Abstract
Exon skipping has been demonstrated to be a successful strategy for the gene therapy of Duchenne muscular dystrophy (DMD): the rational being to convert severe Duchenne forms into milder Becker ones. Here, we show the selection of U1 snRNA-antisense constructs able to confer effective rescue of dystrophin synthesis in a Δ44 Duchenne genetic background, through skipping of exon 45; moreover, we demonstrate that the resulting dystrophin is able to recover timing of myogenic marker expression, to relocalize neuronal nitric oxide synthase (nNOS) and to rescue expression of miRNAs previously shown to be sensitive to the Dystrophin-nNOS-HDAC2 pathway. Becker mutations display different phenotypes, likely depending on whether the shorter protein is able to reconstitute the wide range of wild-type functions. Among them, efficient assembly of the dystrophin-associated protein complex (DAPC) and nNOS localization are important. Comparing different Becker deletions we demonstrate the correlation between the ability of the mutant dystrophin to relocalize nNOS and the expression levels of two miRNAs, miR-1 and miR29c, known to be involved in muscle homeostasis and to be controlled by the Dys-nNOS-HDAC2 pathway.
Introduction
Duchenne muscular dystrophy (DMD) is a rare disorder due to mutations in the dystrophin gene, the largest human gene, made of 79 exons alternatively arranged in tissue-specific isoforms.1
In skeletal muscles, dystrophin is localized on the inner face of the sarcolemma, where it interacts with a complex of proteins named dystrophin-associated protein complex (DAPC).2 In the absence of dystrophin, the DAPC complex is destabilized and muscle fibres become more sensitive to mechanical damage leading to muscle degeneration, chronic inflammation, susceptibility to oxidative stress and increased fibrosis. Besides organization of the DAPC, dystrophin was described to be important also for the localization of the neuronal nitric oxide synthase (nNOS). Specific dystrophin domains were described to be required for this activity3,4,5 which also involves syntrophin interaction.6,7 Delocalization of nNOS was shown to impair nitric oxide (NO) production in dystrophic muscles.8 The relevance of this pathway on muscle pathology was demonstrated by the fact that rescue of nNOS expression in mdx animals prevented exercise-related fatigue9 and ameliorated the dystrophic phenotype.10,11 In addition NO has recently emerged as a key player in mediating epigenetic changes during neuronal development by repressing histone deacetylase 2 (HDAC2) activity through S-nitrosylation.12 Along this line, in the dystrophin-deficient mdx mice, defective for the NO-pathway, the activity of HDAC2 resulted to be specifically increased13 and deacetylase inhibitors were shown to confer a strong morphofunctional benefit to dystrophic muscle fibres.14 Finally, lower HDAC2 nitrosylation state in Duchenne versus healthy conditions was shown to reduce the expression of a specific subset of microRNAs with a relevant role in muscle differentiation and homeostasis.15
Therefore, these data indicate that dystrophin is able to control gene expression through nNOS localization and NO-mediated HDAC2 activity (DYS-nNOS pathway). This feature should be considered when planning dystrophin rescue of different DMD mutations.
The majority of Duchenne mutations are substitutions, deletions and duplications leading to frame shift and premature translation termination.16 On the contrary, if mutations do not perturb the reading frame, shorter and partially functional proteins are produced giving rise to the Becker muscular dystrophy (BMD) phenotype. BMD patients with large deletions can show very mild symptoms17,18 when spectrin-like repeat domains are involved; on the contrary rare in frame deletions that affect crucial domains for dystrophin function result in severe DMD-like phenotypes.3 BMD-like molecules associated with the mildest phenotypes are those in which nNOS is efficiently retained at the sarcolemma.19 For these reasons exon skipping, which is based on the principle of converting DMD into BMD, can show different outcomes depending on the location of the mutation to be rescued. Exon skipping uses antisense sequences against splice junctions and/or exonic splicing enhancers (ESEs) in order to induce the exclusion of a target exon from the mature mRNA.20,21,22 Antisense sequences can be administered as chemically modified antisense oligonucleotides,23 whose safety and local efficacy have been recently demonstrated in phase I and II clinical trials24,25,26,27, or as part of a small nuclear RNA (snRNA), which provide persistent expression, in vivo stability and correct cellular compartmentalization.28 U1 and U7 snRNA-derived antisense molecules have been shown to be effective in inducing exon skipping both in human DMD myoblasts28,29,30,31 and in the mdx mouse32,33,34,35 where a life-long beneficial effect was obtained36 together with recovery of serum biomarkers diagnostic of muscle damage.37
In this article, we tested in a DMD Δ44 genetic background, different U1 snRNA-antisense sequences for the ability to induce exon 45 skipping and recovery of dystrophin. According to several large databases (Leiden muscular dystrophy database; http://www.dmd.nl; UMD-DMD France mutations database38) skipping of this exon should be applicable to ~8% of all known DMD mutations.39
Eight different antisense constructs were tested for exon 45 skipping activity in lentiviral-transduced Δ44 human DMD myoblasts. Several effective molecules were obtained that produced dystrophin rescue and nNOS relocalization to the membrane. Moreover, following in vitro myoblasts differentiation, exon skipping achieved restoration of the timing of myogenic markers expression towards control levels. Notably, miRNAs previously shown to depend on the DYS-nNOS pathway were also recovered in their expression. Finally, the analysis of biopsies from BMD patients with different mutations confirmed the correlation between the ability of dystrophin to relocalize nNOS and the expression of several molecular markers linked to the DYS-nNOS pathway.
Results
Design and expression analysis of antisense molecules against exon 45 of the dystrophin gene
Eight constructs, in the backbone of the U1 snRNA, were designed and produced for the skipping of exon 45 of the dystrophin pre-mRNA (Figure 1a). Nucleotides from position 3 to 10 at the 5′ end of U1 snRNA, required for the recognition of 5′ splice sites40 were substituted with antisense sequences complementary to different portions of exon 45 and splice sites (Supplementary Table S1). The initial construct (#1) contained antisense sequences against both splice junctions since we previously showed that both splice sites have to be targeted in order to induce efficient exon skipping when using U1 snRNA.28 Additional constructs were produced that contained also antisense sequences against putative ESEs, known to represent effective target substrates for efficient exon skipping.21,41,42
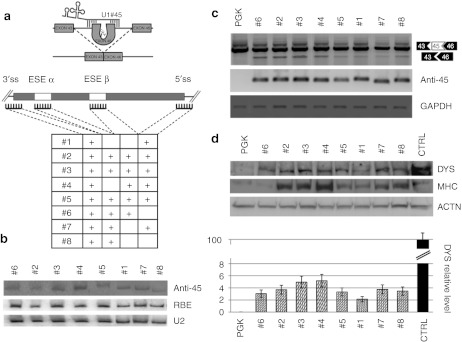
Figure 1.
Exon 45 skipping. (a) Schematic representation of the exon-skipping strategy for the human Δ44 Duchenne muscular dystrophy (DMD) mutation. The table summarizes the eight different constructs produced together with the corresponding target regions on exon 45 and flanking intron (ss, splice site; ESE, exonic splicing enhancer); sequences are indicated in Supplementary Table S1. (b) C27 myoblasts were transfected with the different antisense constructs together with the U16-RBE plasmid [expressing a 143-nucleotide (nt) long modified U16 snoRNA43] Northern blot analysis was performed with probes against the 3′ splice site (anti-45), U16-RBE (RBE), and U2 small nuclear RNA (snRNA) (U2). The two latter hybridizations are used to normalize for transfection efficiency and as loading control, respectively. (c) Exon skipping activity. The amount of exon skipping was calibrated by Nested reverse transcription-PCR (RT-PCR) on RNA extracted from Δ44 cells infected with the antisense-expressing lentiviruses. Unskipped and skipped products are indicated on the right. Anti-45 refers to the expression of the antisense molecules; GAPDH is used as an internal control. (d) Rescue of dystrophin synthesis. Upper panel: western blot on proteins from CTRL (10 µg + 40 µg PGK), mock infected Δ44 (PGK- 50 µg) and exon-skipping Δ44 treated cells (#1 to #8–50 µg) probed with anti-dystrophin (DYS), anti-myosin heavy chain (MHC) and anti-actinin (ACTN) antibodies. Lower panel: the histogram indicates dystrophin levels normalized on actinin signals and expressed as percentage of CTRL. Error bars: means ± SD.
The chimeric U1-antisense sequences were cloned under the control of the strong polII U1 snRNA gene promoter and termination sequences, and subsequently inserted in the dU3 portion of the 3′ long-terminal repeat region of a lentiviral vector plasmid.43
The expression and stability of the different constructs were tested by transfection into the murine myogenic C27 cell line. The relative expression was measured by co-transfection with the U16-RBE plasmid44 and normalized for the endogenous U2 snRNA. Northern blot analysis indicated that all chimeric molecules accumulated at fairly similar levels (Figure 1b).
Study of exon skipping activity in human DMD myoblasts
Lentiviral particles for each construct were produced in 293T cells and used to transduce DMD myoblasts carrying the deletion of exon 44 (Δ44, provided by the Telethon Neuromuscular Biobank). In this case, skipping of exon 45 allows restoration of the correct reading frame and the production of a dystrophin protein 106 aminoacids shorter than the wild type. Δ44 cells were infected with comparable amounts of the different recombinant lentiviruses. After infection, they were induced to differentiate and samples were collected after 10 days for RNA and protein analysis. Exon 45 skipping (Figure 1c, upper panel) and antisense expression (Figure 1c, middle panel) were assessed by RT-PCR analysis, utilizing GAPDH as normalizer (Figure 1c, lower panel). Dystrophin rescue was assessed by Western blot (Figure 1d, upper panel) and the relative values, normalized on actinin levels, are reported in the histogram. Due to the low efficiency of exon skipping and in order to have more comparable signals, 50 µg of proteins from Δ44-treated cells (PGK and #1–#8) were loaded in parallel with 10 µg of proteins from control cells (CTRL). Myosin heavy chain (MHC) detection was used as a marker of muscle terminal differentiation. This analysis revealed that constructs #6 and #8, targeting only the 3′ splice junction and ESE sequences, are poorly active in terms of dystrophin recovery. Moreover, #1, targeting the two splice junctions, provided very low skipping activity and dystrophin rescue. The importance of the simultaneous targeting of splice sites and ESE is shown by constructs #3 and #4 that provided the highest rescue of dystrophin synthesis (see the histogram of Figure 1d). Interestingly to note that higher levels of dystrophin correlated with increase in the MHC terminal differentiation marker (Figure 1d, panel MHC).
Dystrophin levels affect myogenic differentiation
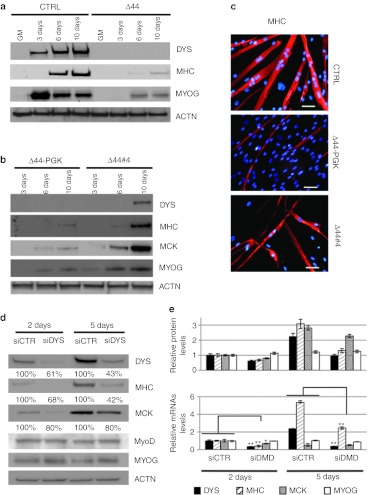
In vitro cultured DMD myoblasts display a delay in the appearance of typical myogenic markers;45 this can be easily appreciated when comparing in vitro differentiation of Δ44 myoblasts with control ones derived from a healthy individual: Figure 2a shows that the terminal differentiation marker MHC, already present at day 6 of differentiation in control cells, is poorly detectable in Δ44 cells even at day 10. Notably, when exon skipping was applied, the treated DMD myoblasts showed an effective recovery of the timing of myogenic marker appearance: Figure 2b shows a time course (3, 6, and 10 days of differentiation) on mock infected (Δ44-PGK) and exon-skipping treated (Δ44#4) Δ44 cells. Dystrophin rescue in treated cells was paralleled by a relevant temporal restoration of late/terminal differentiation markers; in particular, MHC appeared at day 6 similarly to control cells, even if at lower levels, and muscle creatine kinase (MCK) at day 3. A direct comparison on the same gel of MHC and MCK expression levels in healthy control (CTRL), Δ44 (Δ44), mock-infected Δ44 (Δ44-PGK), and exon-skipping treated Δ44 (Δ44#4) at latest time points (6 and 10 days after shift to differentiation medium) is shown in Supplementary Figure S1a. These results clearly demonstrate that, with respect to healthy myoblasts, the delay of appearance of late myogenic markers (MHC and MCK) in untreated (Δ44) and mock infected (Δ44-PGK) DMD cells is recovered in exon-skipping treated (Δ44#4) cells. Moreover, immunostaining analysis of Figure 2c (and more examples in Supplementary Figure S1b) indicates that the terminal differentiation marker MHC, already present at day 8 of differentiation in control cells (CTRL), is almost undetectable in mock infected Δ44 cells (Δ44-PGK) at day 12. At this time point, efficient recovery of MHC staining was detected when exon skipping was applied (Δ44#4).
Figure 2.
Analysis of muscle differentiation markers. (a) Western blot on proteins (10 µg) extracted from CTRL and Δ44 cells probed with anti-dystrophin (DYS), anti-myosin heavy chain (MHC), and anti-myogenin (MYOG) antibodies in growth medium (GM) and at different days upon shift to differentiation conditions (3, 6, and 10 days). Actinin (ACTN) was used as a loading control. (b) Western blot on proteins (50 µg) extracted from Δ44 cells, infected with lentivirus expressing GFP (Δ44-PGK) or with the #4 construct (Δ44#4), at different days upon shift to differentiation conditions (3, 6, and 10 days). Western blot was probed with anti-dystrophin (DYS), anti-MHC, anti-muscle creatine kinase (MCK), and anti-myogenin (MYOG) antibodies. Actinin (ACTN) was used as a loading control. (c) Immunofluorescence for MHC localization (red) on control myoblasts (CTRL) at 8 days of differentiation, in parallel with mock infected (Δ44-PGK) and exon-skipping treated (Δ44#4) Δ44 Duchenne muscular dystrophy (DMD) cells at 12 days of differentiation. DAPI staining for nuclei detection is also shown. Original magnification, ×20. Bar = 50 µm. (d) RNA interference against dystrophin. Control human myoblasts were transfected with scramble (siCTR) or with anti-dystrophin (siDYS) siRNAs and differentiated for 2 and 5 days. Western blot was performed with anti-DYS, anti-MHC, anti-MCK, anti-MyoD, and anti-MYOG antibodies. Actinin (ACTN) was used as loading control. (e) Upper panel: The histogram indicates DYS, MHC, MCK, and MyoG protein levels normalized on actinin signals. Error bars: means ± SD. Lower panel: DYS, MHC, MCK, and MyoG mRNAs relative expression in mock (siCTR) MCK, and DMD-siRNA treated (siDYS) human myoblasts after 2 and 5 days of differentiation assessed by quantitative reverse transcriptase-PCR (qRT-PCR). GAPDH was used as internal control. Relative expressions are shown with respect to mock (siCTR) cells, set to a value of 1. *P < 0.05, **P < 0.01.
To further analyze the link between dystrophin expression and muscle differentiation, RNA interference against dystrophin was performed on human myoblasts from a healthy control. siRNAs were transfected in myoblasts in growth medium and one day after shift to differentiation medium. RNA and protein samples were collected at 2 and 5 days of differentiation. After 2 days of treatment, dys-siRNA–treated cells (siDYS) showed a significant reduction of dystrophin mRNA (residual 61%) that, after 5 days, reached a residual value of 43% (Figure 2d). Dystrophin decrease paralleled that of mRNA levels, even if at a lower extent (Figure 2e), probably due to the well known long half-life of the dystrophin protein. Notably, in RNAi-treated cells, both MHC and MCK accumulated at lower levels, with MHC being reduced to 42% of control levels at day 5 of differentiation; in contrast, early myogenic factors such as myogenin and myoD, were not affected by dystrophin reduction both in terms of protein and mRNA (Figure 2d, e).
Altogether, these data support the hypothesis that dystrophin is a crucial myogenic factor regulating the progression from early to late phases of differentiation.
Dystrophin rescue and miRNAs expression levels
In order to test whether the dystrophin protein produced in Δ44 myoblasts through skipping of exon 45, relocalizes and stabilizes nNOS, an immunofluorescence assay was performed with nNOS antibodies on myoblasts from: healthy control (CTRL), mock-infected (Δ44-PGK), and exon-skipping treated (Δ44#4) Δ44 cells. Figure 3a shows the correct localization of nNOS in control myotubes, revealed by well-defined increase in sarcolemmal labelling. Indeed, this striated staining is almost absent in Δ44-untreated myoblasts (Δ44-PGK), where only faint diffused cytoplasmic staining is visible. These results are in line with the well-known decrease and delocalization of nNOS in DMD patients.8,9,46 Notably, when Δ44 cells were treated with exon skipping (Δ44#4), they displayed a well visible nNOS signal distributed along the fibers. These results indicate that the recovery of dystrophin synthesis allows the correct relocalization and stabilization of nNOS at the sarcolemma and that exons 44 and 45 are dispensable for this function.
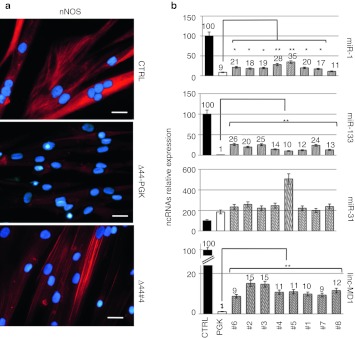
Figure 3.
Analysis of neuronal nitric oxide synthase (nNOS) localization and ncRNAs expression upon exon-skipping treatment. (a) nNOS localization analyzed by immunofluorescence with nNOS antibodies on CTRL, mock-infected Δ44 (Δ44-PGK) and exon-skipping Δ44-treated cells (Δ44#4) after 10 days of differentiation. Original magnification, ×40. Bar = 25 µm. (b) miRNAs and linc-MD1 expression analysis performed by quantitative reverse transcriptase-PCR (qRT-PCR) on RNA extracted from CTRL, mock-infected Δ44 (PGK), and Δ44 cells treated with the different antisense constructs (#1–#8) after 10 days of differentiation. U6 small nuclear RNA (snRNA) is used as endogenous control. Relative expressions are shown with respect to CTRL cells, set to a value of 100 and ncRNA relative quantifications are shown on the top of each lane. *P < 0.05, **P < 0.01.
It has been previously shown15 that the DYS-nNOS pathway regulates, through the control of HDAC2 activity, the expression of a specific subset of miRNAs with specific function in muscle terminal differentiation (miR-1 and miR-133) or in muscle-degenerative processes such as fibrosis (miR-29 and miR-30). Conversely, miRNAs expressed in regenerating fibres, such as miR-31 and miR-206, were shown not to be controlled through this pathway. In order to test the dependence of these miRNAs from nNOS rescue in exon-skipping treated Δ44 myoblasts, quantitative reverse transcriptase-PCR was performed on total RNA extracted from WT and Δ44 cells either mock infected (PGK) or treated with the different antisense constructs (#1 through #8). Figure 3b indicates that in Δ44 myoblasts miR-1 and miR-133 levels are strongly reduced with respect to healthy myoblasts (CTRL), in agreement with previous observation in Δ48–50 DMD myoblasts;15 however, when exon skipping recovered dystrophin synthesis, increase in the levels of both miRNAs was obtained.
At difference with these miRNAs, miR-31 was upregulated in Δ44 cells and, upon dystrophin rescue, its expression remained high (Figure 3b). Recently, it has been shown that Duchenne myoblasts contain reduced levels of linc-MD1, a long noncoding RNA required for muscle differentiation.47 Figure 3b (lower panel) indicates that also Δ44 myoblasts have low levels of linc-MD1; notably, recovery of dystrophin synthesis induced increase in linc-MD1 levels. ChIP experiments on linc-MD1 promoter in mouse myoblasts indicated that this transcript is not under the control of HDAC2 (C. Pinnarò, personal communication, 5 December 2011); therefore, the increase of linc-MD1 upon dystrophin restoration does not directly depend on the DYS-nNOS pathway but it is likely due to an indirect effect on muscle differentiation, as previously discussed.
Altogether, these data confirm that the recovery of dystrophin in Δ44 cells restores nNOS sarcolemmal localization and in turn the biosynthesis of those miRNAs that depend on the DYS-nNOS pathway.
BMD
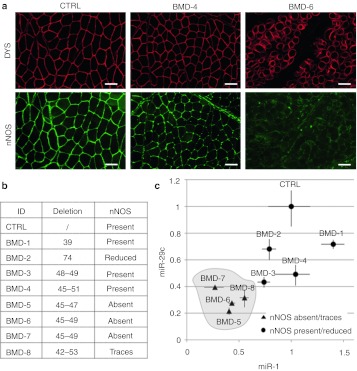
To further correlate the beneficial effects of dystrophin with the integrity of the DYS-nNOS pathway, we analyzed 14 biopsies from Becker patients carrying mutations differently affecting nNOS localization (representative examples are shown in Figure 4a and Supplementary Figure S2). The expression of miR-1 and miR-29, previously shown to respond to the DYS-nNOS pathway, was analyzed from total RNA obtained from eight of such biopsies. Figure 4c shows that BMD samples lacking nNOS (BMD-5, -6, -7, and -8) display low levels of miR-1 and miR-29 if compared to BMD samples with reduced or normal levels of nNOS (BMD-1, -2, -3, and -4), confirming the direct correlation between nNOS and specific miRNA expression.
Figure 4.
Neuronal nitric oxide synthase (nNOS) localization and miRNAs expression in Becker biopsies. (a) Dystrophin (DYS) and neuronal NOS localization analyzed by immunofluorescence on control (CTRL), BMD-4, and BMD-6 human biopsies. Original magnification, ×10. Bar = 100 µm. (b) Table summarizes exon deletions of the different Becker muscular dystrophy (BMD) patients and corresponding nNOS levels. (c) Scatterplot showing the expression levels of miR-29c (Y axis) and miR-1 (X axis) analyzed by quantitative reverse transcriptase-PCR (qRT-PCR) on RNAs extracted from the different BMD biopsies. U6 small nuclear RNA (snRNA) is used as endogenous control. Relative expressions are shown with respect to healthy individual (CTRL), set to a value of 1. Error bars: means ± SD.
Interestingly, in agreement with previous work,19 biopsies lacking or having trace levels of nNOS corresponded to patients displaying an overall more severe clinical phenotype (Supplementary Table S2), measured as Hammersmith functional motor scale.48 This scale, that is routinely performed in the clinical follow-up of patients, consist in 20 consecutive motor activities each scored on a 3-point scale (2 to 0) and the total test score can range from 0 if the child cannot perform any of the items to 40 if all the items are fully achieved. The table indicates that in BMD patients the absence of nNOS correlates with a worse outcome (patients BMD-5, -6, -9, and -14) with the only exception of BMD-7 that show traces of nNOS, low miRNA levels but unaffected HFMS score.
Becker mutations completely lacking nNOS are those missing exons 45–47 and 45–49 as they more severely disrupt spectrin-like repeats 16 and 17. However, the finding that deletion 45-51 has visible levels of nNOS indicates that a direct relationship between primary sequence and protein activity cannot be simply made.
Discussion
Inducing exon skipping as a therapeutic approach of DMD relies on the concept of converting severe Duchenne phenotypes into the milder Becker ones. However, the latter display a large range of phenotypes likely reflecting the ability of shorter forms of dystrophin to accomplish only a subset of the different functions of the wild-type full-length protein. Multiple variables play a role in determining the efficiency of internally shortened dystrophins produced in Becker patients: these relate to structural properties of the residual dystrophin molecules as well as to their capacity to correctly assemble DAPC complexes and to relocalize nNOS.19
Regarding the former, the spectrin-like repeat region is composed of 24 modular repeats found in the rod domain. While removing an integral repeat is mostly dispensable, deletions that affect the phasing of the repeats are associated with less stable and functional dystrophin molecules.49 Concerning the role that different internal dystrophin deletions have on assembly of DAPC proteins, we have recently demonstrated that BMD dystrophins of patients with deletions located around exons 51 and 53 are more efficient in relocalising at the sarcolemma β-destroglycan and α-sarcoglycan compared to other deletions that remove spectrin-like repeats 16 and 17.19 There is also a strong correlation between deletions of spectrin-like repeats 16 and 17 and ability to localize nNOS at the sarcolemma.4,5
These are crucial points when thinking of possible therapeutic benefits of exon skipping in different DMD mutations and suggest that dystrophin rescue should be tested together with correct recovery of the entire complex of proteins at the sarcolemma. We have recently shown that in DMD mutants lacking exons 48–50, further skipping of exon 51 is able to relocalize α-sarcoglycan and nNOS.50
The relevance of nNOS was previously shown to rely on its ability to control gene expression through nitrosylation of HDAC2. The rescue of this pathway in dystrophic animals has been proved to ameliorate the dystrophic phenotype and to influence the expression of a specific subset of genes playing a crucial role in the control of muscle terminal differentiation and tissue homeostasis:15 among them, those coding for relevant myogenic miRNAs (miR-1 and miR-133) and for species controlling fibro-adipocitic degeneration (miR-29 and miR-30). These data suggested that nNOS activity plays an important role in muscle homeostasis via its ability to directly regulate the expression of specific HDAC2 target genes and to have a positive effect on differentiation.
In this work, we produced antisense molecules, raised in the U1 snRNA backbone, able to induce effective skipping of exon 45 that is one of the most common, after exon 51, in terms of number of patients that can be cured (8.1% of known DMD mutations). We showed that rescue of dystrophin provided a considerable enhancement of differentiation in DMD myoblasts, well characterized for their delay in reaching a mature molecular and morphological phenotype. This evidence was further supported by experiments of RNAi against dystrophin in myoblasts from healthy individuals. In this case an evident delay in timing of myogenic markers synthesis was observed. Notably, while having such a clear effect on late differentiation markers, dystrophin did not show any specific effect on timing and levels of early myogenic markers such as Myogenin and MyoD.
Notably, the dystrophin protein, obtained through skipping of exon 45 in a Δ44 genetic background, produced efficient rescue of nNOS localization at the membrane, together with recovery of miR-1 and miR-133 expression, indicating the reactivation of the DYS-nNOS pathway. In line with the relevance of this pathway, nNOS analysis of different Becker individuals indicated that those lacking correct localization of the enzyme had a more severe clinical phenotype as well as reduction of those marker miRNAs depending on the DYS-nNOS pathway.
Altogether, these data demonstrate a crucial role of dystrophin in regulating the switch between early and late phases of muscle differentiation and point to the relevance of the DYS-nNOS pathway in DMD and BMD.
Materials and Methods
Antisense clones construction. Clones U1-45 were obtained by inverse PCR on the construct pCCL-5′3′Esx28.
The oligos utilized are:
U1-45#1:
F: (5′-CAGAAAAAAGAGgtagggcgacagGGCAGGGGA GA TACCATGATC-3′)
R: (5′-CCTGGAGTTCctgtaagataccaaaaaggcATGAG AT CTTGGGCCTCTGC-3′)
U1-45#2:
F: (5′-GCCCAATGCCATCCTGGAGTTCctgGGCAGGGG AGA TACCATGATC-3′)
R: (5′-TCCTCAAAAACAGATGCCAGAAGAGgtagggcgATGAGATCTTGGGCCTCTGC-3′)
U1-45#3:
F: (5′-GCTGCCCAATGCCATCCTGGAGTTCc tgGGCAGGGG AGATACCATGATC-3′)
R: (5′-CAGCAATCCTCAAAAACAGATGCGAGgta gggATGA GATCTT GGGCCTCTGC-3′)
U1-45#4:
F: (5′-GAGGTTGCTGCCTGGAGTTCctgtaagGGCAGGGGAGATACCATGATC-3′)
R: (5′-AAAAACAGAGAAAAAAGAGgtagggcgATGAGAT C TTGGGCCTCTGC-3′)
U1-45#5:
F: (5′-ATTGCCCAATGCCATCCTGGAGTTCctgtGGCAGGGGAGATACCATGATc-3′)
R: (5′-CCTCAAAAACAGAGAAAAAAGAGgtagggATGAGATCTTGGGCCTCTGC-3′)
U1-45#6:
F: (5′-GCTGCCCAATGCCATCCTGGAGTTCctgtaaGGGCAGGGGAGAT ACCATGATC-3′)
R: (5′-CAGCAATCCTCAAAAACAGATGCATGAGATCTTGGGCCTCTGC-3′)
U1-45#7:
F: (5′-GCTGCCCTGCCTCCTGGAGTTCctgtaagGGC AGGGG AGATACCATGATC-3′)
R: (5′-CAGAAAAAAGAGgtagggcgacagATGAGATCTTGGGCCTCTGC-3′)
U1-45#8:
F: (5′-CCTGGAGTTCctgtaagataccaaaaaggcGGCAGGGGAGATACCATGATC-3′)
R: (5′-ATGGCATTGGGCAG CATGAGATCTTGGGCCTCTGC-3′)
C27 transfections. C27 cells were plated on 3.5-cm diameter plates and cotransfected with 3 µg of the lentiviral vector carrying the antisense expression cassette and 1 µg of U16-RBE plasmid used as a transfection efficiency control. Transfection was performed according to the Lipofectamine 2000 protocol (Invitrogen, Carlsbad, CA). Cells were grown in Dulbecco's modified Eagle's medium 10% fetal bovine serum for 36 hours and then harvested with 1 ml of QIAzol Lysis Reagent (Qiagen, West Sussex, UK).
RNA preparation and analysis. Cells were harvested with 1 ml of QIAzol Lysis Reagent (Qiagen) and biopsies were homogenized with a rotor-stator homogenizer in the presence of QIAzol Lysis Reagent (Qiagen). RNAs were extracted by miRNeasy (Qiagen), following manufacturer's specifications; concentration was assessed with Nanodrop ND-1000 Spectrophotometer (CELBIO; Pero, Milan, Italy). Quantitative reverse transcriptase-PCR were performed using miScript System (Qiagen).
Northern blot. Antisense expression was analyzed by northern blot as previously described28 using the following probe: anti-45 (5′-cagGAA CTCCAGG-3′).
Primary myoblasts cultures. Cultures of primary myoblasts (WT-9808 and Δ44-9981 from the Telethon Neuromuscular Biobank) were first pre-plated in order to separate fibroblasts from the primary line, then seeded in Human skeletal muscle growth medium (PromoCell, Heidelberg, Germany) and grown in a humidified incubator, at 5% CO2 and 37 °C.
Virus preparation and cell transduction. Viruses were prepared as described.28 The day before transduction, myoblasts were seeded in growth medium, on 6-cm plates (at least two for each different virus), at a density of 5 × 105 cells per plate. The next day cells were infected once with lentiviruses and polybrene (4 mg/ml). Two days after infection, cells were induced to differentiate with human skeletal muscle differentiation medium (PromoCell). After 10 days of differentiation, cells were washed twice with complete phosphate-buffered saline (PBS) buffer (PromoCell) and collected with 300 ml of protein buffer (100 mmol/l Tris–HCl pH 7.4, 1 mmol/l EDTA, 2% SDS, 1× Complete EDTA-free Protease Inhibitor Cocktail (Roche, Applied Science, Mannheim, Germany) for protein extraction, or with 1 ml of QIAzol Lysis Reagent (Qiagen) for RNA extraction.
RT-PCR. Dystrophin mRNA was analyzed by RT-PCR (Access RT-PCR system—Promega, Madison, WI) on 200 ng of total RNA with oligos E42F (5′-GAAGACATGCCTTTGGAAATTTCT-3′) and E48R (5′-CTGAA CGTCAAATGGTCCTTC-3′). Four microlitres of the RT-PCR products were then used as template for a nested reaction performed with oligo E43F (5′-CTACAACAAAGCTCAGGTCG-3′) and E46R (5′-CTC TTTTCCAGGTTCAAGTGG-3′).
Antisense expression was analyzed by RT-PCR (SuperScript VILO cDNA Synthesis Kit; Invitrogen) on 50 ng of total RNA with oligos U1primer (5′- CAGGGGAAAGCGCGAACG-3′) and RT45 (5′-atcc tggagttcctgtaa-3′). GAPDH was used as loading control (oligos: F 5′-GGAAGGTGAAGGTCGGAGTC-3′ R 5′-TTACCAGAGTTAAAA GCAGCCC-3′).
Ten microlitres of the reactions were run on a 2% agarose-ethidium bromide gel and signals were revealed on a UV transilluminator.
Western blot analysis. Western blot analyses were carried out as previously described.28 Primary antibodies: anti-dystrophyn [NCL-DYS1; Novocastra Laboratories, New Castle upon Tyne, UK, 1:40 in 3% milk); anti-myosin (anti-MF-20, 1:20 in TBST), anti-MCK (sc-15161; Santa Cruz Biotechnology, Santa Cruz, CA) 1:500 in TBST] anti-myogenin (MyoG sc-12732, 1:1,000 in TBST); anti-MyoD (DAKO, Glostrup, Denmark, 1:500 in 3% milk); anti-actinin (ACTN sc-15335, 1:1000 in TBST). Secondary antibodies: ImmunoPure Goat Anti-Rabbit IgG Peroxidase-Conjugated (Pierce, Rockford, IL, 1:5,000 in 5% milk); ImmunoPure®Goat Anti-Mouse IgG Peroxidase Conjugated (Pierce, 1:10,000 in 5% milk); donkey anti-goat IgG-HRP (sc-2020, diluted 1:5,000 in 3% milk).
RNA interference against dystrophin. The WT-9808 cell line, obtained from Telethon Genetic Biobank Network, was grown to 70% confluence on 6-cm plates in Dulbecco's modified Eagle's medium with 18% fetal bovine serum. Transfection was performed twice with 200 pmol of siRNA oligonucleotides targeting the dystrophin gene (Qiagen, target sequence: 5′-AATAACTTGCCATTTCTTTAT-3′) or 200 pmol of siRNA negative control using lipofectamine (Invitrogen) according to manufacturer's specification. After 18 hours, the medium was replaced. RNA and protein samples were collected after 2 and 5 days of differentiation.
Patients. Fourteen BMD patients, aged from 2 to 15 years, were assessed by physiotherapists to quantify muscular strength through Hammersmith motor ability score.47
Skeletal muscle biopsies were obtained, with informed consent, from all BMD patients along with non-myopathic control. BMD-3, BMD-4, BMD-6, BMD-9, BMD-10, BMD-11, BMD-12, BMD-13, BMD-14, and control were obtained from Laboratory of Molecular Medicine, Department of Neuroscience, Bambino Gesù Children's Hospital, Rome whereas BMD-1, BMD-2, BMD-5, BMD-7, BMD-8, BMD-15 biopsies were obtained from Dubowitz Neuromuscular Centre, Institute of Child Health and Great Ormond Street Hospital, London.
Immunohistochemistry. Muscle biopsies were mounted in OCT medium. Serial 7-µm transverse cryosections were fixed for 15 minutes at 4 °C in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), permeabilized with Triton 0.2%/1%BSA/PBS for 15 minutes and subsequently blocked in 10% goat serum/1%BSA/PBS for 1 hour at room temperature. Sections were incubated at 4 °C with primary antibodies diluted in 5% Goat Serum/PBS (incubation buffer) for 16 hours. After serial washes in 0.2% Triton /PBS, secondary antibodies (goat anti-Mouse IgG AlexaFluor 488; Invitrogen or Cy3 conjugated; Jackson ImmunoResearch, West Grove, PA) diluted 1:500 with incubation buffer were added for 1h at room temperature. The specificity of immunolabeling was verified in control samples prepared with the incubation buffer alone, followed by the second conjugated antibody. The sections were counterstained with 1.5 mg/ml 4′, 6-diamidino-2 phenylindole in Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA). Cultured cells were fixed in methanol/acetone for 20 minutes at –20 °C, subsequently air-dried and after brief rehydration with PBS processed for immunostaining with primary antibodies (diluted in 1% goat serum/PBS) as described above.
Primary monoclonal antibodies used in this study were: diluted as follows: anti-Dystrophin NCL-DYS2, Novocastra Laboratories (diluted 1:12.5); anti-Nos-1 sc-55521 (diluted 1:50), anti-MHC from hybridoma supernatants.
Image acquisition and analysis. Immunostained cells and muscle sections were examined using a Zeiss AxioObserver A1 inverted fluorescence microscope equipped with Axiocam MRM R camera and Plan-Neofluar EC 10×/0.3 M27 and LD 40×/0.6 M27 objectives. The images were acquired with AxioVision Rel.4.8 imaging software.
Statistical analyses. Each data shown in quantitative reverse transcriptase-PCR is the result of at least three independent experiments. Data are shown as mean ± SD. Unless specifically stated, statistical significance of differences between means was assessed by two-tailed t-test and a P < 0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Analysis of muscle differentiation markers. Figure S2. Dystrophin and nNOS localization in BMD biopsies. Table S1. Description of antisense constructs. Table S2. Clinical informations of BMD patients.
Acknowledgments
We thank Marina Mora, the Telethon Neuromuscular Biobank, and MRC Neuromuscular Centre Biobank for cell lines and biopsies. This work was partially supported by grants from: Telethon (GGP11149), Parent Project Italia, IIT “SEED,” PRIN and BEMM to I.B.; Great Ormond Street Children's Charity and AFM (which supports V.R.) to F.M. The authors declared no conflict of interest.
Supplementary Material
Analysis of muscle differentiation markers.
Dystrophin and nNOS localization in BMD biopsies.
Description of antisense constructs.
Clinical informations of BMD patients.
REFERENCES
- Blake DJ, Weir A, Newey SE., and, Davies KE. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta. 2007;1772:108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Chao DS, Gorospe JR, Brenman JE, Rafael JA, Peters MF, Froehner SC.et al. (1996Selective loss of sarcolemmal nitric oxide synthase in Becker muscular dystrophy J Exp Med 184609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells KE, Torelli S, Lu Q, Brown SC, Partridge T, Muntoni F.et al. (2003Relocalization of neuronal nitric oxide synthase (nNOS) as a marker for complete restoration of the dystrophin associated protein complex in skeletal muscle Neuromuscul Disord 1321–31. [DOI] [PubMed] [Google Scholar]
- Lai Y, Thomas GD, Yue Y, Yang HT, Li D, Long C.et al. (2009Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy J Clin Invest 119624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameya S, Miyagoe Y, Nonaka I, Ikemoto T, Endo M, Hanaoka K.et al. (1999alpha1-syntrophin gene disruption results in the absence of neuronal-type nitric-oxide synthase at the sarcolemma but does not induce muscle degeneration J Biol Chem 2742193–2200. [DOI] [PubMed] [Google Scholar]
- Adams ME, Mueller HA., and, Froehner SC. In vivo requirement of the alpha-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Xia H, Aldape K., and, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi YM, Rader EP, Crawford RW, Iyengar NK, Thedens DR, Faulkner JA.et al. (2008Sarcolemma-localized nNOS is required to maintain activity after mild exercise Nature 456511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling M, Spencer MJ., and, Tidball JG. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli S, Sciorati C, D'Antona G, Innocenzi A, Covarello D, Galvez BG.et al. (2007Nitric oxide release combined with nonsteroidal antiinflammatory activity prevents muscular dystrophy pathology and enhances stem cell therapy Proc Natl Acad Sci USA 104264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L., and, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S.et al. (2008HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment Proc Natl Acad Sci USA 10519183–19187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V.et al. (2006Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors Nat Med 121147–1150. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M.et al. (2010MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway Cell Metab 12341–351. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H., and, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE.et al. (1990Very mild muscular dystrophy associated with the deletion of 46% of dystrophin Nature 343180–182. [DOI] [PubMed] [Google Scholar]
- Love DR, Flint TJ, Genet SA, Middleton-Price HR., and, Davies KE. Becker muscular dystrophy patient with a large intragenic dystrophin deletion: implications for functional minigenes and gene therapy. J Med Genet. 1991;28:860–864. doi: 10.1136/jmg.28.12.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony K, Cirak S, Torelli S, Tasca G, Feng L, Arechavala-Gomeza V.et al. (2011Dystrophin quantification and clinical correlations in Becker muscular dystrophy: implications for clinical trials Brain 134Pt 123547–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington SJ, Mann CJ, Fletcher S., and, Wilton SD. Target selection for antisense oligonucleotide induced exon skipping in the dystrophin gene. J Gene Med. 2003;5:518–527. doi: 10.1002/jgm.361. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Heemskerk JA, De Winter CL, Van Ommen GJ., and, Van Deutekom JC. Therapeutic modulation of DMD splicing by blocking exonic splicing enhancer sites with antisense oligonucleotides. Ann N Y Acad Sci. 2006;1082:74–76. doi: 10.1196/annals.1348.058. [DOI] [PubMed] [Google Scholar]
- Manzur AY., and, Muntoni F. Diagnosis and new treatments in muscular dystrophies. Postgrad Med J. 2009;85:622–630. doi: 10.1136/jnnp.2008.158329. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A., and, van Ommen GJ. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13:1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M.et al. (2007Local dystrophin restoration with antisense oligonucleotide PRO051 N Engl J Med 3572677–2686. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C.et al. (2009Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study Lancet Neurol 8918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K.et al. (2011Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study Lancet 378595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N.et al. (2011Systemic administration of PRO051 in Duchenne's muscular dystrophy N Engl J Med 3641513–1522. [DOI] [PubMed] [Google Scholar]
- Incitti T, De Angelis FG, Cazzella V, Sthandier O, Pinnarò C, Legnini I.et al. (2010Exon skipping and duchenne muscular dystrophy therapy: selection of the most active U1 snRNA antisense able to induce dystrophin exon 51 skipping Mol Ther 181675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E.et al. (2002Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48-50 DMD cells Proc Natl Acad Sci USA 999456–9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Suter D, Pauli C, Dunant P, Lochmüller H, Burgunder JM.et al. (2003U7 snRNAs induce correction of mutated dystrophin pre-mRNA by exon skipping Cell Mol Life Sci 60557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchaouir R, Meregalli M, Farini A, D'Antona G, Belicchi M, Goyenvalle A.et al. (2007Restoration of human dystrophin following transplantation of exon-skipping-engineered DMD patient stem cells into dystrophic mice Cell Stem Cell 1646–657. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Vulin A, Fougerousse F, Leturcq F, Kaplan JC, Garcia L.et al. (2004Rescue of dystrophic muscle through U7 snRNA-mediated exon skipping Science 3061796–1799. [DOI] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C.et al. (2006Body-wide gene therapy of Duchenne muscular dystrophy in the mdx mouse model Proc Natl Acad Sci USA 1033758–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti MA, Rosa A, D'Antona G, Sthandier O, De Angelis FG, Nicoletti C.et al. (2006Chimeric adeno-associated virus/antisense U1 small nuclear RNA effectively rescues dystrophin synthesis and muscle function by local treatment of mdx mice Hum Gene Ther 17565–574. [DOI] [PubMed] [Google Scholar]
- Goyenvalle A, Babbs A, Wright J, Wilkins V, Powell D, Garcia L.et al. (2012Rescue of severely affected dystrophin/utrophin-deficient mice through scAAV-U7snRNA-mediated exon skipping Hum Mol Genet 212559–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denti MA, Incitti T, Sthandier O, Nicoletti C, De Angelis FG, Rizzuto E.et al. (2008Long-term benefit of adeno-associated virus/antisense-mediated exon skipping in dystrophic mice Hum Gene Ther 19601–608. [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D, Legnini I, Martone J, Cazzella V, D'Amico A, Bertini E.et al. (2011miRNAs as serum biomarkers for Duchenne muscular dystrophy EMBO Mol Med 3258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Béroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L.et al. (2009Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase Hum Mutat 30934–945. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ.et al. (2009Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations Hum Mutat 30293–299. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., and, Weiner AM. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ., and, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, De Winter CL, Janson AA, Kaman WE, Van Ommen GJ, Den Dunnen JT.et al. (2005Functional analysis of 114 exon-internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites Oligonucleotides 15284–297. [DOI] [PubMed] [Google Scholar]
- Bonci D, Cittadini A, Latronico MV, Borello U, Aycock JK, Drusco A.et al. (2003'Advanced' generation lentiviruses as efficient vectors for cardiomyocyte gene transduction in vitro and in vivo Gene Ther 10630–636. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Michienzi A, De Angelis FG., and, Bozzoni I. The Rev protein is able to transport to the cytoplasm small nucleolar RNAs containing a Rev binding element. RNA. 1999;5:993–1002. doi: 10.1017/s1355838299990064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaporte C, Dehaupas M., and, Fardeau M. Comparison between the growth pattern of cell cultures from normal and Duchenne dystrophy muscle. J Neurol Sci. 1984;64:149–160. doi: 10.1016/0022-510x(84)90033-9. [DOI] [PubMed] [Google Scholar]
- Piétri-Rouxel F, Gentil C, Vassilopoulos S, Baas D, Mouisel E, Ferry A.et al. (2010DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis EMBO J 29643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M.et al. (2011A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA Cell 147358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott OM, Hyde SA, Goddard C., and, Dubowitz V. Quantitation of muscle function in children: a prospective study in Duchenne muscular dystrophy. Muscle Nerve. 1982;5:291–301. doi: 10.1002/mus.880050405. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF.et al. (2002Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy Nat Med 8253–261. [DOI] [PubMed] [Google Scholar]
- Cirak S, Feng L, Anthony K, Arechavala-Gomeza V, Torelli S, Sewry C.et al. (2012Restoration of the dystrophin-associated glycoprotein complex after exon skipping therapy in Duchenne muscular dystrophy Mol Ther 20462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of muscle differentiation markers.
Dystrophin and nNOS localization in BMD biopsies.
Description of antisense constructs.
Clinical informations of BMD patients.