Abstract
Many solid malignant tumors arise on a background of inflamed and/or fibrotic tissues, features which are found in more than 80% hepatocellular carcinomas (HCC). Activated hepatic stellate cells (HSC) play a critical role in fibrogenesis associated with HCC onset and progression, yet their functional impact on hepatocyte fate remains largely unexplored. Here, we used a coculture model to investigate the crosstalk between hepatocytes (human hepatoma cells) and activated human HSC. Unsupervised genome-wide expression profiling demonstrated that hepatocyte-HSC crosstalk is bidirectional and results in the deregulation of functionally relevant gene networks. Notably, coculturing increased the expression of pro-inflammatory cytokines and modified the phenotype of hepatocytes toward motile cells. Hepatocyte-HSC crosstalk also generated a permissive pro-angiogenic microenvironment, particularly by inducing VEGFA and MMP9 expression in HSC. An integrative genomic analysis revealed that the expression of genes associated with hepatocyte-HSC crosstalk correlated with HCC progression in mice and was predictive of a poor prognosis and metastasis propensity in human HCC. Interestingly, the effects of crosstalk on migration and angiogenesis were reversed by the histone deacetylase inhibitor trichostatin A. Our findings therefore indicate that the crosstalk between hepatoma cells and activated HSC is an important feature of HCC progression, which may be targeted by epigenetic modulation.
Keywords: hepatocellular carcinoma, microenvironment, hepatic stellate cell, inflammation, integrative genomics, epigenetic
Introduction
Extensive evidence from genetics, genomics and cell biology demonstrated that cancer onset and progression is not only determined by tumor cells but is also influenced by the microenvironment (1–3). Microenvironment is a complex system which largely consists of extracellular matrix (ECM) proteins and proteoglycans, soluble factors and small signaling molecules such as cytokines and chemokines, along with a variety of cell types such as fibroblasts, immune cells and endothelial cells. Under normal conditions, the microenvironment constitutes an important modulator of epithelium cell fate and a barrier to cell transformation (4). Dynamic communications between the epithelium and the microenvironment notably modulate cell growth, apoptosis, and maintain epithelial cell polarity and differentiation (4). In cancer, the microenvironment which is also referred to as stroma, experiences drastic changes, including the recruitment and the activation of stromal cells and the remodeling of ECM. Importantly, co-evolution of tumor cells with their microenvironment during tumorigenesis suggests that tumor-stroma crosstalk may likely influence the phenotype of tumor cells and may provide a selective pressure for tumor initiation, progression and metastasis (1, 5, 6).
Hepatocellular carcinoma (HCC) is the most common primary tumor of the liver. The incidence of HCC is rising in many countries and its prognosis is typically poor. Thus, with 550,000 cases newly diagnosed and 600,000 deaths annually, HCC ranks among the deadliest forms of human malignancies worldwide (7). The remodeling of liver microenvironment is a hallmark of HCC pathogenesis (8). Indeed, more than 80% HCC develop in the setting of chronic hepatitis, fibrosis and cirrhosis, conditions in which inflammation and ECM deposition profoundly alter the hepatic microenvironment (9). However, the functional impact of the disrupted microenvironment on hepatocyte biology remains poorly understood (10). Over the last decade, high-throughput genomic studies provided important insights to classify HCC at a molecular level and to identify deregulated gene networks and signaling pathways (11). However, most of gene expression signatures in HCC have been derived from whole tumor tissues consisting of both the cancer epithelial cells and their surrounding microenvironment, a strategy which rendered elusive to evaluate the specific contribution of each compartment.
Activation of hepatic stellate cells (HSC) is a key feature of liver fibrosis and cirrhosis (12). Following liver injury, quiescent HSC become activated and convert into highly proliferative myofibroblast-like cells which express inflammatory and fibrogenic mediators responsible for ECM accumulation within microenvironment (12, 13). Therefore, the present study was specifically designed to address the functional impact and the clinical relevance of the crosstalk between tumor hepatocytes and activated HSC. By using a coculture model system, genome-wide expression profiling and functional assays we demonstrated that the crosstalk between hepatocytes and activated HSC i) is bidirectional; ii) induces an alteration of hepatocyte phenotype toward migration; iii) generates a permissive pro-inflammatory and pro-angiogenic microenvironment; iv) is predictive of a poor prognosis in human HCC; and v) could be targeted by epigenetic modulation.
Materials and Methods
Cell lines and coculture experiments
HepaRG and HuGB cell lines were established in our lab and maintained as previously described (14, 15). HepaRG cells were grown in William's E medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 µg/ml streptomycin, 5 µg/ml insulin, and 50 µM hydrocortisone hemisuccinate. Differentiation of HepaRG from progenitors to mature well-differentiated hepatocytes was achieved in 4 weeks by culturing the cells in the supplemented medium in presence of 2% dimethyl sulfoxide (DMSO) for the last 2 weeks as previously described (16) (Supporting Fig. 1A). All experiments hereinafter referred to as HepaRG were conducted using mature hepatocytes selectively isolated by mild trypsinization from DMSO-treated cultures. LX2 cells (Supporting Fig. 1B) were provided by S.L. Friedman and were maintained in supplemented DMEM medium as described (17). HepaRG/LX2 cocultures were conducted in serum- and DMSO-free William's E medium using 6-well plates and 1 µm pore size transwell inserts which allow diffusion of media components but prevent cell migration (BD Biosciences, San Jose, CA) (Supporting Fig. 1C). Huh7 and HepG2 cell lines were obtained from the European Collection of Cell Cultures (ECACC) which performed cell lines authentication by DNA barcoding. Primary human umbilical vein endothelial cells (HUVEC) were purchased from Invitrogen (Carlsbad, CA, USA) and were maintained in 200PRF medium supplemented with a low serum growth supplement (LSGS). All cell cultures were performed at 37°C in a 5% CO2 atmosphere. Trichostatin A was purchased from Sigma-Aldrich (St. Louis, MO, USA). Independent culture experiments were performed at least in triplicate.
Microarray analysis
Total RNA was purified from cells at 80% confluence with a RNAeasy kit (Qiagen, Valencia, CA, USA). Genome-wide expression profiling was performed using the low-input QuickAmp labeling kit and human SurePrint G3 8×60K pangenomic microarrays (Agilent Technologies, Santa Clara, CA, USA) as previously described (18). Starting from 150 ng total RNA, amplification yield was 9.7±0.6 µg cRNA and specific activity was 20.3±1.3 pmol Cy3 per µg cRNA. Gene expression data were processed using Feature Extraction and GeneSpring softwares (Agilent Technologies) and further analyzed using R-based BRB-ArrayTools. Briefly, differentially expressed genes were identified by a two-sample univariate t-test and a random variance model (P<0.01; false discovery rate <1%) as described (19). Permutation P-values for significant genes were computed based on 10,000 random permutations. Class prediction was performed using 7 algorithms and misclassification rate was computed using a leave-one-out cross-validation method (20). Clustering analysis was done using Cluster 3.0 and TreeView 1.6 using uncentered correlation and average linkage options. MIAME compliant microarray data have been deposited into gene expression omnibus (GEO) database (GSE32565).
Data mining and integrative genomics
Gene annotation was based on gene ontology and enrichment for specific biological functions or canonical pathways was evaluated using FuncAssociate 2.0 program (21). Ingenuity pathway analysis (IPA) was used to examine the functional association between differentially expressed genes and to generate the highest significant gene networks (Mountain View, CA, USA). Relevant networks were identified using the scoring system provided by IPA. Gene set enrichment analysis (GSEA) was performed by using the Java-tool developed at the Broad Institute (Cambridge, MA, USA) as previously described (22). Unsupervised GSEA was done with the whole C2 collection of curated gene sets from the molecular signatures database (MSigDB). Enrichment score was determined after 1,000 permutations. Connectivity map algorithm was used to link gene expression signatures with putative therapeutic molecules (23). Integration of genomic data was performed as previously described (20) using publicly available gene expression datasets downloaded from GEO.
Real time RT-PCR
Expression of relevant genes was measured by quantitative real time PCR as previously described (22). Quantitative analysis of PCR data was performed with the 2−ΔΔCt method using GAPDH Ct values for normalization. Melting analysis was conducted to validate the specificity of PCR products. PCR and microarray analysis were performed using RNA extracted from independent culture experiments (n=3).
Cell proliferation
HepaRG (10,000 cells/well) were seeded onto 96-well plates. Following 4 hrs incubation at 37°C the medium was replaced by serum-free medium supplemented with 50% (v/v) conditioned medium (CM) derived from culture and coculture of HepaRG and LX2. Proliferation was evaluated after 24, 48 and 72 hrs using a CyQUANT cell proliferation assay kit (Invitrogen). Experiments were performed in triplicate.
Cell migration
Influence of CM on HepaRG migration was determined using a 2D gap-closure radius 96-well migration assay, according to manufacturer’s instructions (Cell Biolabs, San Diego, CA, USA). Cell migration was independently evaluated from scratch-wounded confluent monolayers of HepaRG incubated in presence of serum-free medium supplemented with 50% CM as above. Migration was evaluated up to 72 hrs in triplicate.
In vitro angiogenesis
HUVEC (30,000 cells/well) were seeded onto 48-well plates previously coated with Geltrex reduced growth factor basement membrane matrix (100 µL/cm2) using non-supplemented 200PRF medium (Invitrogen). Endothelial tube formation was monitored after 6 hrs in the presence of 50% (v/v) serum-free CM from culture/coculture of LX2 and HepaRG. LSGS-supplemented HepaRG medium (2% FBS; 3 ng/mL bFGF) was used as a positive inducer control and non-supplemented HepaRG medium was used as a negative control. Triplicate experiments were performed.
Gel zymography
MMP activity in CM was evaluated in triplicate by gelatine zymography as described (8). Recombinant human MMP2 and MMP9 were used as positive controls. After scanning, images were analyzed by densitometry using ImageJ (NIH, Bethesda, USA).
Statistical analysis
Quantitative results were expressed as mean and SD and the significance was evaluated by Student’s t-test.
Results
Coculturing hepatocytes with activated hepatic stellate cells results in a bidirectional crosstalk
HepaRG and LX2 cells were used as a paradigm to model the crosstalk between transformed but differentiated human hepatocytes and activated HSC in the context of liver cancer (Supporting Fig. 1). HepaRG cell line was established in our lab from a well-differentiated Edmondson grade I HCC and was shown to possess the unique property to spontaneously differentiate into functional mature hepatocytes and biliary cells (14, 16). LX2 cells were reported to greatly recapitulate the in vivo phenotype of primary human activated HSC (17). However, since gene expression profiles during HSC activation may differ in culture and in vivo (24), we first conducted a gene expression profiling to validate the molecular phenotype of LX2 cells in our culture conditions. IPA demonstrated that highly expressed genes in LX2 (top 1%, 163 genes) were significantly linked to hepatic fibrosis and HSC activation (Supporting Fig. 2A). As expected, these genes included major regulators of ECM synthesis and degradation (e.g. COL1A1, MMP2), markers of HSC activation (e.g. ACTG2, VIM) as well as pro-inflammatory cytokines (e.g. IL1B). Further validating the choice of this cell line, GSEA demonstrated that highly expressed genes in LX2 were significantly enriched in a rat model of hepatic fibrosis and were characteristic of the transformation of quiescent HSC into myofibroblasts (Supporting Fig. 2B).
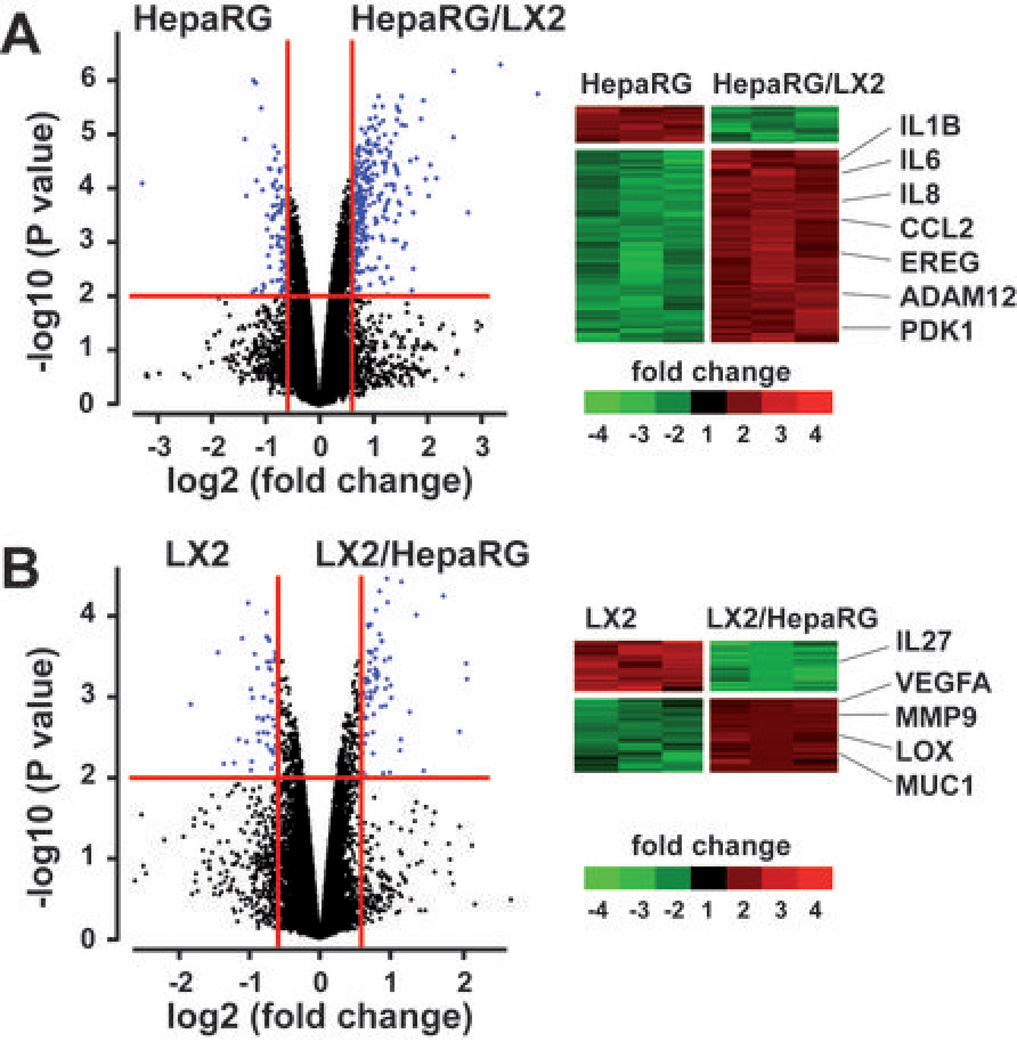
Hepatocyte-HSC crosstalk was next addressed by analyzing cocultures of HepaRG and LX2 separated by a transwell insert (Supporting Fig. 1C). Microarray experiments were conducted after 48 hrs, when cells reached 80% confluence. In HepaRG, the analysis of gene expression profiles by means of class comparison and class prediction algorithms identified 212 genes which expression was significantly modulated (P<0.01; 1.5 fold-change) by the presence of LX2 (Fig. 1A and Supporting Table 1). More than 83% genes were up-regulated suggesting that coculture with LX2 induced a global shift toward transcriptional activation in HepaRG (Fig. 1A). In LX2, the expression of 123 genes was significantly altered by the coculture condition (Fig. 1B and Supporting Table 2), including the up-regulation of master genes involved in ECM remodeling and angiogenesis (e.g. MMP9, VEGFA). Importantly, the unsupervised analysis of genes differentially expressed in coculture vs culture condition revealed a bidirectional crosstalk between HepaRG and LX2.
Figure 1.
Genome-wide expression profiles changes in cocultures of HepaRG and LX2. HepaRG and LX2 cell lines were cultured alone or side by side using transwell inserts (n=3 independent culture experiments). After 48hrs, total RNA was extracted from culture and coculture experiments and subjected to a microarray analysis. (A) Volcano plot (left) and clustering analysis (right) of 212 genes differentially expressed in three independent experiments using HepaRG cultured alone (HepaRG) or in presence of LX2 (HepaRG/LX2). (B) Volcano plot (left) and clustering analysis (right) of 123 genes differentially expressed in three independent experiments using LX2 cultured alone (LX2) or in presence of HepaRG (LX2/HepaRG). In (A) and (B), RNAs were selected based on the significance of the differential gene expression in coculture vs culture conditions (horizontal red line; P<0.01) and the level of induction or repression (vertical red lines; fold-change >1.5); examples of main changes in steady-state levels of mRNAs are indicated on the right.
Coculture with LX2 induces an inflammatory response and a motile phenotype in HepaRG
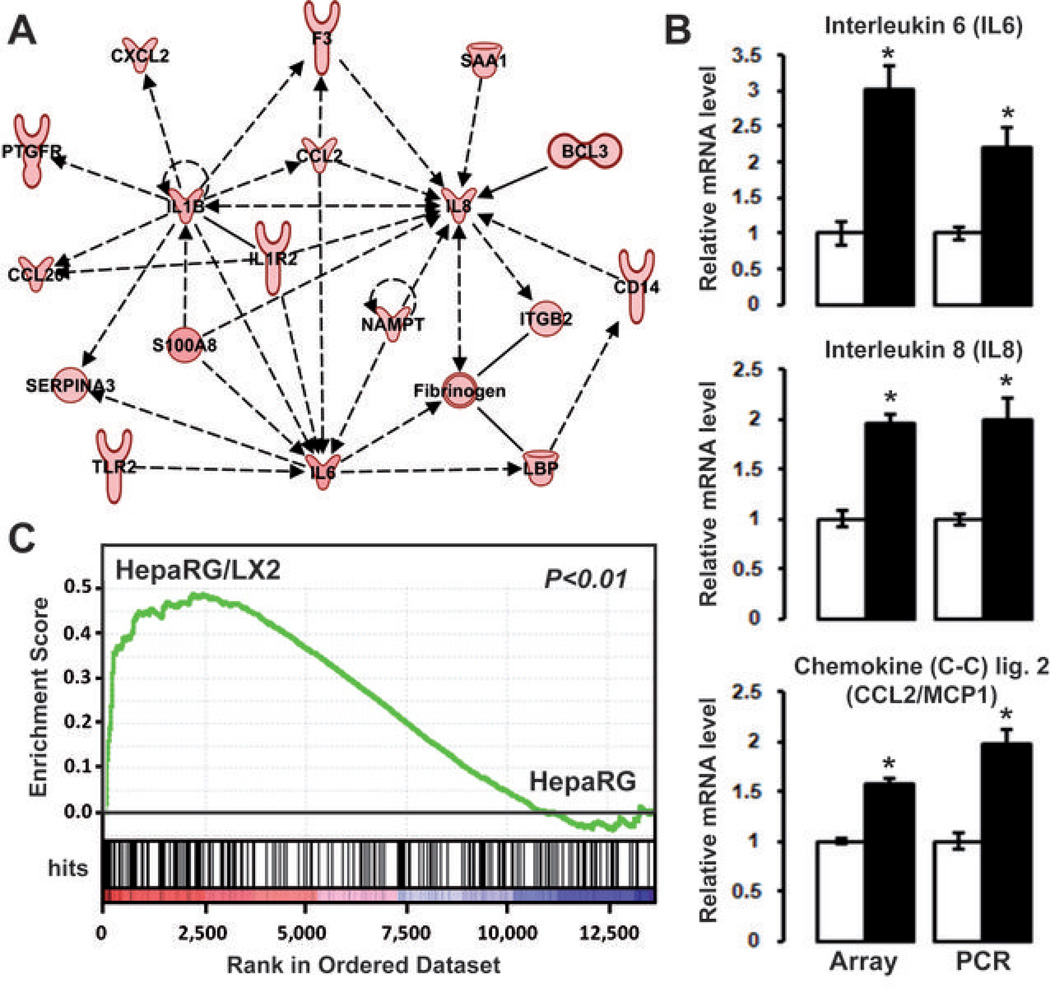
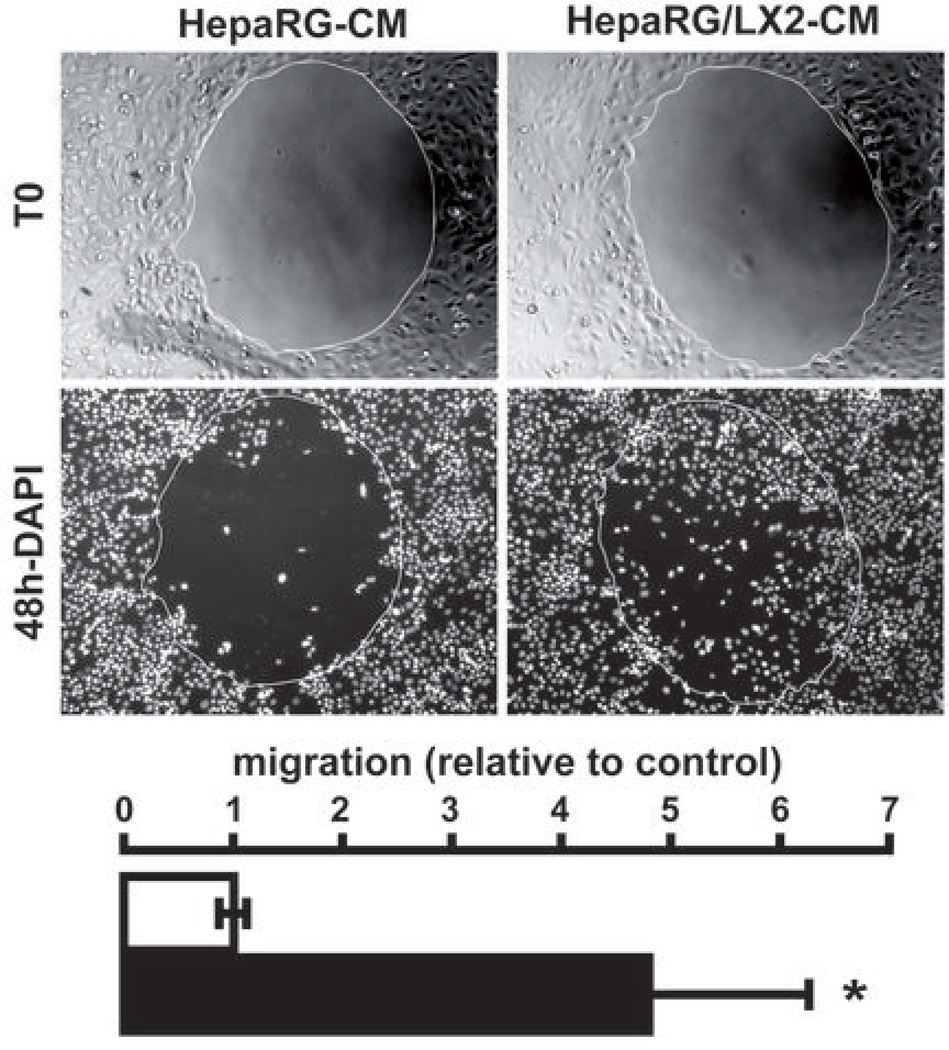
Gene ontology and ingenuity analysis showed that the genes related to cell chemotaxis, motility and inflammation were significantly enriched in the HepaRG/LX2 signature (Supporting Table 3). Notably, several important pro-inflammatory and pro-fibrogenic cytokines (e.g. IL1B, IL6, IL8), acute phase proteins (e.g. CP, SAA1), and growth factors (e.g. AREG, EREG) were up-regulated in HepaRG when the two cell types were cultured together (Fig. 1A and Supporting Table 1). Accordingly, a well-organized gene network linked to IL1B, IL6, IL8 and CCL2, which is also known as the monocyte chemoattractant protein 1 (MCP1) was identified by IPA (Fig. 2A and Supporting Fig. 3). The expression of IL6, IL8 and CCL2 was further evaluated by Q-RT-PCR using RNA extracted from independent cell culture experiments. As shown in Fig. 2B, all genes were significantly up-regulated in HepaRG after 48 hrs coculture with LX2. Together these data suggested that the crosstalk between HepaRG and LX2 resulted in the establishment of a pro-inflammatory microenvironment. To validate this observation, we performed a GSEA using an independent gene set which covered the whole response of Hep3B-hepatocytes to pro-inflammatory cytokines (25). This approach unambiguously demonstrated that coculture with LX2 induced a prominent inflammatory response in HepaRG (Fig. 2C). Inflammation is thought to play a key role in cancer initiation and progression by fostering multiple hallmarks of cancer including tumor cell proliferation and motility (1). To evaluate whether the coculture condition had any impact on the phenotype of HepaRG, mature hepatocytes were isolated from new HepaRG cultures and were exposed to conditioned media (CM) derived from the initial cultures and cocultures of HepaRG and LX2. Gap closure assay demonstrated that exposing fresh HepaRG-hepatocytes to CM derived from HepaRG/LX2 coculture significantly induced cell migration (Fig. 3). Of note cell proliferation remained unaffected by CM treatment suggesting that gap closure was not secondary to enhanced cell proliferation (data not shown). Collectively, these data indicated that coculturing hepatocytes with activated HSC resulted in the production of soluble factors, including pro-inflammatory signals, which were able to modify the phenotype of hepatocytes toward migration.
Figure 2.
The HepaRG/LX2 gene signature is related to inflammation. (A) Ingenuity analysis of up-regulated genes identified a gene network centered on IL1B, IL6, IL8 and CCL2. (B) Comparison of IL6, IL8, and CCL2/MCP1 mRNA levels detected by microarray and Q-RT-PCR in HepaRG using independent 48 hrs cultures (white bar, HepaRG cultured alone; black bar, HepaRG cocultured with LX2; n=3). Both microarray and Q-RT-PCR showed an up-regulation of these cytokines in cocultured HepaRG (* P<0.01). (C) GSEA analysis using the gene expression profiles of HepaRG cultured alone (HepaRG; right side) or in presence of LX2 (HepaRG/LX2; left side) and a proinflammatory gene signature established in Hep3B cell line (25). GSEA demonstrated a significant enrichment of the pro-inflammatory gene signature in HepaRG/LX2 gene expression profiles (P<0.01).
Figure 3.
Conditioned medium from HepaRG/LX2 cocultures induces HepaRG migration. Cell migration was analyzed by a gap closure assay. After seeding (T0), HepaRG were cultured in presence of conditioned medium derived from HepaRG cultures (HepaRG-CM) or HepaRG/LX2 cocultures (HepaRG/LX2-CM). After 48 hrs, cells were fixed and nuclei were stained with a DAPI fluorescent dye (48h-DAPI). Image analysis demonstrated that HepaRG/LX2-CM induces a migration of HepaRG (white bar, HepaRG-CM; black bar, HepaRG/LX2-CM; n=3; * P<0.01).
HepaRG-LX2 crosstalk generates a permissive pro-angiogenic microenvironment by modulating MMP9 and VEGFA in LX2 cells
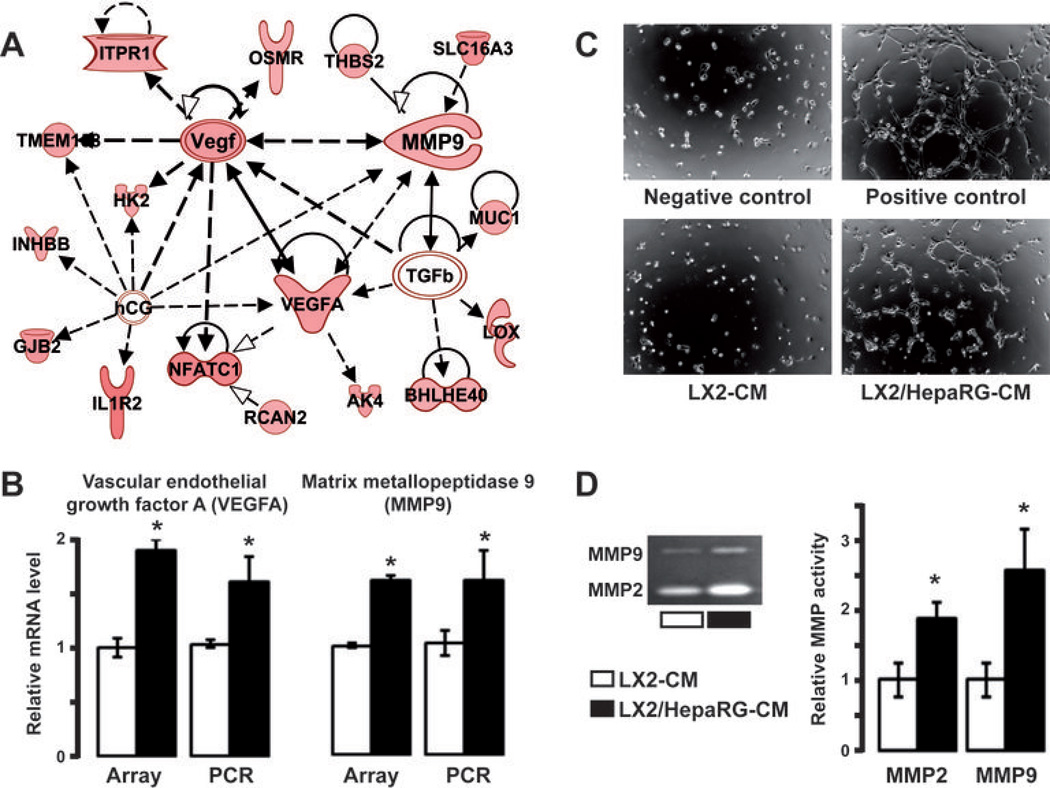
Data mining of 112 genes differentially expressed when LX2 were cocultured with HepaRG (LX2/HepaRG signature, Fig. 1B) identified a gene network linked to VEGFA and MMP9 (Fig. 4A). Upregulation of VEGFA and MMP9 genes was validated by Q-RT-PCR using RNA derived from independent experiments (Fig. 4B). Together with gene ontology and ingenuity analysis (Supporting Table 3), these results suggested that HepaRG-LX2 crosstalk may have profound impact on angiogenesis and ECM remodeling. This hypothesis was first tested by exposing HUVEC to CM derived from the culture or the coculture of LX2 and HepaRG. As shown in Fig. 4C, CM issued from coculture experiments (LX2/HepaRG-CM) induced the formation of tubule complexes by HUVEC. In contrast, CM issued from LX2 cultured alone (LX2-CM) failed to induce in vitro angiogenesis (Fig. 4C). Absence of tube formation was also noticed when HUVEC were exposed to CM derived from HepaRG cultured alone (data not shown). Next, MMP activity in CM was evaluated by gelatin zymography. In agreement with the increased expression of MMP9 (Fig. 4B), we showed that MMP9 activity was significantly higher (P<0.01) in LX2/HepaRG-CM than in LX2-CM (Fig. 4D). Similar observation was made for MMP2. These data suggested that hepatocyte-HSC crosstalk resulted in the establishment of a permissive proangiogenic microenvironment which may facilitate the migration of tumor cells.
Figure 4.
HepaRG-LX2 crosstalk induces in vitro angiogenesis and MMP expression. (A) Ingenuity pathway analysis of genes up-regulated in LX2 cocultured with HepaRG identified a gene network connected to VEGFA and MMP9. (B) Both VEGFA and MMP9 mRNA levels were up-regulated in LX2 after 48 hrs coculture with HepaRG (black bar) as compared to culture alone (white bar); gene expression analysis was performed in genome-wide array and Q-RT-PCR using independent culture experiments (n=3). (C) In vitro angiogenesis assay using HUVEC grown on a Geltrex matrix in presence of conditioned medium (CM) derived from the culture of LX2 (LX2-CM) or the coculture of LX2 with HepaRG (LX2/HepaRG-CM). After 6hrs, tube formation by HUVEC was observed in wells corresponding to positive control and treatment with LX2/HepaRG-CM (n=3; representative images are shown). (D) Detection of MMPs by gelatin zymography in culture-CM and coculture-CM. Significant increased in MMP2 and MMP9 expression was measured in LX2/HepaRG-CM. B,D; n=3; * P<0.01.
HepaRG/LX2 crosstalk signs a poor prognosis in human HCC and correlates with tumor progression
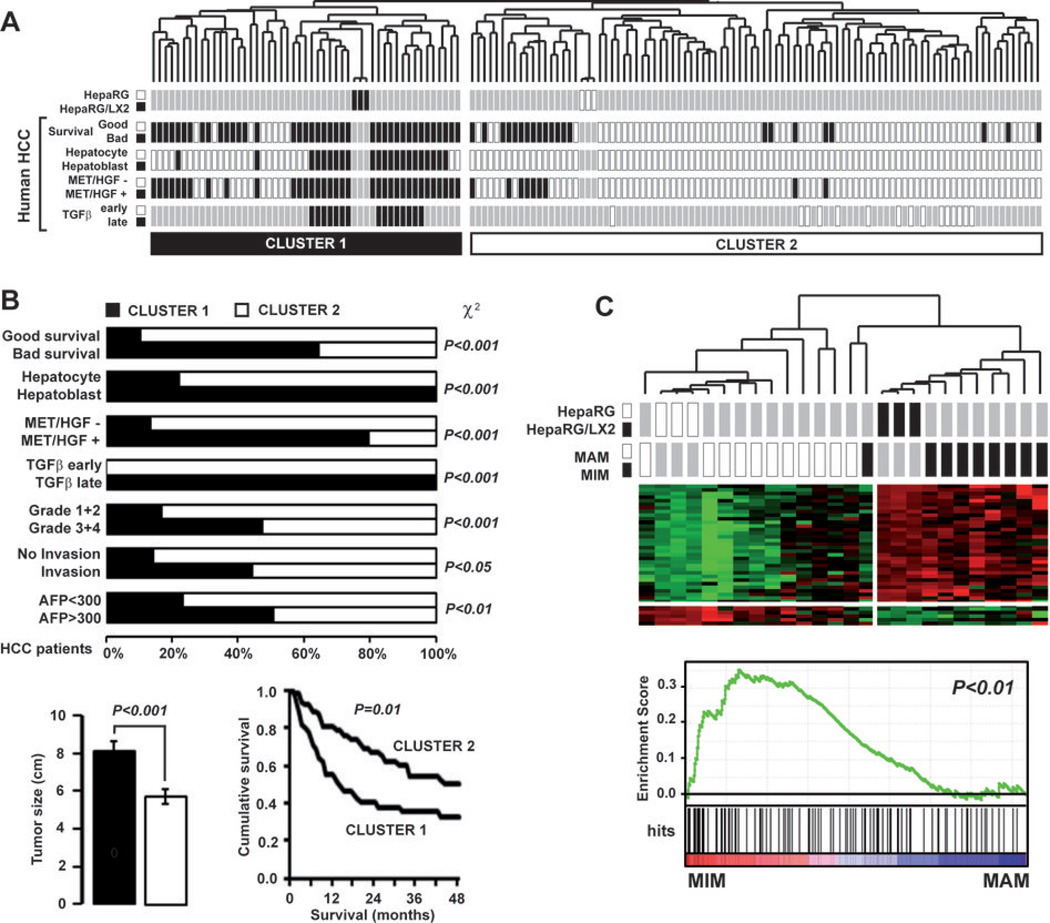
Integrative genomics was next used to evaluate the clinical relevance of the crosstalk between cultured hepatocytes and activated HSC. The HepaRG/LX2 signature (i.e. 212 genes differentially expressed in HepaRG by the coculture condition, Fig. 1A) was first integrated with the gene expression profiles of 139 cases of human HCC which were extensively characterized (20, 26–28). Hierarchical clustering analysis of the integrated dataset identified two robust clusters which organization was driven by the culture condition of HepaRG: cluster 1, HepaRG/LX2 coculture; cluster 2, HepaRG culture (Fig. 5A). Interestingly, we observed that clinical and biological parameters of HCC were not randomly distributed between the clusters 1 and 2. Strikingly, cluster 1 included significantly more tumors which were previously assigned to a poor prognosis group than cluster 2 (Fig. 5A). As shown in Fig. 5B, cluster 1 HCC were previously defined by a bad survival (27), hepatoblast traits (28), along with the activation of oncogenic MET/HGF (26) and TGFβ pathways (20). In addition, they exhibited a poorly differentiated phenotype (Edmondson grade >3), signs of vascular invasion and were significantly bigger in size (Fig. 5B). More importantly, the survival of patients included in cluster 1 was significantly reduced (P<0.01). Interestingly, if these observations demonstrated that the HepaRG/LX2 crosstalk signature is predictive of a poor prognosis when evaluated as a unique gene set, we also reported that 85% of genes included in the signature and expressed in primary HCC were clinically relevant when analyzed individually (Supporting Table 4). Unsupervised GSEA further supported the relevance of HepaRG/LX2 signature in predicting a poor prognosis phenotype, not only in HCC but also in other cancers. In the context of HCC, we found that specific signatures for recurrence (29), c-Myc/Tgfα aggressive mouse model of HCC (30) or TGFβ (20) were significantly enriched in the HepaRG/LX2 gene profiles (Supporting Table 5). This approach also demonstrated that gene signatures associated with a bad prognosis in cancers other than HCC (e.g. invasive breast cancer, advanced gastric cancer, metastatic stromal cells, and highly metastatic pancreatic cancer cells) were significantly (P<0.01) enriched both in HepaRG and LX2 under coculture conditions (Supporting Fig. 4). To further investigate the relevance of the HepaRG/LX2 crosstalk signature in HCC progression, we performed a cross-species integrative genomic approach as described previously (20). First, we integrated the HepaRG/LX2 signature with the gene expression profiles of 80 cases of HCC derived from 4 transgenic mouse models of HCC (c-Myc, E2f1, c-Myc/E2f1 and c-Myc/Tgfα) (19, 30). Consistent with the unsupervised GSEA, the analysis of the integrated datasets demonstrated that the HepaRG/LX2 signature clustered specifically with HCC derived from c-Myc/Tgfα (Supporting Fig. 5A). This observation pointed out c-Myc/Tgfα transgenic mice as the best in vivo model for evaluating the HepaRG/LX2 signature in HCC onset and progression. This was investigated by using the gene expression profiles characteristic of c-Myc/Tgfα induced hepatocarcinogenesis. These profiles were established previously using liver samples collected at various time-points of tumor onset and progression in transgenic mice, ranging from 3 weeks (moderate dysplasia), 3 months (severe dysplasia) and 9 months (HCC) (30). By using a multidimensional scaling approach, we demonstrated that the HepaRG/LX2 signature effectively discriminated the mouse samples based on HCC progression (Supporting Fig. 5B).
Figure 5.
Clinical relevance of HepaRG/LX2 signature in human HCC. (A) Dendrogram overview of HepaRG and HepaRG/LX2 experiments integrated with 139 cases of human HCC. Clustering analysis was based on the expression of 212 genes differentially expressed in HepaRG cocultured with LX2. Two major clusters (1 and 2) were identified. Distribution of human HCC samples between previously described subgroups with respect to survival (27) (good vs bad prognosis), cell origin (28) (hepatoblast vs hepatocytes), activation of MET/HGF (26) (− vs +) and TGFβ signaling pathway (20) (early vs late) is indicated on the left. (B) Statistical analysis of HCC distribution between clusters 1 and 2 based on previous gene signatures and clinical parameters. Cluster 1, which is defined by the HepaRG/LX2 coculture signature, shows a significant enrichment in HCC with the following features: bad survival, hepatoblast traits, activation of MET/HGF and late TGFβ pathways, higher differentiation grade and serum AFP level. Tumors size was significantly higher for HCC included in cluster 1. Kaplan-Meier plots and log-rank statistics analysis revealed a significant decreased in overall survival for patients included in cluster 1. (C) Integrative genomics using HepaRG/LX2 signature and gene expression profiles of peri-tumoral cirrhotic tissues from patient with (MIM) or without (MAM) metastasis (31). Clustering analysis (upper panel) and GSEA (lower panel) shows that the HepaRG/LX2 signature was significantly enriched in the gene expression profiles of cirrhotic tissues from patients with metastasis (MIM).
Because HSC activation is an early event in the pathogenesis of HCC, we asked whether the HepaRG/LX2 signature within cirrhosis tissues from patients with HCC could predict any specific clinical outcome. To test this hypothesis, we used a publicly available gene expression dataset established from the noncancerous hepatic tissue of patients with primary HCC (GSE5093 in GEO database). In this cohort, patients were divided in two groups based on the presence or absence of venous metastasis (31). Accordingly, cirrhosis tissues isolated from patients with or without metastasis were respectively termed metastasis-inclined microenvironment (MIM) and metastasis-averse microenvironment (MAM) (31). Based on the HepaRG/LX2 signature, we showed that MIM and MAM samples coclustered with HepaRG/LX2 and HepaRG samples respectively (Fig. 5C). GSEA confirmed that the genes which signed the crosstalk between HepaRG and LX2 were significantly enriched in the expression profiles of cirrhosis tissue from patients with metastasis (Fig. 5C). Altogether, these results demonstrated that the expression of genes embedded in the HepaRG/LX2 signature, either in cirrhosis or in HCC, was predictive of a poor prognosis and was associated with metastasis propensity.
Epigenetic modulation of HepaRG/LX2 crosstalk by trichostatin A
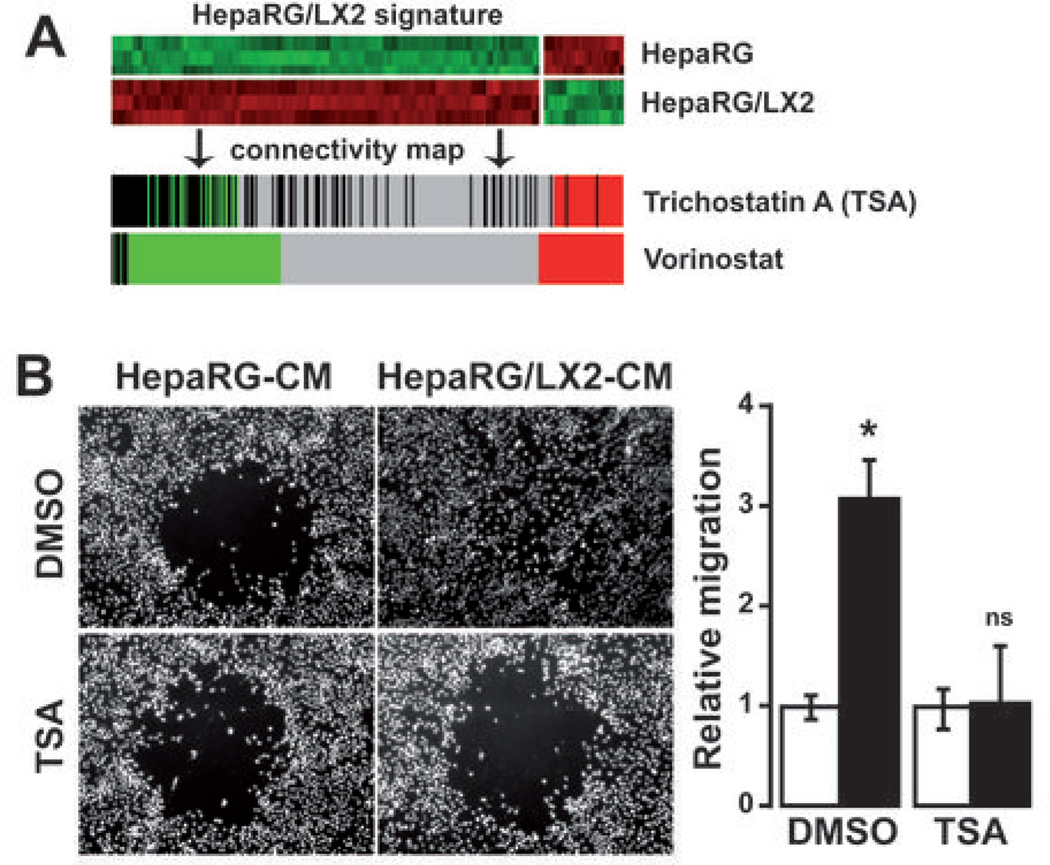
The clinical relevance of HepaRG/LX2 signature in predicting a poor prognosis in human HCC suggested that molecules targeting hepatocyte-HSC crosstalk may represent a promising therapeutic strategy. In this context, connectivity map was used to identify molecules that could reverse the global gene expression profile induced by LX2 on HepaRG. Interestingly, the top-10 ranked molecules identified by this approach included three inhibitors of histone deacetylases, vorinostat, trichostatin A (TSA) and valproic acid (Supporting Table 6). TSA was chosen for further experiments on the basis of the higher number of hits obtained with connectivity map (n=182 hits, Fig. 6A) and the results of the unsupervised GSEA which demonstrated that genes silenced by TSA in pancreatic cancer were significantly (P<0.01) enriched in the gene profiles of both HepaRG and LX2 under coculture conditions (Supporting Fig. 4). To test whether TSA could modulate the HepaRG/LX2 crosstalk, we produced CM from culture and coculture of HepaRG and LX2 cells exposed to 500nM TSA or DMSO control. As shown in Fig. 6B, the migration of freshly isolated HepaRG-hepatocytes in response to HepaRG/LX2-CM was completely abrogated in presence of TSA. We further showed in HepaRG that LX2-induced up-regulation of amphiregulin and epiregulin, two genes which have been linked to invasion and metastasis, was abolished by TSA (Supporting Fig. 6A). Our data also suggested that TSA was able to inhibit coculture-induced angiogenesis as evidenced by the absence of tube formation by endothelial cells and the reduced VEGFA expression by LX2 cells under coculture condition (Supporting Fig. 6B).
Figure 6.
Inhibition of LX2-induced migration of HepaRG by trichostatin A. (A) Connectivity map identification of trichostatin A (TSA) and vorinostat as candidate molecules to target HepaRG/LX2 crosstalk. (B) Cell migration was analyzed by a gap closure assay. After seeding, HepaRG were cultured in presence of conditioned medium derived from the culture (HepaRG-CM) or the coculture (HepaRG/LX2-CM) of HepaRG and LX2 exposed to either 500 nM TSA or DMSO control. After 72 hrs, cells were fixed and nuclei were stained with a DAPI fluorescent dye. Image analysis confirmed that HepaRG/LX2-CM induced migration of HepaRG (upper 2 micrographs) and demonstrated that migration was abolished in presence of TSA (lower 2 micrographs). Histogram: quantification of HepaRG migration (white bar, HepaRG-CM; black bar, HepaRG/LX2-CM; n=3; * P<0.01).
Discussion
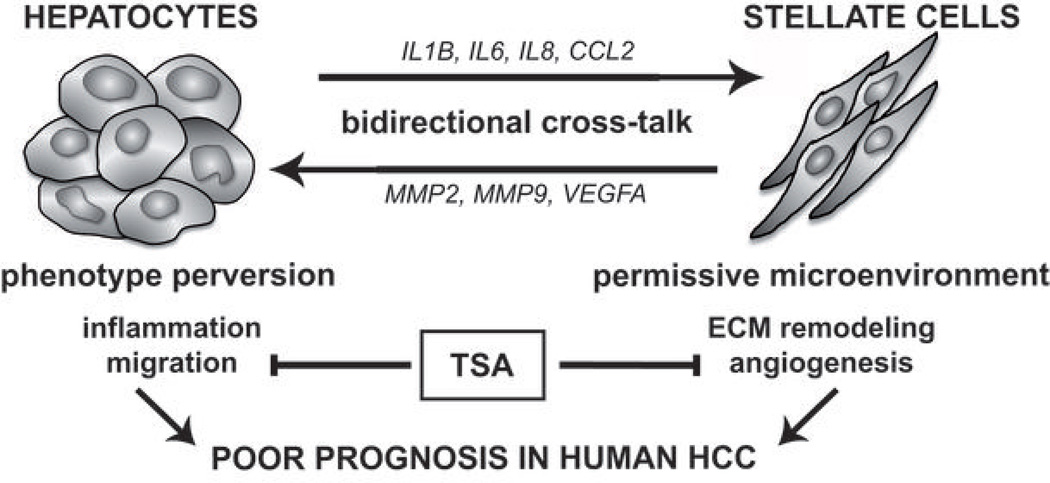
The tumor microenvironment contributes in the acquisition of multiple hallmarks of cancer (1). However, the underlying molecular mechanisms involved in the interactions between the tumor cells and the microenvironment remain poorly understood. We investigated the molecular mechanisms involved in the crosstalk between tumor cells and their microenvironment in liver cancer, by analyzing cocultures of hepatoma cells and activated HSC. We propose a model in which this crosstalk is bidirectional and leads to a permissive microenvironment through ECM remodeling and angiogenesis, along with the alteration of hepatocyte phenotype toward motile cells (Fig. 7). Thus, the data suggest that the dynamic interactions between hepatocytes and activated HSC through soluble mediators play an important role in the progression of hepatocellular carcinoma. Supporting this hypothesis, we further established by integrative genomics that hepatocyte-HSC crosstalk in vitro is clinically relevant and is associated with a poor prognosis in human HCC. More investigations will be needed to fully establish the contribution of each specific genes and/or pathways in HepaRG/LX2 crosstalk. Interestingly, analyzing the crosstalk between LX2 and other hepatoma cells, namely Huh7 and HepG2, showed that the regulation of some important factors such as IL8 was conserved (Supporting Fig. 7). More broadly, the results also suggested that the crosstalk with LX2 may depend on the differentiation status of the cells. Indeed, HepaRG cell line was established from an Edmonson grade I, well differentiated HCC, and showed a unique property to differentiate into mature hepatocytes. Taking this feature into account was particularly relevant given that several studies reported an accumulation of activated HSC in dysplastic nodules at early stages of HCC development.
Figure 7.
Proposed model for molecular crosstalk between hepatocytes and activated HSC in hepatocellular carcinoma.
Unsupervised analysis of genes deregulated in HepaRG in response to the coculture with LX2 highlighted pro-inflammatory cytokines (e.g. IL1B, IL6) and chemokines (e.g. IL8, CCL2) as key orchestrators of the crosstalk between hepatocytes and activated HSC, consistent with previous observations (32, 33). Inflammatory cells and mediators are frequent in the local environment of tumors and several lines of evidences suggested that inflammation contributes to the acquisition of core hallmark capabilities in cancer (1). Notably, epidemiological studies suggested that inflammatory diseases predispose individuals to cancer (34). Thus, an increase expression of IL6 was reported in the serum of most cancer patients. In HCC, IL6 expression was found to correlate with a rapid progression from hepatitis to HCC (35), and an activation of IL6 pathway was reported in HCC with a poor prognosis (36). The invasive capacity of malignant cells has been shown to be increased in presence of IL1B and IL6 (34). Recently, somatic alterations of gp130 and STAT3, which are required for IL6 signaling, have been reported in inflammatory hepatocellular adenomas (37, 38).
Previous studies by Omenetti et al. described a similar bidirectional crosstalk between HSCs and cholangiocytes and provided evidence supporting the importance of the Hedgehog signaling (39). This pathway has been shown also to promote the viability of HSC (40). In the HepaRG/LX2 coculture model, no significant difference was observed in the expression of Hedgehog ligands (e.g. SHH), receptors (e.g. PTCH1), inducible transcription factors (e.g. GLI2) or inhibitors (e.g. HHIP) (Supporting Fig. 8), suggesting that the Hedgehog is not the prominent signaling pathway involved in the crosstalk between mature hepatocytes and activated HSC. Given the fundamental role of this pathway in the crosstalk between immature liver epithelial cells and HSC (39), it is plausible that Hedgehog signaling may play a role in the fate of progenitor HepaRG cells.
Consistent with previous observations using human or rat HSC (32, 33), our results also demonstrate that hepatocyte-HSC crosstalk may greatly impact the local tumor microenvironment, particularly ECM turnover by MMPs and angiogenesis, two main mechanisms promoting tumor growth and metastasis. Previously, we have shown that hepatocyte-HSC interplays induces enhanced extracellular matrix remodeling though MMP2 activation (41). In the present study, in addition to the increased expression of VEGF and MMPs, we report that the enhanced expression of chemoattractant chemokines may also contribute to the establishment of a permissive microenvironment by recruiting other cell types than endothelial cells, notably immune cells. Particularly, we show that mRNA levels for IL8, CCL2, CCL20, and CXCL2 genes were significantly induced in the coculture condition. These soluble mediators are potent chemoattractants for monocytes and lymphocytes. IL8 is also a potent inducer of angiogenesis and metastasis. Interestingly, emerging evidences indicate that CCL2 may also modulate the Th1/Th2 immune responses. Indeed, CCL2 has been shown to induce the expression of IL4, a potent Th2 cytokines, while decreasing the expression of IL12, a major Th1 cytokine (42). Thus, the induction of CCL2, IL6 and IL8 suggests that the crosstalk between hepatocytes and HSC may compromise a Th1 polarization and switch the expression of cytokines toward a Th2 profile. Differential expression of Th1 and Th2 cytokines has been reported in the microenvironment of various types of cancer in vivo. In breast cancer, analysis of gene expression profiles of tumor stroma identified a gene signature which predicted clinical outcome independently of existing standard clinical prognosis factors (43). Importantly, tumor stroma derived from patients with a poor outcome was characterized by an increased expression of genes linked to hypoxia and angiogenesis along with a decreased expression of genes characteristic of a Th1 response (43). Further supporting the hypothesis that a Th2 signature within tumor stroma was predictive of a poor prognosis, we show that the HepaRG/LX2 signature recapitulates the MIM group in HCC which was characterized by the presence of venous metastasis and reported to be significantly associated with an increase in Th2 cytokines and a decrease in Th1 cytokines (31).
Treatment of HCC represents an important clinical challenge. Here, we provide evidences that targeting tumor-stroma crosstalk by epigenetic modulation may represent a promising therapeutic strategy (Fig. 7). Notably, TSA was able to inhibit the coculture-induced migration of HepaRG-hepatocytes as well as angiogenesis and VEGFA expression in LX2. Interestingly, TSA has been also reported to abrogate TGFβ1-induced epithelial-mesenchymal transition (EMT) in hepatocytes and to reverse EMT-induced fibrosis by epigenetic modulation of type I collagen (44). Besides acting on hepatocytes, TSA could also mediate beneficial effects through HCS. Indeed, TSA was shown to strongly suppress the proliferation of rat HSC and to inhibit their conversion into myofibroblasts (45).
In conclusion, the study highlights the central role of the crosstalk between hepatocytes and their microenvironment in HCC and suggests that targeting tumorstroma crosstalk by epigenetic modulation may represent a promising therapeutic strategy.
Supplementary Material
Acknowledgements
The authors thank Dr S.L. Friedman, Mount Sinai School of Medicine, NY, for his generous gift of LX2 cells, Dr M. Ravache and K. Jarnouen, Inserm UMR991, for their help in cell migration assays and Q-RT-PCR, and the microarray core facility team from plateforme génomique santé, IFR140, Rennes.
Financial support
This research was supported by Inserm, Cnrs, University of Rennes 1, Institut National du Cancer and Association pour la Recherche sur le Cancer, France.
Abbreviations
- CM
conditioned medium
- ECM
extracellular matrix
- GSEA
gene set enrichment analysis
- HCC
hepatocellular carcinoma
- HSC
hepatic stellate cell
- HUVEC
human umbilical vein endothelial cells
- IPA
ingenuity pathway analysis
- LSGS
low serum growth supplement
- MMP
matrix metallopeptidase
- TSA
trichostatin A
Footnotes
Author contributions
Conception and design (CC, AC and BC); data acquisition (CC, DG, IG); data analysis and interpretation (CC, AC, BC); writing and review of the manuscript (CC, AC, SST, BC).
Contributor Information
Cédric Coulouarn, Email: cedric.coulouarn@univ-rennes1.fr.
Anne Corlu, Email: anne.corlu@inserm.fr.
Denise Glaise, Email: denise.glaise@inserm.fr.
Isabelle Guenon, Email: isabelle.guenon@univ-rennes1.fr.
Snorri S. Thorgeirsson, Email: snorri_thorgeirsson@nih.gov.
Bruno Clement, Email: bruno.clement@inserm.fr.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmick NA, Chytil A, Plieth D, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 3.Trimboli AJ, Cantemir-Stone CZ, Li F, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson CM, Bissell MJ. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–38. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature reviews. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 8.Theret N, Musso O, Turlin B, et al. Increased extracellular matrix remodeling is associated with tumor progression in human hepatocellular carcinomas. Hepatology. 2001;34:82–88. doi: 10.1053/jhep.2001.25758. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 10.Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin Cancer Biol. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006;43:S145–S150. doi: 10.1002/hep.21063. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 13.Loreal O, Levavasseur F, Fromaget C, Gros D, Guillouzo A, Clement B. Cooperation of Ito cells and hepatocytes in the deposition of an extracellular matrix in vitro. Am J Pathol. 1993;143:538–544. [PMC free article] [PubMed] [Google Scholar]
- 14.Gripon P, Rumin S, Urban S, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–15660. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rumin S, Loreal O, Drenou B, et al. Patterns of intermediate filaments, VLA integrins and HLA antigens in a new human biliary epithelial cell line sensitive to interferon-gamma. J Hepatol. 1997;26:1287–1299. doi: 10.1016/s0168-8278(97)80464-3. [DOI] [PubMed] [Google Scholar]
- 16.Cerec V, Glaise D, Garnier D, et al. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology. 2007;45:957–967. doi: 10.1002/hep.21536. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Hui AY, Albanis E, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavergne E, Hendaoui I, Coulouarn C, et al. Blocking Wnt signaling by SFRP-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active beta-catenin. Oncogene. 2011;30:423–433. doi: 10.1038/onc.2010.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulouarn C, Gomez-Quiroz LE, Lee JS, et al. Oncogene-specific gene expression signatures at preneoplastic stage in mice define distinct mechanisms of hepatocarcinogenesis. Hepatology. 2006;44:1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- 20.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 24.De Minicis S, Seki E, Uchinami H, et al. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 25.Coulouarn C, Lefebvre G, Daveau R, et al. Genome-wide response of the human Hep3B hepatoma cell to proinflammatory cytokines, from transcription to translation. Hepatology. 2005;42:946–955. doi: 10.1002/hep.20848. [DOI] [PubMed] [Google Scholar]
- 26.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 29.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–2064. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 30.Coulouarn C, Factor VM, Conner EA, Thorgeirsson SS. Genomic modeling of tumor onset and progression in a mouse model of aggressive human liver cancer. Carcinogenesis. 2011;32:1434–1440. doi: 10.1093/carcin/bgr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 32.Sancho-Bru P, Juez E, Moreno M, et al. Hepatocarcinoma cells stimulate the growth, migration and expression of pro-angiogenic genes in human hepatic stellate cells. Liver Int. 2010;30:31–41. doi: 10.1111/j.1478-3231.2009.02161.x. [DOI] [PubMed] [Google Scholar]
- 33.Xia Y, Chen R, Song Z, et al. Gene expression profiles during activation of cultured rat hepatic stellate cells by tumoral hepatocytes and fetal bovine serum. J Cancer Res Clin Oncol. 2010;136:309–321. doi: 10.1007/s00432-009-0666-5. [DOI] [PubMed] [Google Scholar]
- 34.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 35.Wong VW, Yu J, Cheng AS, et al. High serum interleukin-6 level predicts future hepatocellular carcinoma development in patients with chronic hepatitis B. Int J Cancer. 2009;124:2766–2770. doi: 10.1002/ijc.24281. [DOI] [PubMed] [Google Scholar]
- 36.Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Pilati C, Amessou M, Bihl MP, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–1366. doi: 10.1084/jem.20110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebouissou S, Amessou M, Couchy G, et al. Frequent in-frame somatic deletions activate gp130 in inflammatory hepatocellular tumours. Nature. 2009;457:200–204. doi: 10.1038/nature07475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omenetti A, Porrello A, Jung Y, et al. Hedgehog signaling regulates epithelial-mesenchymal transition during biliary fibrosis in rodents and humans. J Clin Invest. 2008;118:3331–3342. doi: 10.1172/JCI35875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, Wang Y, Mao H, et al. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol. 2008;48:98–106. doi: 10.1016/j.jhep.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theret N, Musso O, L'Helgoualc'h A, Clement B. Activation of matrix metalloproteinase-2 from hepatic stellate cells requires interactions with hepatocytes. Am J Pathol. 1997;150:51–58. [PMC free article] [PubMed] [Google Scholar]
- 42.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 44.Kaimori A, Potter JJ, Choti M, Ding Z, Mezey E, Koteish AA. Histone deacetylase inhibition suppresses the transforming growth factor beta1-induced epithelial-to-mesenchymal transition in hepatocytes. Hepatology. 2010;52:1033–1045. doi: 10.1002/hep.23765. [DOI] [PubMed] [Google Scholar]
- 45.Niki T, Rombouts K, De Bleser P, et al. A histone deacetylase inhibitor, trichostatin A, suppresses myofibroblastic differentiation of rat hepatic stellate cells in primary culture. Hepatology. 1999;29:858–867. doi: 10.1002/hep.510290328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.