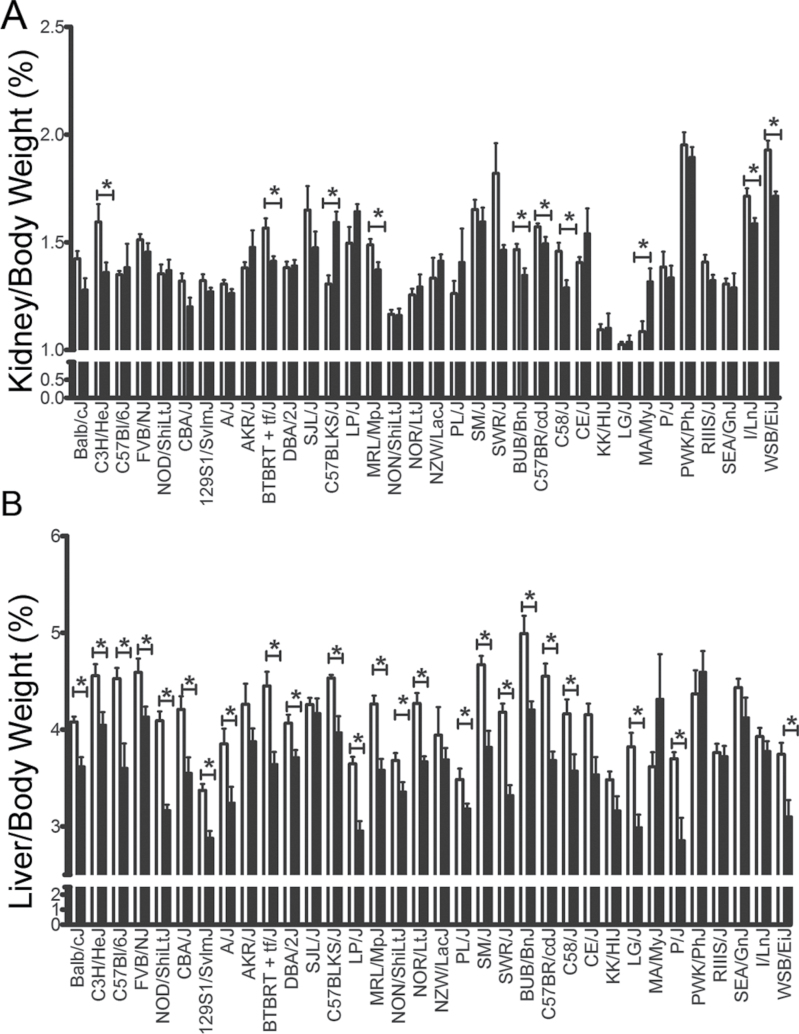
Kidney weights (relative to the body weight) were affected by DB289 treatment in 10/34 (29%) strains (Fig. 1A). There was a significant decrease in strains C3H/HeJ, BTBR T + tf/J, BUB/BnJ, C57BR/cdJ, C58/J, MRL/MpJ, I/LnJ, and WSB/EiJ (P < 0.05). A significant increase in kidney to body weight in strains C57BLKS/J and MA/MyJ was observed.
FIG. 1.
Organ-to-body weight ratios (%) are shown for (A) kidney weight and (B) liver weight. Values are expressed as the average ± SEM with white bars and black bars, indicating values for vehicle-treated and DB289-treated animals, respectively.
*Significant difference between treated and control animals within a strain (p < 0.05).
DB289 exposure resulted in decreased liver weight with respect to the body weight in the majority (24/34; 71%) of strains tested (Fig. 1B). More than half (55%) of the mouse strains exhibited a significant decrease in body weight between the first day and last day of dosing with DB289 (data not shown). However, of the eight strains for which there was a significant decrease in the kidney:body weight ratio, only two exhibited a treatment-related decrease in body weight (C58/J and MRL/MpJ). There were seven mouse strains for which a decrease in the liver:body weight ratio was not accompanied by a significant decrease in body weight during the dosing interval; these strains included A/J, BTBR + tf/J, BUB/BnJ, C3H/HeJ, C57BR/cdJ, DBA/2J, and FVB/NJ. Taken together, these data suggest that in selected strains a decrease in liver or kidney weight was unrelated to changes in body weight.
Liver Injury in Mice Following DB289 Exposure
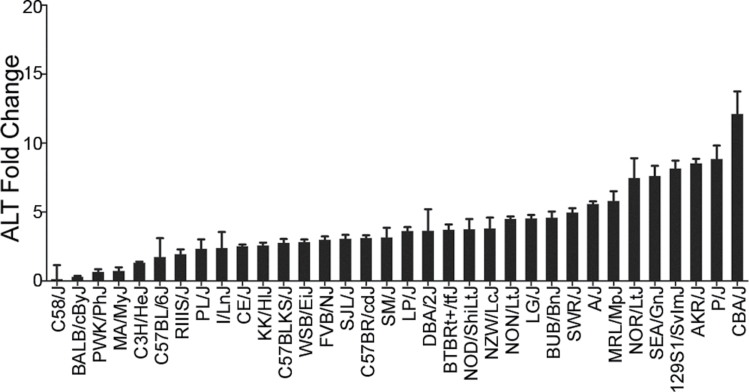
To determine the extent to which the liver was affected across the MDP following DB289 exposure, alanine aminotransferase (ALT) was quantified in serum collected during necropsy. The majority (29/34 or 85%) of strains tested exhibited a significant increase in ALT levels as a result of DB289 treatment; however, response varied across strains. ALT fold increases from baseline ranged from none in strain C58/J (0.1±1.1 fold increase) to greater than 10 in strain CBA/J (12.1±1.7 fold increase) (Fig. 2). Histologic examination of the livers indicated that the ALT increases were associated with hepatocellular degeneration and necrosis. Inflammatory cells were also found to be centered around necrotic hepatocytes in both periportal and centrilobular areas (data not shown). In strain C3H/HeJ, these findings were accompanied by mild-to-moderate midzonal hepatocellular vacuolation.
FIG. 2.
Fold changes are shown as the average of DB289-treated animals over the average of vehicle-treated animals within a strain for serum ALT levels collected at necropsy ± SEM.
Assessment of Renal Injury in Mice Using Standard Biomarkers
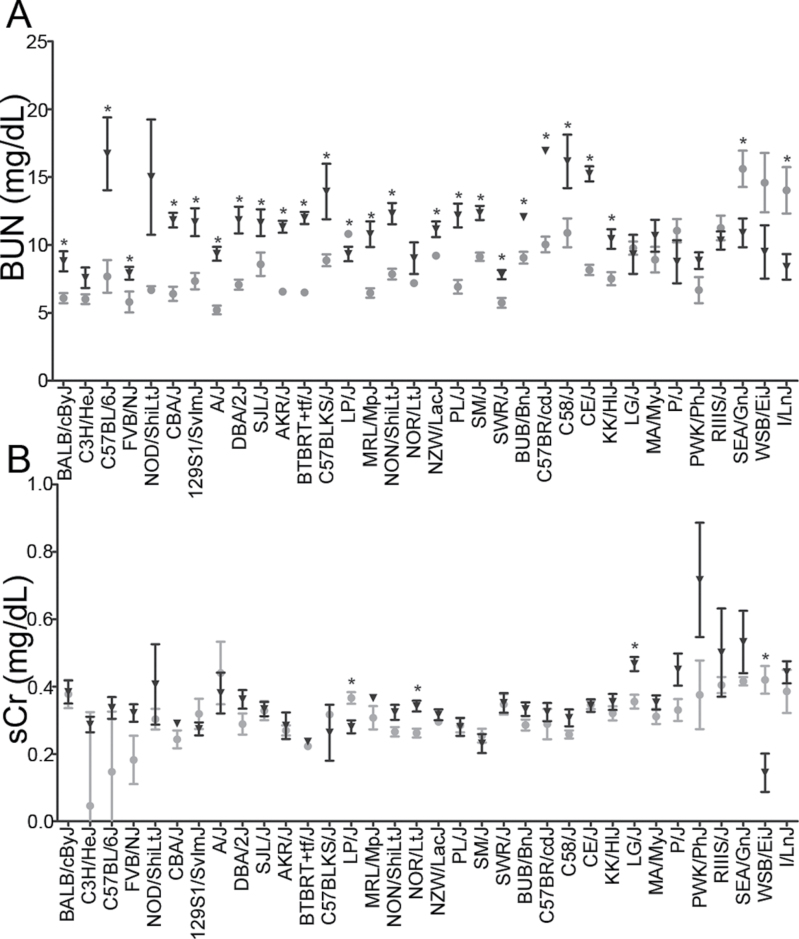
A statistically significant, yet mild, elevation (p < 0.05) in serum BUN was observed in 23 (68%) mouse strains (Fig. 3A). Similarly, four strains exhibited changes in sCr, with two strains exhibiting a minor increase (NOR/LtJ and LG/J) and two strains exhibiting a minor decrease (LP/J and WSB/EiJ) with DB289 treatment (Fig. 3B). However, the increase in both BUN and sCr was minor and all values were well within the standard mouse reference range for the analytes.
FIG. 3.
Serum levels of (A) BUN and (B) sCr are shown across mouse strains. Gray circles indicate average values for vehicle-treated animals and black triangles indicate average values for DB289-treated animals (± SEM).
*Significant difference between treated animals within a strain (p < 0.05).
Assessment of Renal Injury in Mice Using Urinary Biomarker, KIM-1
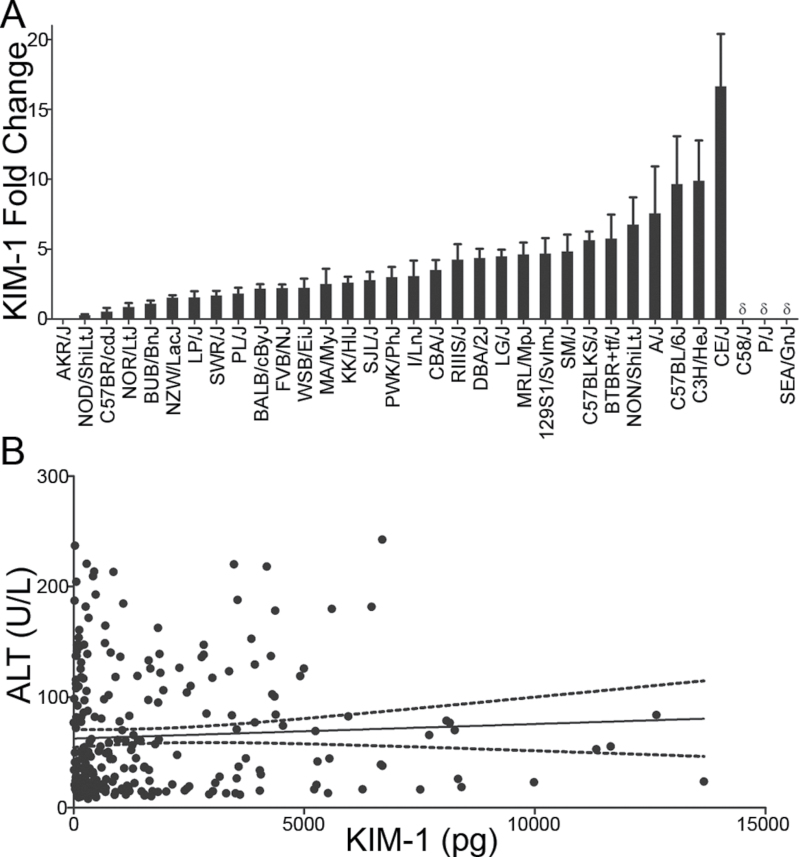
To better characterize the renal response to DB289 in the MDP, a sensitive biomarker of regeneration due to proximal tubule injury, KIM-1, was measured in urine collected overnight prior to necropsy. A marked interstrain variation was observed in urinary elimination of KIM-1 with fold increases in the DB289-treated animals ranging from 0.03±0.02 (AKR/J) to 16.7±8.4 (CE/J) (Fig. 4A). Of the strains tested, 38% demonstrated a significant KIM-1 increase with respect to control values. Neither BUN nor sCr values were correlated with KIM-1 (p > 0.05). The strains that displayed the most sensitivity to DB289-related kidney responses were C57BLKS/J, BTBR T + tf/J, NON/ShiLtJ, A/J, C57BL/6J, C3H/HeJ, and CE/J; these strains exhibited a KIM-1 fold change greater than five times the average of the vehicle-treated animals. The majority of KIM-1 increases were not associated with observable histologic lesions, supporting the increased sensitivity of KIM-1 as a biomarker for detecting renal injury over traditional histologic methods. In addition, there was no correlation between extent of renal injury, as assessed by KIM-1 and liver toxicity, as assessed by serum ALT (R 2 = 0.003, p = 0.36, Fig. 4B).
FIG. 4.
(A) Fold changes are shown as the average (± SEM) of DB289-treated animals over the average of vehicle-treated animals within a strain for urinary KIM-1 levels collected 18h prior to necropsy. δ indicates that the KIM-1 levels were below the quantifiable limit for strains C58/J, SEA/GnJ, and P/J. (B) A correlation plot is shown for ALT measured in the serum on day 11 and KIM-1 accumulation in the urine for 18h between days 10 and 11. The solid line indicates the regression line and dotted lines indicate the 95% confidence interval.
Identification of Genetic Variation Associated With Response to DB289
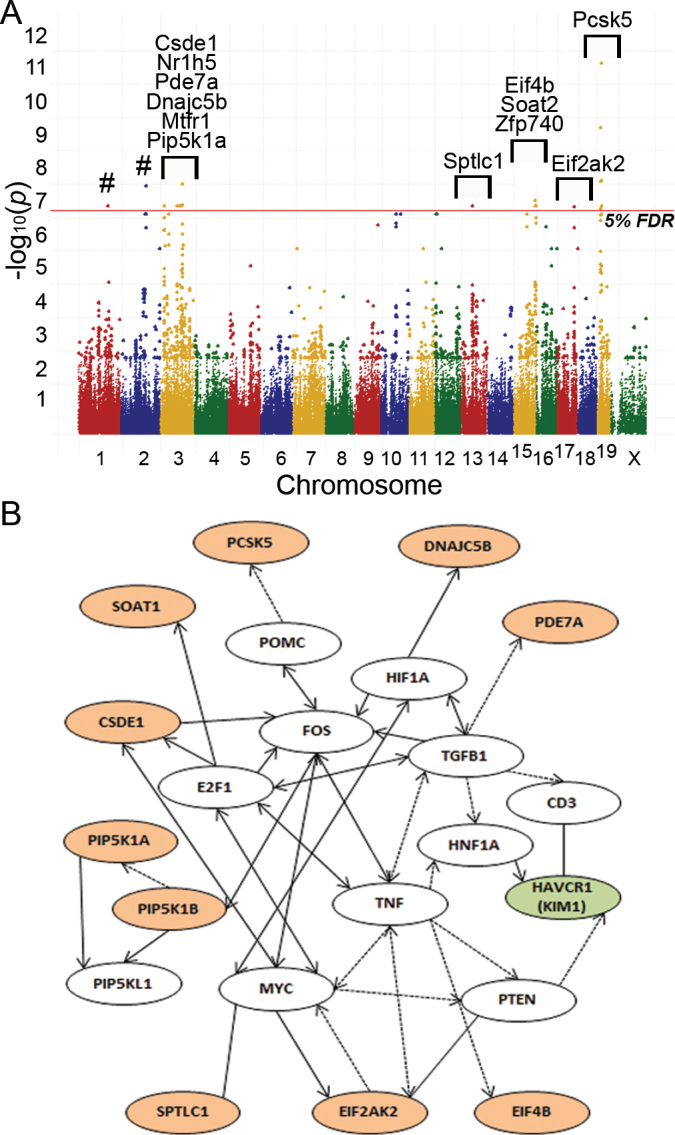
To determine genetic variants that are associated with kidney responses due to DB289, GWA mapping was performed. There were no significant QTL associated with an increase in ALT (data not shown). However, several SNPs were associated with urinary KIM-1 elimination, and the strength of association in some instances far exceeded the correction for multiple comparisons (Fig. 5A and Table 1). Of the significant SNPs that were associated with a known gene, the majority were associated with genes involved in the regulation of lipids (Pcsk5, Pip5k1a, Soat2, and Sptlc1; Table 2). Polymorphisms in Pde7a, a protein involved in cytokine production in natural killer T (NKT) cells, were significantly associated with the kidney response. Interestingly, genes involved in oxidative stress and heat/cold shock were also implicated, including Dnajc5b, Mtfr1, and Csde1. Regulatory genes that were implicated in the response included Nr1h5, Eif2ak3, Eif4b, and Zfp740. The majority of the association was driven by genetic variation in high responder strains A/J, C3H/HeJ, and CE/J, as well as midresponder strain CBA/J (Supplemental table 3).
FIG. 5.
(A) GWA mapping of DB289-induced elevations in KIM-1. The fold change increase in urinary KIM-1 was used to identify genomic intervals significantly associated with the renal response. Dots above the 5% false discovery rate (FDR) threshold (red line) indicate a significant −log P association score at the indicated SNP. Marker colors indicate the chromosome number within the mouse genome. Labels denote genes that include significant SNPs. # indicates that significant SNPs are within intergenic regions and not associated with a specific gene. (B) The network map of GWA significant genes is shown. Nodes highlighted in orange represent significant QTL. The gene for KIM-1 (Havcr1) is highlighted in green color. Solid arrows between nodes indicate a direct effect, dashed arrows indicate an indirect effect, and solid lines indicate direct binding.
TABLE 1.
Genomic Locations and Significance Scores for Candidate QTL Markers Within Genes
| Significance (−log10 P) | Chromosome | SNP genome location (Build 37) | Gene symbol | SNP function class |
|---|---|---|---|---|
| 9.20 | 19 | 17850776 | Pcsk5 | Intron |
| 7.60 | 19 | 17659581 | Pcsk5 | Intron |
| 6.77 | 19 | 17586468 | Pcsk5 | Intron |
| 6.77 | 19 | 17643240 | Pcsk5 | Intron |
| 6.77 | 19 | 17748435 | Pcsk5 | Intron |
| 6.77 | 19 | 17882060 | Pcsk5 | Intron |
| 7.50 | 3 | 102860565 | Csde1 | Intron |
| 7.50 | 3 | 102753254 | Nr1h5 | Exon (coding nonsynonymous) |
| 6.84 | 3 | 19168471 | Pde7a | Intron |
| 6.84 | 3 | 19198078 | Pde7a | Intron |
| 6.84 | 3 | 19425662 | Dnajc5b | Intron |
| 6.81 | 17 | 79266139 | Eif2ak2 | Intron |
| 6.84 | 15 | 101906580 | Eif4b | Intron |
| 6.84 | 15 | 101912637 | Eif4b | Intron |
| 6.84 | 3 | 19101103 | Mtfr1 | Intron |
| 6.84 | 3 | 94876525 | Pip5k1a | Intron |
| 6.84 | 15 | 101981667 | Soat2 | Intron |
| 6.84 | 13 | 53468361 | Sptlc1 | Intron |
| 6.84 | 15 | 102043864 | Zfp740 | 3′ untranslated region |
TABLE 2.
Genes Associated With Genetic Differences in KIM-R1 Response and Reported Functions
| Gene symbol | Gene name | Function |
|---|---|---|
| Pcsk5 | Proprotein convertase subtilisin/kexin type 5 | Critical role in cholesterol metabolism by controlling levels of circulating LDL |
| Csde1 | Cold shock domain containing E1, RNA binding | Unknown |
| Nr1h5 | Nuclear receptor subfamily 1, group H, member 5 | DNA binding, receptor activity, metal ion binding |
| Pde7a | Phosphodiesterase 7A | Cellular proliferation and cytokine production in NKT cells |
| Dnajc5b | DnaJ (Hsp40) homolog, subfamily C, member 5 beta | Heat shock protein binding |
| Eif2ak2 | Eukaryotic translation initiation factor 2-alpha kinase 2 | Serine/threonine protein kinase that phosphorylates translation initiation factor EIF2S1, inhibiting protein synthesis |
| Eif4b | Eukaryotic translation initiation factor 4B | mRNA helicase |
| Mtfr1 | Mitochondrial fission regulator 1 | Mitochondrial protein that protects cells from oxidative stress |
| Pip5k1a | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | Phospholipid biosynthesis |
| Soat2 | Sterol O-acyltransferase 2 | Produces cholesterol esters from long-chain fatty acyl CoA and cholesterol; cholesterol absorption and secretion of very LDLs |
| Sptlc1 | Serine palmitoyltransferase, long chain base subunit 1 | Key enzyme in sphingolipid biosynthesis |
| Zfp740 | Zinc finger protein 740 | Transcription factor |
Ten of the GWA significant genes were associated in a genetic network with several genes associated with tissue development and cellular proliferation, including a link to the gene encoding KIM-1 (Havcr1; Fig. 5B). Genes at the center of the network, myelocytomatosis oncogene (Myc) and FBJ osteosarcoma oncogene (Fos), are key regulators of cellular proliferation.
Kidney Tissue Exposure to DB289
To investigate whether strain-specific differences in the kidney response to DB289 were due to differences in kidney exposure to the parent or active metabolite (DB75), kidney tissue from each mouse was measured (Fig. 6A). Parent drug (DB289) was not detected. There was no observed correlation between the urinary KIM-1 concentrations for each mouse and the concentration of DB75 in the kidney tissue (R 2 = 0.002, p = 0.65, Fig. 6B).
FIG. 6.
(A) The concentration of DB75 in kidney tissue at time of necropsy is shown (average ± SEM) with strains ordered as in Figure 4A (increasing KIM-1 fold changes). (B) A correlation plot is shown for DB75 concentration in the kidney tissue and KIM-1 accumulation in the urine on day 10–11. The solid line indicates the regression line and dotted lines indicate the 95% confidence interval.
DISCUSSION
The observation of severe renal injury in the clinic led to an abrupt termination of the development program for DB289, which was an otherwise promising treatment for HAT. The hypothesis tested in this study was that the failure to detect this serious liability prior to entering the clinic may have resulted from an insufficient genetic diversity in the species utilized for standard nonclinical testing.
In each inbred strain studied, levels of the standard serum biomarkers of renal injury, BUN, and sCr remained within the standard reference ranges following DB289 treatment and would not have been considered significant in standard toxicity screens. However, DB289 treatment resulted in a marked increase in the urinary elimination of the renal proximal tubule injury marker KIM-1 among some but not all mouse strains. KIM-1 is present in low levels in healthy kidney tissue, but is expressed at high levels in regenerating proximal tubule epithelial cells following toxic injury (Vinken et al., forthcoming). The histological lesions observed in association with DB289 treatment were exclusively located in the proximal tubule, further supporting the use of KIM-1 in studies to assess degree of toxic response to DB289. That histological evidence of proximal tubule injury was not observed in all strains with elevations in urinary KIM-1 is consistent with the high sensitivity of KIM-1 to detect kidney injury in rodents (Vaidya et al., 2010) and in man (Liangos et al., 2009). KIM-1 therefore enabled detection and quantification of potentially injurious responses occurring below the detection capabilities of standard approaches. The data presented here indicate that an MDP approach to early phase toxicity testing using KIM-1 would have identified the kidney as a potential target organ for toxicity in the clinical trials. Urinary KIM-1 might then have been used as a biomarker to detect and monitor for renal toxicity in the clinical trials. Importantly, increased levels of KIM-1 in genetically sensitive strains enabled the use of GWA mapping to identify genetic variants that may contribute to the kidney response to DB289.
Most of the genes identified by GWA for KIM-1 levels are linked to cellular proliferation via regulatory gene networks (Fig. 5). This may not be entirely surprising because KIM-1 levels should directly reflect the rate of proliferation of the renal proximal tubular epithelia. Functional variations in the identified genes may therefore affect the kidney’s ability to repair after a toxic injury. The predominance of associations with cellular proliferation genes may indicate that the major factor underlying susceptibility to kidney injury due to DB289 is variation in the ability to repair after injury. It may be important that many of the associated genes are involved in lipid metabolism. Disruptions in lipid metabolism following acute kidney injury in mice can precipitate the onset of progressive renal disease and, ultimately, renal failure (Zager et al., 2011). It should be noted that the association with genes involved in lipid homeostasis may not solely reflect membrane synthesis during cell proliferation. It has also been reported that diverse forms of acute kidney injury are mediated by “lipotoxic” factors that include lipid dysregulation within both mouse and human kidney cells (Johnson et al., 2005; Zager et al., 2001, 2005). It may therefore be possible that perturbations in these pathways could lead to increased susceptibility to proximal tubular injury. Unfortunately, subsequent in vivo studies to investigate the mechanisms of Pcsk5 are not possible using genetic knockout animals as the loss of this essential gene causes embryonic lethality (Essalmani et al., 2006).
The gene with the greatest degree of association to KIM-1 levels, as well as the largest number of associated SNPs contained within the gene, was proprotein convertase subtilisin/kexin type 5 (Pcsk5), a serine protease that cleaves and inactivates endothelial and lipoprotein lipases. In addition to PCSK5’s involvement in cell proliferation networks (Fig. 5), PCSK5-regulated lipases influence circulating levels of high-density lipoprotein, low-density lipoprotein (LDL), and chylomicrons (Jin et al., 2005). Overexpression of PCSK9 protein, which is regulated by PCSK5 (Seidah et al., 2008; Zaid et al., 2008), in mice results in a marked decrease in circulating LDL coincident with a decrease in hepatic LDL receptors (Lalanne et al., 2005; Maxwell and Breslow, 2004). PCSK5 is expressed in the kidney and may help regulate kidney concentrations of cholesterol and other lipids (Essalmani et al., 2008). It may be relevant that following acute sublethal injury in the kidney, the kidney has been shown to be protected from subsequent toxic insult; furthermore, this property may be unique to proximal tubule cells due to increased cholesterol content that occurs subsequent to the initial injury (Zager et al., 2001). It should be noted that another gene involved in cholesterol absorption and secretion, sterol O-acyltransferase 2 (Soat2), was associated with urinary KIM-1 elimination. To date, there has been no direct connection reported between Soat2 expression and kidney injury. However, inhibition of SOAT2 has been shown to have significant impact on hypercholesteremia and atherosclerosis owing to its functional role in the esterification of cholesterol with its substrate, fatty acyl coenzyme A, to facilitate both intracellular storage and intercellular transport (An et al., 2006). Therefore, as with Pcsk5, functional variation in Soat2 may contribute to injury susceptibility by influencing kidney content of lipids.
One additional possibility is that DB289 or its metabolites are trafficked in blood or in tissues by lipid carriers and that variation in Pcsk5 and/or Soat2 may thereby influence proximal tubule exposure to DB289. However, concentration of DB75, the active metabolite, in the kidney across mouse strains was not correlated with KIM-1 levels, suggesting that this may be an unlikely basis for the association observed.
Pde7a (phosphodiesterase 7A), a gene not linked to the cell proliferation network, was identified as a gene that may also play a role in the response to DB289-mediated renal injury. The function of the PDE7A protein is to regulate cytokine production in NKT cells. NKT cells have been shown to play an essential role in the innate immune response of renal ischemia-reperfusion injury by mediating production of interferon-γ, which propagates the injury (Li et al., 2007). It was demonstrated that the absence of NKT cells in CD1d(−/−) or Jα18(−/−) genetic knockout mice accentuated the severity of renal injury following ischemia-reperfusion injury, indicating that NKT cells are protective (Yang et al., 2011). It is therefore possible that NKT cells or cytokine signaling cascades may modulate the extent of renal injury during treatment with DB289.
Proximal tubule cell toxicity may also be potentiated by variation in mitochondrial fission regulator 1 (Mtfr1), a gene that is involved in antioxidant activity (Monticone et al., 2010). Loss of function of this gene has been shown to cause reactive oxygen species-mediated DNA damage in the mouse, particularly in the testes where expression is the greatest (Monticone et al., 2007). The specific role that this protein may play in the kidney response to DB289 requires further study.
In summary, our approach with the MDP detected the renal injury potential of DB289 where traditional nonclinical testing failed, and identified urinary KIM-1 as a biomarker that could potentially be translated to the clinic to detect this target organ toxicity at an early stage. Variation in susceptibility to renal injury as assessed by KIM-1 was associated with sequence variation in genes related to cell proliferation, lipid biosynthesis and transport, oxidative stress, and cytokine signaling. The implicated genes may control the rate of cell proliferation in response to tubular injury, but may also have a role in susceptibility to initial injury and progression. Unfortunately, DNA samples from clinical DB289 studies were not available to test the hypotheses generated by the mouse models in humans. Further studies with human primary cells and in animal models are necessary to elucidate the mechanisms by which these gene variants may modulate the renal outcome. Finally, the MDP mouse strains identified as susceptible to DB289 kidney toxicity should be useful to test renal toxicity potential for next-in-class diamidine analogs now in development.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
This work was supported by the Bill and Melinda Gates Foundation.
Supplementary Material
ACKNOWLEDGMENTS
The authors extend their gratitude to Drs. Timothy Wiltshire and Oscar Suzuki at The University of North Carolina at Chapel Hill for the contribution of R code to facilitate EMMA analysis.
References
- An S., Cho K. H., Lee W. S., Lee J. O., Paik Y. K., Jeong T. S. (2006). A critical role for the histidine residues in the catalytic function of acyl-CoA:cholesterol acyltransferase catalysis: Evidence for catalytic difference between ACAT1 and ACAT2. FEBS Lett. 580 2741–2749 [DOI] [PubMed] [Google Scholar]
- Bogue M. A., Grubb S. C. (2004). The mouse phenome project. Genetica 122 71–74 [DOI] [PubMed] [Google Scholar]
- Boorman G. A., Blackshear P. E., Parker J. S., Lobenhofer E. K., Malarkey D. E., Vallant M. K., Gerken D. K., Irwin R. D. (2005). Hepatic gene expression changes throughout the day in the Fischer rat: Implications for toxicogenomic experiments. Toxicol. Sci. 86 185–193 [DOI] [PubMed] [Google Scholar]
- Boykin D. W., Kumar A., Spychala J., Zhou M., Lombardy R. J., Wilson W. D., Dykstra C. C., Jones S. K., Hall J. E., Tidwell R. R. (1995). Dicationic diarylfurans as anti-Pneumocystis carinii agents. J. Med. Chem. 38 912–916 [DOI] [PubMed] [Google Scholar]
- Bradford B. U., Lock E. F., Kosyk O., Kim S., Uehara T., Harbourt D., DeSimone M., Threadgill D. W., Tryndyak V., Pogribny I. P., et al. (2011). Interstrain differences in the liver effects of trichloroethylene in a multistrain panel of inbred mice. Toxicol. Sci. 120 206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespaux V., de Koning H. P. (2007). Drugs and drug resistance in African trypanosomiasis. Drug Resist. Updat. 10 30–50 [DOI] [PubMed] [Google Scholar]
- Essalmani R., Hamelin J., Marcinkiewicz J., Chamberland A., Mbikay M., Chrétien M., Seidah N. G., Prat A. (2006). Deletion of the gene encoding proprotein convertase 5/6 causes early embryonic lethality in the mouse. Mol. Cell. Biol. 26 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essalmani R., Zaid A., Marcinkiewicz J., Chamberland A., Pasquato A., Seidah N. G., Prat A. (2008). In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc. Natl. Acad. Sci. U.S.A. 105 5750–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Ross P. K., Gatti D. M., Threadgill D. W., Rusyn I. (2009a). Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 110 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrill A. H., Watkins P. B., Su S., Ross P. K., Harbourt D. E., Stylianou I. M., Boorman G. A., Russo M. W., Sackler R. S., Harris S. C., et al. (2009b). Mouse population-guided resequencing reveals that variants in CD44 contribute to acetaminophen-induced liver injury in humans. Genome Res. 19 1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Fuki I. V., Seidah N. G., Benjannet S., Glick J. M., Rader D. J. (2005). Proprotein convertases [corrected] are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 280 36551–36559 [DOI] [PubMed] [Google Scholar]
- Johnson A. C., Stahl A., Zager R. A. (2005). Triglyceride accumulation in injured renal tubular cells: Alterations in both synthetic and catabolic pathways. Kidney Int. 67 2196–2209 [DOI] [PubMed] [Google Scholar]
- Kang H. M., Zaitlen N. A., Wade C. M., Kirby A., Heckerman D., Daly M. J., Eskin E. (2008). Efficient control of population structure in model organism association mapping. Genetics 178 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalanne F., Lambert G., Amar M. J., Chétiveaux M., Zaïr Y., Jarnoux A. L., Ouguerram K., Friburg J., Seidah N. G., Brewer H. B., Jr, et al. (2005). Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. J. Lipid Res. 46 1312–1319 [DOI] [PubMed] [Google Scholar]
- Li L., Huang L., Sung S. S., Lobo P. I., Brown M. G., Gregg R. K., Engelhard V. H., Okusa M. D. (2007). NKT cell activation mediates neutrophil IFN-gamma production and renal ischemia-reperfusion injury. J. Immunol. 178 5899–5911 [DOI] [PubMed] [Google Scholar]
- Liangos O., Tighiouart H., Perianayagam M. C., Kolyada A., Han W. K., Wald R., Bonventre J. V., Jaber B. L. (2009). Comparative analysis of urinary biomarkers for early detection of acute kidney injury following cardiopulmonary bypass. Biomarkers 14 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. H., Lu P., Guo Y., Farrell E., Zhang X., Zheng M., Bosano B., Zhang Z., Allard J., Liao G., et al. (2010). An integrative genomic analysis identifies Bhmt2 as a diet-dependent genetic factor protecting against acetaminophen-induced liver toxicity Genome Res. 20, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K. N., Breslow J. L. (2004). Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. U.S.A. 101 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg P., Janes J., Wu C., Delano D. L., Walker J. R., Batalov S., Takahashi J. S., Shimomura K., Kohsaka A., Bass J., et al. (2007). Genomewide association analysis in diverse inbred mice: Power and population structure. Genetics 176 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux D. H., Malecela M. N. Neglected tropical diseases and the millennium development goals: Why the “other diseases” matter: Reality versus rhetoric. Parasit. Vectors. (2011);4:234. doi: 10.1186/1756-3305-4-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticone M., Panfoli I., Ravera S., Puglisi R., Jiang M. M., Morello R., Candiani S., Tonachini L., Biticchi R., Fabiano A., et al. (2010). The nuclear genes Mtfr1 and Dufd1 regulate mitochondrial dynamic and cellular respiration. J. Cell. Physiol. 225 767–776 [DOI] [PubMed] [Google Scholar]
- Monticone M., Tonachini L., Tavella S., Degan P., Biticchi R., Palombi F., Puglisi R., Boitani C., Cancedda R., Castagnola P. (2007). Impaired expression of genes coding for reactive oxygen species scavenging enzymes in testes of Mtfr1/Chppr-deficient mice. Reproduction 134 483–492 [DOI] [PubMed] [Google Scholar]
- Saulter J. Y., Kurian J. R., Trepanier L. A., Tidwell R. R., Bridges A. S., Boykin D. W., Stephens C. E., Anbazhagan M., Hall J. E. (2005). Unusual dehydroxylation of antimicrobial amidoxime prodrugs by cytochrome b5 and NADH cytochrome b5 reductase. Drug Metab. Dispos. 33 1886–1893 [DOI] [PubMed] [Google Scholar]
- Seidah N. G., Mayer G., Zaid A., Rousselet E., Nassoury N., Poirier S., Essalmani R., Prat A. (2008). The activation and physiological functions of the proprotein convertases. Int. J. Biochem. Cell Biol. 40 1111–1125 [DOI] [PubMed] [Google Scholar]
- Thuita J. K., Karanja S. M., Wenzler T., Mdachi R. E., Ngotho J. M., Kagira J. M., Tidwell R., Brun R. (2008). Efficacy of the diamidine DB75 and its prodrug DB289, against murine models of human African trypanosomiasis. Acta Trop. 108 6–10 [DOI] [PubMed] [Google Scholar]
- Vaidya V. S., Ozer J. S., Dieterle F., Collings F. B., Ramirez V., Troth S., Muniappa N., Thudium D., Gerhold D., Holder D. J., et al. (2010). Kidney injury molecule-1 outperforms traditional biomarkers of kidney injury in preclinical biomarker qualification studies. Nat. Biotechnol. 28 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken P., Starckx S., Barale-Thomas E., Looszova A., Sonee M., Goeminne N., Vermissen L., Buyens K., Lampo A. Tissue Kim-1 and urinary clusterin as early indicators of cisplatin-induced acute kidney injury in rats. Toxicol. Pathol. (forthcoming) doi: 10.1177/0192623312444765. [DOI] [PubMed] [Google Scholar]
- Yang S. H., Lee J. P., Jang H. R., Cha R. H., Han S. S., Jeon U. S., Kim D. K., Song J., Lee D. S., Kim Y. S. (2011). Sulfatide-reactive natural killer T cells abrogate ischemia-reperfusion injury. J. Am. Soc. Nephrol. 22 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeramian P., Meshnick S. R., Krudsood S., Chalermrut K., Silachamroon U., Tangpukdee N., Allen J., Brun R., Kwiek J. J., Tidwell R., et al. (2005). Efficacy of DB289 in Thai patients with Plasmodium vivax or acute, uncomplicated Plasmodium falciparum infections. J. Infect. Dis. 192 319–322 [DOI] [PubMed] [Google Scholar]
- Zager R. A., Andoh T., Bennett W. M. (2001). Renal cholesterol accumulation: A durable response after acute and subacute renal insults. Am. J. Pathol. 159 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager R. A., Johnson A. C., Becker K. (2011). Acute unilateral ischemic renal injury induces progressive renal inflammation, lipid accumulation, histone modification, and “end-stage” kidney disease. Am. J. Physiol. Renal Physiol. 301 F1334–F1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zager R. A., Johnson A. C., Hanson S. Y. (2005). Renal tubular triglyceride accumulation following endotoxic, toxic, and ischemic injury. Kidney Int. 67 111–121 [DOI] [PubMed] [Google Scholar]
- Zaid A., Roubtsova A., Essalmani R., Marcinkiewicz J., Chamberland A., Hamelin J., Tremblay M., Jacques H., Jin W., Davignon J., et al. (2008). Proprotein convertase subtilisin/kexin type 9 (PCSK9): Hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology 48 646–654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.