Abstract
The wild ancestor of cultivated barley, Hordeum vulgare subsp. spontaneum (K. Koch) A. & Gr. (H. spontaneum), is a source of wide genetic diversity, including traits that are important for malting quality. A high β-amylase trait was previously identified in H. spontaneum strains from Israel, and transferred into the backcross progeny of a cross with the domesticated barley cv Adorra. We have used Southern-blot analysis and β-amy1 gene characterization to demonstrate that the high β-amylase trait in the backcross line is co-inherited with the β-amy1 gene from the H. spontaneum parent. We have analyzed the β-amy1 gene organization in various domesticated and wild-type barley strains and identified three distinct β-amy1 alleles. Two of these β-amy1 alleles were present in modern barley, one of which was specifically found in good malting barley cultivars. The third allele, linked with high grain β-amylase activity, was found only in a H. spontaneum strain from the Judean foothills in Israel. The sequences of three isolated β-amy1 alleles are compared. The involvement of specific intron III sequences, in particular a 126-bp palindromic insertion, in the allele-dependent expression of β-amylase activity in barley grain is proposed.
The endosperm of barley (Hordeum vulgare L.) and other cereals is a storage organ in which starch and protein accumulate during grain development and are later degraded to provide energy and N2 during germination and seedling growth (for review, see MacGregor and Fincher, 1993). Starch degradation in the cereal grain requires the concerted action of several enzymes (MacGregor, 1987), including limit dextrinase, β-amylase, α-glucosidase (Sun and Henson, 1990), and α-amylases. β-Amylase is a 1,4-α-d-glucan maltohydrolase (EC 3.2.1.2.), which catalyzes the liberation of maltose and limit dextrins from the nonreducing ends of starch (Robyt and Whelan, 1968; Sopanen and Laurière, 1989).
β-Amylase is synthesized during grain development (Kreis et al., 1987) and is one of the major proteins found in the starchy endosperm (Hejgaard and Boisen, 1980). In dry barley grains it is found in two forms: a free, active form and a bound, less-active form, the latter accounting for about 75% of the total β-amylase. Both forms may be aggregated or disulfide bonded via a Cys residue near the C terminus to other grain proteins such as protein Z (Hejgaard, 1978). The free form of β-amylase is easily extractable in saline solutions and comprises many forms separable by IEF (Shewry et al., 1988). The release and activation of β-amylase during germination is accompanied by the accumulation of additional β-amylase isoforms (Sopanen and Laurière, 1989; Guerin et al., 1992), which could be caused by a progressive endoproteolytic removal of C-terminal peptides (Lundgard and Svensson, 1987).
Synthesis of β-amylase during barley grain development is regulated by nutritional N2 (Giese and Hopp, 1984), and high levels of β-amylase are generally correlated with increased grain protein content (Santos and Riis, 1996). Modern plant breeding has reduced the genetic variability among domesticated barley cultivars (Thompson et al., 1990; Forster et al., 1991). This in turn limits the potential for selecting cultivars with high β-amylase levels independent of grain protein content, which is an important quality parameter for malt barley. However, a large genetic diversity is found in H. vulgare subsp. spontaneum (K. Koch) A. & Gr. (abbreviated to H. spontaneum), the wild ancestor of cultivated barley (Wiberg, 1974; Zhang et al., 1993; Saghai Maroof et al., 1995). H. spontaneum strains from Israel and Jordan show a wide variation in β-amylase, β-glucanase, and other hydrolase activities (Ahokas and Naskali, 1990a, 1990b). We have previously shown that a high β-amylase trait in selected H. spontaneum strains is inherited in the backcrossed progeny of H. spontaneum and domesticated barley (Ahokas and Erkkilä, 1992), which may be linked to inheritance of a β-amy allele of H. spontaneum.
To study the biochemical and molecular mechanisms underlying the different levels of β-amylase activity in domesticated barley and its wild ancestors, we have isolated and characterized the β-amylase cDNAs and genes from the H. spontaneum × domesticated barley backcross and the respective parental lines generated by Ahokas and Erkkilä (1992). We report here that the observed differences in β-amylase activity in the grains are correlated with sequence differences in the β-amy1 gene, which involve the insertion/deletion of a palindromic 126-bp sequence and a 39-bp sequence in intron III, as well as two amino acid substitutions in the ORF. We have exploited the sequence variation in intron III to develop a PCR method to explore the genetic diversity of the β-amy1 locus among domesticated and wild barley and to develop a screening tool for breeding lines with high β-amylase activity.
MATERIALS AND METHODS
Barley (Hordeum vulgare) plants were grown either in a greenhouse or in fields in the southern part of Finland. Grains of various barley cultivars were provided by the Agricultural Research Centre of Finland (Jokioinen) and cv Haruna Nijo was provided by Sapporo Brewing, Ltd. (Yaizu, Japan). The grain material analyzed included BC-F6 lines derived from a single backcross of Hordeum spontaneum × Adorra (accession no. PI 296897) from the U.S. Department of Agriculture (Beltsville, MD), followed by six generations of selection for the domesticated growth habit, and the two parents (cv Adorra and H. spontaneum). An additional line, denoted strain 86-H2-64, was selected in the sixth generation from an independent but similar single backcross. The accession PI 296897 originated from the Judean Foothills near Jerusalem, Israel. Other wild barleys used for testing β-amy1 allele diversity were H. spontaneum strains collected from the Israeli localities of Talpiyyot, Mehola, Atlit, Negev, and from Cyprus, and Hordeum murinum subsp. leporinum from Corsica (France).
Assays of β-Amylase Activity
β-Amylase activity and grain protein content were determined in grain samples (2 g) milled in an A/10 analytical mill (Janke and Kunkel, Staufen, Germany). β-Amylase activity was assayed in duplicate samples of 0.5 g of fresh flour or 0.5 g of flour stored at −80°C using the Beta-Amylase Kit from Megazyme (Bray, Ireland), according to the method of Santoz and Riis (1996). One unit of β-amylase activity is defined as the amount of enzyme required to release 1 μmol of p-nitrophenol from p-nitrophenyl-α-d-maltopentaose in 1 min in the presence of excess α-glucosidase under assay conditions defined in the kit. Total soluble protein in flour samples was determined according to the method of Bradford (1976). β-Amylase activity values are expressed per milligram of protein in the flour.
RNA and DNA Protocols
Total RNA was extracted from endosperm tissue using aurintricarboxylic acid as the RNase inhibitor (Leah et al., 1991) and further purified using the RNeasy Plant Total RNA kit (Qiagen, Chatsworth, CA).
DNA was extracted from 14-d-old leaves according to the method of Maniatis et al. (1982), or from 7-d-old leaves using FastPrep (BIO 101, Vista, CA) for PCR reactions. DNA sequencing was performed (model 373A sequencer, Applied Biosystems). The reaction mixtures contained 0.5 μg of DNA, 10 pmol of primer (20 bp), and the Ampli Cycle Sequencing Kit (Perkin-Elmer). Sequences were analyzed with the Micro Genie Sequence software package (Beckman).
Southern blotting and conditions for hybridizations were performed as described previously (Leah et al., 1991). Blots were hybridized with the 1750-bp β-amylase cDNA insert of pcβC51 (Kreis et al., 1987) labeled with [32P]dCTP using the Megaprime DNA-labeling system (Amersham).
Allele-Specific β-Amy1 cDNA Cloning
cDNA was synthesized in two parts by RNA-PCR according to the method of Dyanov and Dzitoeva (1995) with Moloney murine leukemia virus reverse transcriptase (Boehringer Mannheim) from total RNA isolated from endosperm tissue (approximately 26 d after anthesis) of the different barley strains. The primers were based on the sequence of the β-amylase cDNA (Kreis et al., 1987; Yoshigi et al., 1994) or the cv Haruna Nijo β-amy gene (Yoshigi et al., 1995).
The antisense primer for synthesis of the 5′ part of the transcript was 5′-AGCAGATGAATTCTCCGATGCCTGGGAACGACC-3′, having an EcoRI site, together with the sense primer 5′-TAGCCAGGATCCACAATGGAGGTGAACGTGAAAGGC-3′, having a BamHI site. The antisense primer for the 3′ part of the transcript was 5′-TAAACTCGAGGTTCCATTACATGGTGGCAGGGA-3′, having an XhoI site, together with the sense primer 5′-CCCAGGCATCGGAGAATTCATCTGCTATGA-3′, having an EcoRI site. The resulting PCR fragments were restricted with EcoRI, BamHI, and XhoI, purified, and ligated into pBluescript SK− vector (Stratagene) to produce full-length cDNA clones.
PCR Cloning of β-Amy1 Alleles
The promoter and ORF regions of the β-amy1 gene were amplified from genomic DNA of different barley strains using the Expand High-Fidelity PCR system (Boehringer Mannheim), according to the manufacturer's instructions. The PCR primers, based on the sequence of the cv Haruna Nijo β-amy gene (Yoshigi et al., 1995; EMBL accession nos. D63574 and D49999), were as follows: 5′-ATTGGATCCATGGAGGTGAACGTGAAAGGCAACTATGTC-3′ and 5′-AAAGGATCCATTACATGGTGGCAGGGAGCTCCCCACCCA-3′, both with a 5′ BamHI restriction site, for amplification of the gene ORF; and 5′-ACGCGTCGACACATCATCTTGAGAACGTCTTCTC-3′ and 5′-ACGCGTCGACTCGAACCTGTTGTTCACGCTCAC-3′, both with a 5′ SalI restriction site, for amplification of the corresponding promoter region.
The PCR reactions were performed as follows: DNA was denatured for 5 min at 95°C, followed by 30 cycles of 0.5 min at 94°C, 1 min at 61°C, 3 min at 72°C, the last 20 cycles with a 20-s increase in extension time in each cycle, ending with a final polymerization step of 10 min at 72°C. The PCR fragments were cloned into pBluescript SK− vector (Stratagene).
β-amy1 intron III-specific sequences were amplified by PCR from genomic DNA of different barley lines with the following primer pair: 5′-GATGGTCGTTCCCAGGCATC-3′ and 5′-AGGGAACCGCACGTGTGGGGTCAATGA-3′. The reaction mixture contained 0.5 to 0.7 μg of template DNA, 10 pmol of each PCR primer, 0.2 mmol/L deoxyribonucleotide triphosphate, 1.5 mmol/L MgCl2, 50 mmol/L KCl, 10 mmol/L Tris-HCl (pH 9.0), 0.1% Triton, and 1 unit of Taq polymerase (Promega). The mixture was reacted for 2 min at 95°C, then for 1 min at 94°C, for 0.5 min at 60°C, and for 1 min at 72°C for 25 cycles, followed by 10 min at 72°C. The PCR fragments were detected in 5% polyacrylamide gels.
RESULTS
Identification of Three Distinct β-Amy1 Alleles
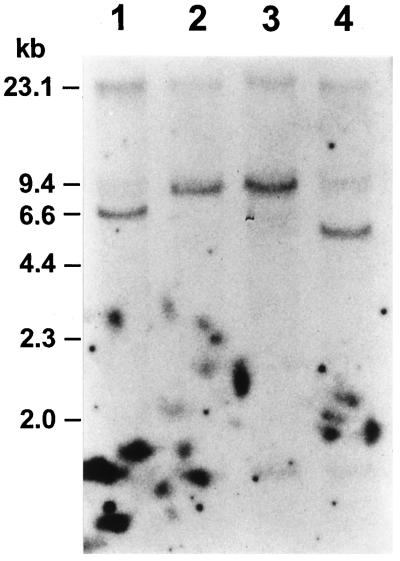
The organization of the β-amy1 structural gene in different barley strains was analyzed by Southern-blot analysis. Genomic DNA isolated from cv Haruna Nijo, cv Adorra, H. spontaneum PI 296897, and line 86-H2-64 was restricted with HindIII, NcoI, SalI, and SpeI and hybridized with the β-amy1 pcβC51 cDNA (Kreis et al., 1987). The size of the hybridizing fragments for each restriction digest was not identical in the four strains. For example, in a HindIII restriction digest (Fig. 1) the hybridizing fragments were approximately 7.0 kb in cv Adorra (lane 1), approximately 9.0 kb in line 86-H2-64 and H. spontaneum PI 296897 (lanes 2 and 3), and approximately 6.0 kb in cv Haruna Nijo (lane 4). An additional hybridizing HindIII fragment of an approximately 1.6-kb fragment was detected in all four barley strains. Most of the hybridizing fragment sizes in cv Haruna Nijo DNA, including the 1.6-kb HindIII fragment, were consistent with the sequence of this β-amy1 gene (Yoshigi et al., 1995).
Figure 1.
Southern-blot analysis of the β-amy1 structural gene in different barley cultivars. Barley genomic DNA was digested with HindIII, fractionated by agarose-gel electrophoresis, Southern blotted, and hybridized with β-amy1 pcβC51 cDNA probe. The genomic DNAs were from cv Adorra (lane 1), line 86-H2-64 (lane 2), H. spontaneum PI 296897 (lane 3), and cv Haruna Nijo (lane 4).
The β-amy1 pcβC51 cDNA hybridizes to two loci in the barley genome, designated Bmy1 and Bmy2, located on chromosomes 4 and 2, respectively (Kreis et al., 1988). The authors assigned a strongly hybridizing HindIII fragment of 6.9 kb in cv Betzes to Bmy1, whereas a weaker hybridizing 4.7-kb fragment was assigned to Bmy2. Under the hybridization conditions used here, hybridizing restriction fragments that might correspond to the Bmy2 locus were not detected. In agreement with Kreis et al. (1988), restriction fragment length polymorphism was observed at the Bmy1 locus, indicating the presence of at least three distinct alleles in cv Haruna Nijo, cv Adorra, and H. spontaneum PI 296897. Furthermore, the strain 86-H2-64, derived from a H. spontaneum PI 296897 × Adorra cross and identified as having a high grain β-amylase activity, is seen to have inherited the H. spontaneum PI 296897-like β-amy1 allele.
Isolation and Characterization of the β-Amy1 Alleles and Their Transcripts
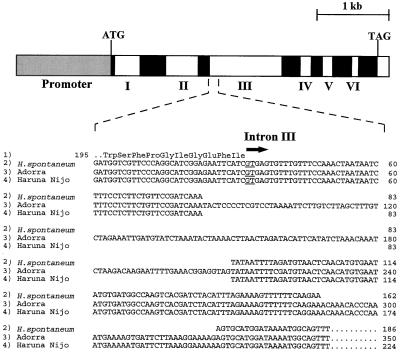
To identify the molecular mechanisms underlying the enhanced β-amylase activity in grain of H. spontaneum PI 296897 and its cv Adorra backcross progeny line 86-H2-64, we isolated and sequenced the β-amy1 genes and transcripts from line 86-H2-64 and its parents. All three β-amy1 genes were PCR amplified from genomic DNA using primers based on the sequence of the β-amy1 gene from cv Haruna Nijo, which is expressed in developing endosperm (Yoshigi et al., 1994, 1995). Single β-amy1 amplification products were generated from each of the three barley strains and their sequences were determined. All four β-amy1 sequences were approximately 5 kb in length, comprising an ORF interrupted by six introns, as shown schematically in Figure 2, top. The sequences of the β-amy1 genes in line 86-H2-64 and H. spontaneum PI 296897 showed complete identity. The H. spontaneum PI 296897 β-amy1 allele showed high homology to the β-amy1 allele of cv Haruna Nijo. Apart from a few single-base substitutions, sequence differences in cv Haruna Nijo β-amy1 were restricted to the 5′-proximal end of intron III and a 25-bp deletion positioned 761 bp upstream of the TATAA box. The cv Adorra β-amy1 allele contained this upstream 25-bp insertion, and in addition to a number of single-base substitutions, showed significant sequence divergence from H. spontaneum PI 296897 β-amy1, confined to intron III.
Figure 2.
The β-amy1 gene organization. Top, Schematic presentation of the β-amy structural gene. The β-amy1 ORF is composed of seven exons (black boxes) interrupted by six introns (numerals I–VI) located within a DNA fragment of approximately 5 kb. ATG, TAG, and gray box indicate the translational start and stop codons and the approximately 1.3-kb promoter, respectively. Bottom, Sequence alignment of the 5′-proximal ends of intron III from β-amy1 alleles of H. spontaneum PI 296897 (2), cv Adorra (3), and cv Haruna Nijo (4). Line 1 shows the amino acid sequence (residues 195–205) encoded by the DNA sequence upstream of intron III. The sequences of intron III from the β-amy1 allele of line 86-H2-64 (not shown) and H. spontaneum PI 296897 (2) are identical. The β-amy1 genomic sequences are in the EMBL-GenBank database under accession numbers AF061204 (H. spontaneum PI 296897), AF061203 (cv Adorra), and D49999 (cv Haruna Nijo).
The cDNAs of the corresponding β-amy1 transcripts were cloned by reverse-transcriptase RNA-PCR. The sequences of the β-amy1 cDNAs of H. spontaneum PI 296897, line 86-H2-64, and cv Haruna Nijo were identical, apart from two silent nucleotide substitutions in the latter. Four of the single-base substitutions found in the cv Adorra β-amy1 allele were located in the ORF of the transcript. Two of these led to amino acid substitutions, namely Ala-233→Val and Ser-347→Leu, which are also seen in the cv Hiproly β-amy1 allele (Kreis et al., 1987).
The major sequence differences between the three β-amy1 alleles were limited to the first 320 bp at the 5′ end of intron III (Fig. 2, bottom). Compared with the published sequence of cv Haruna Nijo, cv Adorra had a 126-bp insertion (Fig. 2, line 4 versus line 3), and H. spontaneum PI 296897 (and line 86-H2-64; data not shown) had a 39-bp deletion (Fig. 2, line 4 versus line 2).
Inheritance of the H. spontaneum PI 296897-Like β-Amy1 Allele Is Linked to Enhanced β-Amylase Activity
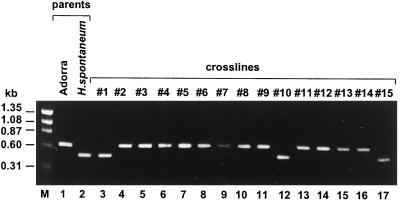
We have shown that the high-β-amylase-activity trait in line 86-H2-64, an offspring of an H. spontaneum and domesticated barley cross (Ahokas and Erkkilä, 1992), is linked to the inheritance of the β-amy1 allele from H. spontaneum. To gain further evidence for a close correlation between the H. spontaneum β-amy1 allele and high β-amylase activity levels, we have studied a population of 15 individual BC-F6 lines from a H. spontaneum PI 296897 × Adorra backcross grown under identical conditions. The two parental β-amy1 alleles inherited in the individual BC-F6 lines were distinguished by the allele-specific sequence of the β-amy1 intron III (Fig. 2). This was accomplished by the design and use of PCR primers to amplify a region of the β-amy1 intron III, so that the cv Adorra allele was identified by a 643-bp PCR fragment, whereas the H. spontaneum PI 296897 allele was identified by a 477-bp PCR fragment (Fig. 3). Three of the offspring had inherited the H. spontaneum PI 296897 allele (Fig. 3, lane 2 versus lanes 3, 12, and 17), whereas the remaining 12 offspring inherited the cv Adorra allele (Fig. 3, lane 1 versus lanes 4–11 and 13–16).
Figure 3.
Identification of the β-amy1 allele in the offspring of the H. spontaneum × Adorra cross. Genomic DNA sequences from the parents cv Adorra (lane 1) and H. spontaneum PI 296897 (lane 2), and from the 15 individual lines #1 to #15 (lanes 3–17) were PCR amplified for β-amy1 intron III-specific sequences. The PCR products were size fractionated by acrylamide-gel electrophoresis. Lane M, Molecular size marker.
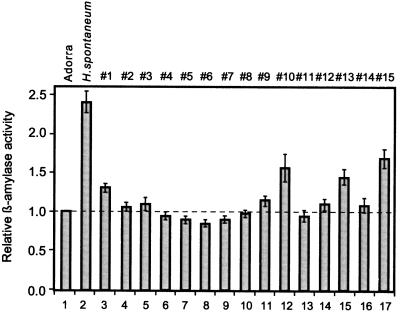
The total β-amylase activity in grain from the 15 BC-F6 lines was also determined and compared with that of the parent lines (Fig. 4). The relative β-amylase activity of 11 BC-F6 lines was lower or similar to that of the parent cv Adorra (Fig. 4, bar 1 versus bars 3–10, 12, 13, and 15), whereas the relative β-amylase activity of 4 BC-F6 lines were 27 to 61% higher (Fig. 4, bar 1 versus bars 2, 11, 14, and 16), although not to the level of H. spontaneum. Three of these four BC-F6 lines had inherited the H. spontaneum PI 296897 β-amy1 allele (Fig. 3, BC-F6 lines #1, #10, and #15). According to the Mann-Whitney U test (Siegel, 1956) (where U = 2; P < 0.01), the levels of β-amylase activity measured in lines carrying the cv Adorra β-amy1 allele or the H. spontaneum PI 296897 β-amy1 allele are statistically different. Hence, the data provide supporting evidence that inheritance of the H. spontaneum β-amy1 allele is linked to a trait for high β amylase activity in grain.
Figure 4.
β-Amylase activity in offspring of the H. spontaneum × Adorra cross. Histogram showing the relative β-amylase activity of the H. spontaneum parent strain (bar 2) and 15 individual lines #1 to #15 (bars 3–17) from the H. spontaneum × Adorra cross normalized to the β-amylase activity of the parent cv Adorra (bar 1), calculated from the mean of six independent determinations. The error bars in the histogram represent the se values. The relative β-amylase activity of cv Adorra of 1.0 is equivalent to a β-amylase activity of approximately 15 units per 1 mg of protein.
The H. spontaneum PI 296897-Like β-Amy1 Allele Is Not Found in Modern Barley Cultivars
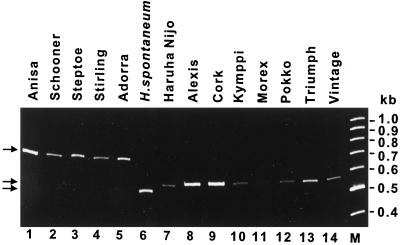
To study the genetic diversity of the β-amy1 locus in domesticated barley, we examined several cultivars and lines with different levels of β-amylase activity and known malting characteristics. The allele-specific region of the β-amy1 intron III, amplified by specific PCR primers from genomic DNA, was used to identify the β-amy1 allele in relation to the characterized cv Adorra, H. spontaneum PI 296897, and cv Haruna Nijo β-amy1 alleles (Fig. 5). Using this approach the cv Adorra-like 643-bp PCR fragment was found in cvs Anisa, Schooner, Steptoe, and Stirling (Fig. 5, lanes 1–4 versus lane 5), whereas the H. spontaneum PI 296897-like 477-bp PCR fragment (Fig. 5, lane 6) was not seen in modern barley. The cv Haruna Nijo-like 516-bp PCR fragment was found in cvs Alexis, Cork, Kymppi, Morex, Pokko, Triumph, and Vintage (Fig. 5, lanes 8–14 versus lane 7).
Figure 5.
Distribution of different β-amy1 alleles in domesticated barley. The β-amy1 allele-specific sequence of intron III was PCR amplified from genomic DNA of various barley cultivars. The fragments were size fractionated by acrylamide-gel electrophoresis. Lane M, Molecular size marker.
We have not determined the β-amylase activity of these domesticated barley lines, since our grain samples were harvested from different locations and years, and their β-amylase activity would thus reflect growth conditions as well as genotype. However, the selected good-malting-quality cultivars (Alexis, Cork, Kymppi, Morex, Pokko, Triumph, and Vintage), known to possess high-β-amylase activity, inherited the cv Haruna Nijo-like β-amy1 allele (Fig. 5), whereas the poorer-malting-quality cultivars (Anisa, Schooner, Steptoe, Stirling, Clipper, Chebec, Bomi, Carlsberg II, Golden Promise, and Bonus) inherited the cv Adorra-like β-amy1 allele (Fig. 5 and data not shown). None of the barley cultivars tested inherited the H. spontaneum-like β-amy1 allele.
Several wild Hordeum spp. collected from different Mediterranean localities were also examined for genetic diversity at the β-amy1 locus, detected as allele-specific sequence differences of the β-amy1 intron III. PCR amplification of the intron III region from genomic DNA samples showed that the wild Hordeum spp. from Cyprus and Corsica and from the areas of Talpiyyot and Mehola had the cv Haruna Nijo-like β-amy1 allele, whereas the H. spontaneum strain from the Negev had the cv Adorra-like β-amy1 allele (data not shown). Both alleles were found in the H. spontaneum strain from Atlit, whereas the H. spontaneum PI 296897-like β-amy1 allele was found only in the H. spontaneum strain from the Judean foothills (data not shown).
DISCUSSION
We are interested in understanding the genetic and molecular mechanisms underlying the variation in β-amylase activity in the grain of modern and wild barley. As a part of this work, we have previously examined 242 accession lines of the wild barley H. spontaneum and found a wide variation in β-amylase activities, which were significantly correlated with the annual rainfall pattern in their territory of origin (Ahokas and Naskali, 1990b). The high-β-amylase-activity trait found in H. spontaneum PI 296897 has been introduced into the domesticated barley cv Adorra by crossing (Ahokas and Erkkilä, 1992). Preliminary evidence suggested that the backcrossed derivative showing increased enzyme activity (86-H2-64) had inherited a novel β-amylase allele from H. spontaneum PI 296897.
To gain a closer insight into the β-amy1 alleles present in the barley gene pool, we performed Southern-blot analysis of the β-amy1 genes from H. spontaneum PI 296897, cv Adorra, and their backcross derivative (86-H2-64), together with the malt barley cv Haruna Nijo. The existence of three distinct β-amy1 alleles present in H. spontaneum PI 296897, cv Adorra, and cv Haruna Nijo was indicated, whereas the backcross was shown to have inherited the H. spontaneum PI 296897 allele.
We have cloned and sequenced the β-amy1 alleles from H. spontaneum PI 296897, cv Adorra, and line 86-H2-64 and confirmed that the high-β-amylase trait in the backcross 86-H2-64 is co-inherited with the β-amy1 allele from H. spontaneum PI 296897. The sequences of the H. spontaneum PI 296897 and cv Adorra β-amy1 genes and their respective transcripts have been compared with each other and with those of cv Haruna Nijo with the aim of identifying the molecular basis for the different β-amylase activities found in their barley grains. The promoter regions of the three alleles, extending approximately 1.3 kb upstream of the ORF, were very similar apart from a 25-bp deletion located at the 5′ end of the cv Haruna Nijo promoter fragment. This promoter sequence, encompassing all essential elements required for tissue-specific expression of the β-amy1 gene (Kihara et al.. 1997), cannot explain the observed differences in β-amylase activity between cv Adorra and the backcross 86-H2-64. Sequence comparisons between the β-amy1 ORFs of cvs Hiproly and Haruna Nijo have previously revealed three amino acid substitutions in the deduced sequence (Yoshigi et al., 1994). The deduced amino acid sequence of the H. spontaneum PI 296897 ORF shows complete identity to that of cv Haruna Nijo, whereas cv Adorra ORF shows two substitutions, Ala-233→Val and Ser-347→Leu, both of which are found in cv Hiproly. Although Val-233 is a conservative substitution, lying in a loop at the protein surface remote from the enzyme catalytic site, the nonconservative Leu-347 substitution may influence β-amylase activity, since it involves a polar/nonpolar substitution near Glu-345, which lies near the catalytic site (Mikami et al., 1993; Totsuka et al., 1994).
The three β-amy1 alleles show significant differences, however, in the length and sequence of intron III. The cv Adorra β-amy1 allele has a unique 126-bp insertion, whereas the H. spontaneum PI 296897 β-amy1 allele has a 39-bp deletion compared with the β-amy1 intron III of cv Haruna Nijo. To investigate the linkage between the high-β-amylase-activity trait and the allelic nature of intron III of the β-amy1 gene, we have screened a population of BC-F6 lines derived from a single H. spontaneum PI 296897 × Adorra backcross. After five generations of strong selection for domesticated morphological features, the remaining population was largely composed of lines with β-amylase activity levels similar to those of cv Adorra. However, three of the four lines exhibiting the high-β-amylase-activity trait had the H. spontaneum PI 296897 intron III, which, according to the Mann-Whitney U test, establishes genetic linkage. The high-β-amylase-activity trait could also be attributed to unknown quality trait loci located close to the β-amy1 locus. Unlinked quality trait loci regulating β-amylase activity may have contributed to the enhanced β-amylase activity in one line (Hayes et., 1993; Oziel et al., 1996).
The molecular mechanism by which the 126- or 39-bp intron III sequence might influence the qualitative or quantitative expression of β-amy1 can only be speculated. Recent reports provide evidence for transcriptional control elements located within the gene-transcription unit, in addition to the promoter elements upstream of the transcriptional start site (Bolle et al., 1996; Flieger et al., 1994; Maas et al., 1997). Thus, elements in the 126-bp sequence in intron III may bind to a factor(s) that modulates β-amy1 transcription efficiency or influences posttranscriptional events such as mRNA maturation and stability. The 126-bp intron III sequence has not previously been found in barley, but shares >85% identity with a part of the chloroplastic phosphoglycerate kinase promoter (−861 to −735 upstream of the transcriptional start) from wheat (Jones et al., 1995). The 126-bp sequence is composed of a long, inverted repeat, which can be modeled into a stem/loop structure with nucleotides 57 to 72 forming an unpaired terminal loop and 14 mismatched nucleotides in the stem. Further studies will be aimed at providing direct evidence for the role of sequence elements within intron III in the regulation of β-amy1 at the level of transcription efficiency, mRNA maturation, and/or gene product accumulation.
In the brewing industry DP is an essential parameter of malt quality, since it is a measure of the capacity of the malt to degrade starch into fermentable sugars. Although DP represents the combined activity of all of the amylolytic enzymes, it is primarily determined by β-amylase activity (Santos and Riis, 1996). The PCR analytical technique, developed to distinguish the three β-amy1 alleles, could provide a valuable breeding tool if it could be used to predict DP in barley at the seedling stage. As a first step in validating this method, a range of poor- and good-malt-quality barley lines were screened for intron III structure by PCR, as a means of predicting their β-amy1 allele identity. Based on their known malting quality and published DP measurements (Santos and Riis, 1996), good-quality malting barleys were found to have the cv Haruna Nijo-like β-amy1 allele, whereas nonmalting barleys had the cv Adorra-like β-amy1 allele.
The identification of three β-amy1 alleles is in agreement with the number of barley β-amy1 alleles distinguished by IEF. Forster et al. (1991) have identified two alleles at the β-amy1 locus in spring and winter barley genotypes, the β-Amy1a and β-Amy1b alleles. Analysis of additional cultivars will reveal whether β-Amy1a and β-Amy1b alleles correspond to the cv Adorra-like and cv Haruna Nijo-like β-Amy1 alleles. The third allele was only found in a H. spontaneum strain originating from Judea and in breeding strains thereof. Chalmers et al. (1992) have analyzed the different β-amylase phenotypes present in the H. spontaneum populations of Israel. They showed that individual phenotypes were found in geographical regions where specific environmental regimes predominate. Thus, the G phenotype is restricted to the Negev Desert and Dead Sea regions, whereas the A phenotype is found in northern parts of Israel (Chalmers et al., 1992). We have shown by the allele-specific PCR approach that H. spontaneum strains collected from the geographic areas associated with the G phenotype contain the cv Adorra-like β-amy1 allele, whereas strains collected from the localities of the A phenotype contain the cv Haruna Nijo-like β-amy1 allele. We found both β-amy1 alleles in strains along the Mediterranean coast, where Chalmers et al. (1992) found a heterozygous population, AB.
The H. spontaneum PI 296897-like β-amy1 allele is not widespread in wild barley, although it is linked to high β-amylase activity in the grain. Use of this β-amy1 allele in the modern barley-breeding program may contribute to increased levels of grain β-amylase activity in malting barley.
ACKNOWLEDGMENTS
We thank Dr. Jacques Rouster, Marja-Riitta Mäkelä, Ella Meiling, Maj-Britt Rask, and Suksawad Vongvisuttikun for genomic DNA samples and technical assistance, and Nina Rasmussen and Ann-Sofi Steinholz for help with the figures. We thank Dr. Finn Lok, Dr. Mikael Blom Sørensen, and Dr. David J. Simpson for helpful discussions and reading the manuscript. We also thank Dr. Yukio Okada (Sapporo Brewing, Ltd.) for cv Haruna Nijo grains; Dr. Barbro Jende-Strid (Carlsberg Research Laboratory) for the pro-ant barley line (Anisa); Reino Aikasalo (Boreal Plant Breeding, Jokioinen, Finland) and Outi Manninen and Timo Turpeinen MSc (Agricultural Research Centre) for providing samples of grains and DNA from selected Finnish cultivars, breeding lines, and H. spontaneum strains.
Abbreviations:
- DP
diastatic power
- ORF
open reading frame
Footnotes
This research was supported by the Nordisk Forskerutdanningsakademi (grant no. 95.30.075-O).
LITERATURE CITED
- Ahokas H, Erkkilä MJ. Barley β-amylase and β-glucanase activities at germination in vulgare-type lines from backcrosses of wild, spontaneum strains with cv. Adorra. Agric Sci Finl. 1992;1:339–350. [Google Scholar]
- Ahokas H, Naskali L. Variation of α-amylase, β-amylase, β-glucanase, pullulanase, proteinase and chitinase activity in germinated samples of the wild progenitor of barley. J Inst Brew. 1990a;96:27–31. [Google Scholar]
- Ahokas H, Naskali L. Geographic variation of α-amylase, β-amylase, β-glucanase, pullulanase and chitinase activity in germinating Hordeum spontaneum barley from Israel and Jordan. Genetica. 1990b;82:73–78. [Google Scholar]
- Bolle C, Herrmann RG, Oelmüller R. Intron sequences are involved in the plastid- and light-dependent expression of the spinach PsaD gene. Plant J. 1996;10:919–924. doi: 10.1046/j.1365-313x.1996.10050919.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chalmers KJ, Waugh R, Watters J, Forster BP, Nevo E, Abbott RJ, Powell W. Grain isozyme and ribosomal DNA variability in Hordeum spontaneum populations from Israel. Theor Appl Genet. 1992;84:313–322. doi: 10.1007/BF00229489. [DOI] [PubMed] [Google Scholar]
- Dyanov HM, Dzitoeva SG. Isolation of DNA-free RNA from a very small number of cells. BioTechniques. 1995;18:559–564. [PubMed] [Google Scholar]
- Flieger K, Wicke A, Herrmann RG, Oelmüller R. Promoter and leader sequences of the spinach PsaD and PsaF genes direct an opposite light response in tobacco cotyledons: PsaD sequences downstream of the ATG codon are required for a positive light response. Plant J. 1994;6:359–368. doi: 10.1046/j.1365-313x.1994.06030359.x. [DOI] [PubMed] [Google Scholar]
- Forster BP, Thompson DM, Watters J, Powell W. Water-soluble proteins of mature barley endosperm: genetic control, polymorphism, and linkage with β-amylase and spring/winter habit. Theor Appl Genet. 1991;81:787–792. doi: 10.1007/BF00224991. [DOI] [PubMed] [Google Scholar]
- Giese H, Hopp HE. Influence of nitrogen nutrition on the amount of hordein, protein Z and β-amylase messenger RNA in developing endosperms of barley. Carlsberg Res Commun. 1984;49:365–383. [Google Scholar]
- Guerin JR, Lance RCM, Wallace W. Release and activation of barley β-amylase by malt endopeptidases. J Cereal Sci. 1992;15:5–14. [Google Scholar]
- Hayes PM, Liu BH, Knapp SJ, Chen F, Jones B, Blake T, Franckowiak J, Rasmusson D, Sorrels M, Ullrich SE and others. Quantitative trait locus effects and environmental interaction in a sample of North American barley germ plasm. Theor Appl Genet. 1993;87:392–401. doi: 10.1007/BF01184929. [DOI] [PubMed] [Google Scholar]
- Hejgaard J. ‘Free’ and ‘bound’ β-amylases during malting of barley: characterization by two-dimensional immunoelectrophoresis. J Inst Brew. 1978;84:43–46. [Google Scholar]
- Hejgaard J, Boisen S. High-lysine proteins in Hiproly barley breeding: identification, nutritional significance and new screening methods. Hereditas. 1980;93:311–320. [Google Scholar]
- Jones PG, Raines CA, Lloyd JC. Nucleotide sequence of a wheat chloroplastic phosphoglycerate kinase gene. Plant Physiol. 1995;107:1483–1484. doi: 10.1104/pp.107.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M, Okada Y, Kuroda H, Saeki K, Yoshigi N, Ito K (1997) Generation of fertile transgenic barley synthesizing thermostable β-amylase. In European Brewing Convention Proceedings 26th Congress Maastricht. Oxford University Press, Oxford, UK, pp 83–90
- Kreis M, Williamson M, Buxton B, Pywell J, Hejgaard J, Svendsen I. Primary structure and differential expression of β-amylase in normal and mutant barleys. Eur J Biochem. 1987;169:517–525. doi: 10.1111/j.1432-1033.1987.tb13640.x. [DOI] [PubMed] [Google Scholar]
- Kreis M, Williamson MS, Shewry PR, Sharp P, Gale M. Identification of a second locus encoding β-amylase on chromosome 2 of barley. Genet Res. 1988;51:13–16. [Google Scholar]
- Leah R, Tommerup H, Svendsen I, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem. 1991;266:1564–1573. [PubMed] [Google Scholar]
- Lundgard R, Svensson B. The four major forms of barley β-amylase: purification, characterization and structural relationship. Carlsberg Res Commun. 1987;52:313–326. [Google Scholar]
- Maas C, Simpson CG, Eckes P, Schickler H, Brown JWS, Reiss B, Salchert K, Chet I, Schell J, Reichel C. Expression of intron modified NTP II genes in monocotyledonous and dicotyledonous plant cells. Mol Breeding. 1997;3:15–28. [Google Scholar]
- MacGregor AW. α-Amylase, limit dextrinase, and α-glucosidase enzymes in barley and malt. CRC Crit Rev Biotechnol. 1987;5:117–128. doi: 10.3109/07388558709086972. [DOI] [PubMed] [Google Scholar]
- MacGregor AW, Fincher GB (1993) Carbohydrates of the barley grain. In AW MacGregor, RS Bhatty, eds, Barley: Chemistry and Technology. American Association of Cereal Chemists, St. Paul, MN, pp 73–130
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Mikami B, Hehre EJ, Sato M, Katsube Y, Hirose M, Morita Y, Sacchettini JC. The 2.0-Å resolution structure of soybean β-amylase complexed with α-cyclodextrin. Biochemistry. 1993;32:6836–6845. doi: 10.1021/bi00078a006. [DOI] [PubMed] [Google Scholar]
- Oziel A, Hayes PM, Chen FQ, Jones B. Application of quantitative trait locus mapping to the development of winter-habit malting barley. Plant Breeding. 1996;115:43–51. [Google Scholar]
- Robyt JF, Whelan WJ. The β-amylases. In: Radley J, editor. Starch and Its Derivatives. London: Chapman & Hall; 1968. pp. 430–476. [Google Scholar]
- Saghai Maroof MA, Zhag Q, Biyashev R. Comparison of restriction fragment length polymorphisms in wild and cultivated barley. Genome. 1995;38:298–306. doi: 10.1139/g95-037. [DOI] [PubMed] [Google Scholar]
- Santos MMM, Riis P. Optimized McCleary method for measurement of total β-amylase in barley and its applicability. J Inst Brew. 1996;102:271–275. [Google Scholar]
- Shewry PR, Parmar S, Buxton B, Gale MD, Liu CJ, Hejgaard J, Kreis M. Multiple molecular forms of β-amylase in seeds and vegetative tissues of barley. Planta. 1988;176:127–134. doi: 10.1007/BF00392488. [DOI] [PubMed] [Google Scholar]
- Siegel S. Nonparametric Statistics for the Behavioural Sciences. New York: McGraw-Hill; 1956. [Google Scholar]
- Sopanen T, Laurière C. Release and activity of bound β-amylase in a germinating barley grain. Plant Physiol. 1989;89:244–249. doi: 10.1104/pp.89.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Henson CA. Degradation of native starch granules by barley α-glucosidases. Plant Physiol. 1990;94:320–327. doi: 10.1104/pp.94.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Powell W, Forster BP. Use of isoelectric-focusing in barley varietal identification. Ann Appl Biol. 1990;117:625–631. [Google Scholar]
- Totsuka A, Nong VH, Kadokawa H, Kim C-S, Itoh Y, Fukazawa C. Residues essential for catalytic activity of soybean β-amylase. Eur J Biochem. 1994;221:649–654. doi: 10.1111/j.1432-1033.1994.tb18777.x. [DOI] [PubMed] [Google Scholar]
- Wiberg A. Genetical studies of spontaneous sources of resistance to powdery mildew in barley. Hereditas. 1974;77:89–148. doi: 10.1111/j.1601-5223.1974.tb01357.x. [DOI] [PubMed] [Google Scholar]
- Yoshigi N, Okada Y, Sahara H, Koshino S. PCR cloning and sequencing of the β-amylase cDNA from barley. J Biochem. 1994;115:47–51. doi: 10.1093/oxfordjournals.jbchem.a124303. [DOI] [PubMed] [Google Scholar]
- Yoshigi N, Okada Y, Sahara H, Tamaki T. A structural gene encoding β-amylase of barley. Biosci Biotechnol Biochem. 1995;59:1991–1993. doi: 10.1271/bbb.59.1991. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Saghai Maroof MA, Kleinhofs A. Comparative diversity analysis of RFLPs and isozymes within and among populations of Hordeum vulgare ssp. spontaneum. Genetics. 1993;134:909–916. doi: 10.1093/genetics/134.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]